Abstract
RNAomics, analogous to proteomics, concerns aspects of the secondary and tertiary structure, folding pathway, kinetics, comparison, function and regulation of all RNA in a living organism. Given recently discovered roles played by micro RNA, small interfering RNA, riboswitches, ribozymes, etc., it is important to gain insight into the folding process of RNA sequences. We describe the web server RNALOSS, which provides information about the distribution of locally optimal secondary structures, that possibly form kinetic traps in the folding process. The tool RNALOSS may be useful in designing RNA sequences which not only have low folding energy, but whose distribution of locally optimal secondary structures would suggest rapid and robust folding. Website: http://clavius.bc.edu/~clotelab/RNALOSS/.
INTRODUCTION
RNA can play an important functional role in catalysis, e.g. ribozymes are RNA enzymes that cleave RNA phosphodiester bonds at specific sites (1); see (2) for an overview of potential therapeutic applications of ribozymes to cleave mRNAs of oncogenes (ras or bcr-abl) and viral transcripts (HIV-1), to overcome drug resistance, control arthritis, etc. Additionally, some small molecules can function as drugs acting on RNA. Such is the case for the aminoglycoside and macrolide families of antibiotics, which disrupt RNA translation in prokaryotes by targeting ribosomal (rRNA) (3).
In contrast to mRNA, noncoding RNA (ncRNA) is transcribed from genomic DNA and plays a biologically important role, although it is not translated into protein. Examples of ncRNA include ribozymes, riboswitches, micro RNA, small interfering RNA (4), tRNA, rRNA, etc. Riboswitches have recently been discovered to interact with small ligands and up- or down-regulate certain genes. Breaker and co-workers (5) report the crystal structures of the add A-riboswitch and xpt G-riboswitch aptamer modules, which distinguish between bound adenine and guanine; see (6) for an overview of bacterial riboswitches, and (7) for the structure, as given in the PDB code 1U8D of a guanine-responsive riboswitch with the metabolite hypoxanthine.
RNAomics (8), analogous to proteomics, concerns aspects of the secondary and tertiary structure, folding pathway, kinetics, comparison, function and regulation of all RNA in a living organism. RNAomics requires the application of numerous existent tools, as well as the development of new computational methods. Well-known RNA computational tools include secondary structure prediction web servers mfold (9) and Vienna RNA Package (10), the Sfold web server (11) to sample secondary structures according to the Boltzmann probability distribution, the tRNAscan-SE gene finder for tRNA (12), multiple sequence alignment for the statistical detection of RNA secondary structure MSARI (13), dynamic programming pairwise sequence-structure alignment Dynalign (14), tertiary structure modeling tool Mc-Sym (15), etc. Only a few of the many important computational tools for RNA structure prediction, gene finding, alignment, etc. have been listed.
In this paper, we describe the web server RNALOSS, based on the algorithm of Clote (16), which computes an aspect of the folding landscape of an RNA nucleotide sequence s = s1,…, sn. Given s, this algorithm runs in time O(n4) and space O(n3), and computes for each k, the number of k-locally optimal secondary structures (explained below). Work by Clote (16) was motivated by the following question, as has been suggested for proteins (17): is it the case that RNA has been under selective pressure to fold rapidly? Using the algorithm of the web server RNALOSS, it appears that structural RNA has a different folding landscape than random RNA of the same dinucleotide frequency; specifically, for small values of k, there appear to be fewer k-locally optimal secondary structures than in random RNA. Related, but distinct work has appeared in (18–21), for discussion see (16).
METHODS
A secondary structure for an RNA sequence s = s1,…,sn is an expression s = s1,…,sn involving dot, left and right parenthesis, which is well balanced, such that nucleotides corresponding to matching parentheses are either Watson–Crick complements or GU wobble pairs.
Definition
A secondary structure S on RNA sequence s = s1,…,sn is defined to be a set of ordered pairs (i, j), such that i +3 < j and the following conditions are satisfied.
Watson–Crick or GU wobble pairs: If (i, j) belongs to S, then pair (ai, aj) must be one of the following canonical base pairs: (A, U), (U, A), (G, C), (C, G), (G, U) and (U, G).
Threshold requirement: If (i, j) belongs to S, then j − i > 3; i.e. there must be at least three unpaired bases in a hairpin loop.
Non-existence of pseudoknots: If (i, j) and (k, l) belong to S, then it is not the case that i < k < j < l.
No base triples: If (i, j) and (i, k) belong to S, then j = k; if (i, j) and (k, j) belong to S, then i = k.
A secondary structure is k-locally optimal if it has k fewer base pairs than the maximum possible number [i.e. than in the Nussinov–Jacobson optimal structure (22,23)], and yet no base pairs can be added without violating the definition of secondary structure (e.g. without introducing a pseudoknot). To illustrate this notion, consider the RNA sequence GGGGCCCCC, which has three as the maximum possible number of base pairs, as given in the structure (((…))). There is only one structure having 3 bp, so the number of 0-locally optimal secondary structures is 1. On the other hand, there are twelve 1-locally optimal secondary structures and three 2-locally optimal secondary structures. The latter are listed as follows: (i) (…)....(ii) (....)..(iii) …(.....). The algorithm of (16) uses dynamic programming to compute, for each i < j and each k, the number of k-locally optimal secondary structures on the subsequence s = si,…,sj. Additionally, the algorithm must keep track of visible nucleotides and positions, i.e. those external to any base pair [for technical details see (16)].
WEB SERVER
The web server RNALOSS implements a new algorithm, described in (16), running in O(n4) time and O(n3) space, which computes for a given RNA sequence s = s1,…,sn and all k ≥ 0, the number of k-locally optimal secondary structures for s. An RNA nucleotide sequence may be input by uploading a FASTA-format file or by entering a nucleotide sequence in the blank provided on the web server form. Three tables are returned by RNALOSS: the number of k-locally optimal secondary structures, the relative density of states (i.e. the ratio of number of k-locally optimal structures over the total number of locally optimal structures) and the minimum free energy (mfe) of a sample k-locally optimal secondary structure (for each value of k, RNALOSS computes a single k-locally optimal secondary structure, denoted here as Sk, among the many possible k-locally optimal structures. Since this feature was implemented for debugging purposes, the current version of RNALOSS does not guarantee that Sk has lowest mfe as evaluated by RNAeval, over all k-locally optimal secondary structures. For this reason, the energy of sample structures Sk does not necessarily increase monotonically with increasing value of k). For the latter, mfe is computed using RNAfold from the Vienna RNA Package http://www.tbi.univie.ac.at/~ivo/RNA/. A screen shot of two of the tables is presented.
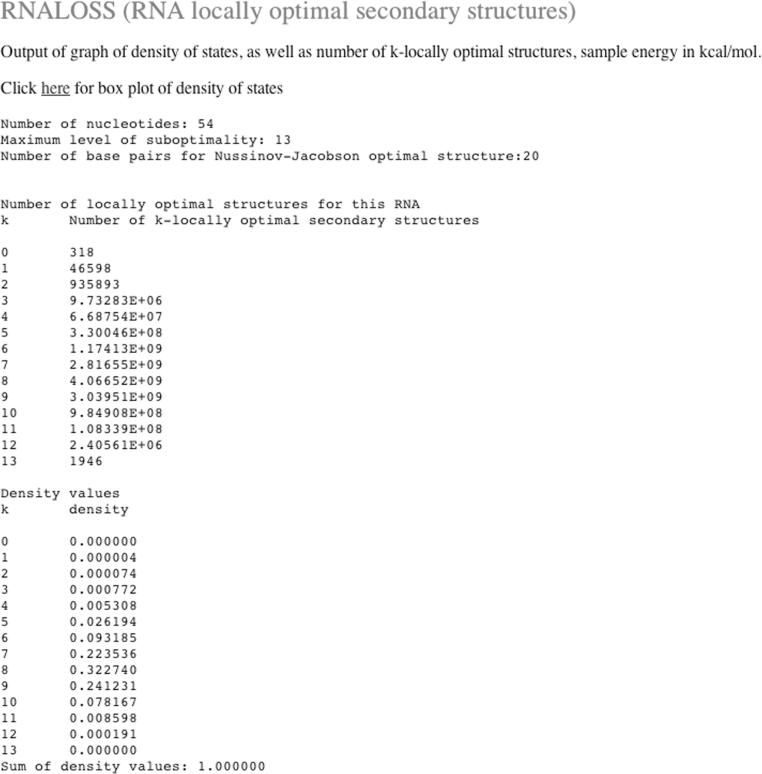
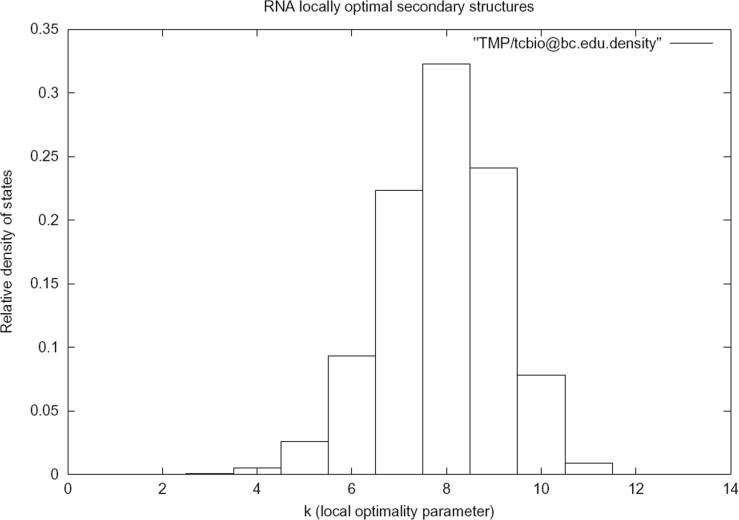
Figure 1 displays a screen shot of the RNALOSS web server form. Figure 2 lists the number of k-locally optimal secondary structures as computed by RNALOSS for type III hammerhead ribozyme AF170517 from Rfam (24). Figure 3 presents the relative density of states for k-locally optimal secondary structures for AF170517.
Figure 1.
Screen image of web server RNALOSS. Users may input an RNA sequence consisting of upper or lower case nucleotides A, C, G, T, U, either by uploading a FASTA-format file (A) or by pasting a nucleotide sequence into (B).
Figure 2.
Output of web server RNALOSS on type III hammerhead ribozyme AF170517 from Rfam (24). The web server RNALOSS outputs a table of number of k-locally optimal secondary structures, for each possible value of k ≥ 0 (shown here). Additionally, tables for the relative density of states and mfe values of sample k-locally optimal secondary structures are displayed in the browser (data not shown).
Figure 3.
Column graph of relative density of states is obtained by clicking a hot link from the previous screen shot. For each value of k ≥ 0, the ratio of number of k-locally optimal secondary structures over the total number of locally optimal secondary structures is displayed.
Owing to algorithmic time and space constraints, the RNALOSS web server immediately processes RNA of length at most 60 nt, while for RNA of length 61–100 nt, the results are emailed to the user. Currently, RNALOSS refuses to process any sequence of length >100 nt. Current hardware supporting RNALOSS web server consists of a Beowulf-style cluster comprising 6 Dell 1650, 2 × 1300 MHz Pentium III, 2 GB RAM with 4 Apple XServe, 2 × 1333 MHz G4, 2 GB RAM and finally 6 Dell 1850, 2 × 2800 MHz Xeon EM64T, 2 GB RAM. Interconnect is 1 Gbit Ethernet. Pentium III nodes are running RedHat Linux 9, Xeon EM64T nodes are running WhiteBox Linux 3 and G4 nodes are running MacOS 10.2.8.
DISCUSSION
Upon testing, structurally important RNA, such as selenocysteine insertion sequence elements, precursor mRNAs, type III hammerhead ribozymes and tRNA, all have a markedly smaller number of k-locally optimal structures than that of random RNA of the same dinucleotide frequency, for small and moderate values of k. Since the free energy of k-locally optimal secondary structures is generally closer to that of the native state for small k, this suggests that structural RNA has been optimized not only to have low folding energy (25), but also to have relatively few potential kinetic traps. This suggests that RNALOSS might be of use in designing RNA sequences for rapid folding.
Acknowledgments
The author would like to thank anonymous referees for helpful criticisms and suggestions. Funding to pay the Open Access publication charges for this article was provided by start-up funds from Boston College.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doudna J.A., Cech T.R. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.James H.A., Gibson I. The therapeutic potential of ribozymes. Blood. 1998;91:371–381. [PubMed] [Google Scholar]
- 3.Sucheck S., Wong A.L., Koeller K.M., Boehr D.D., Draker K., Sears P., Wright G.D., Wong C.-H. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J. Am. Chem. Soc. 2000;122:5230–5231. [Google Scholar]
- 4.Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K., Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–106. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batey R.T., Gilbert S.D., Montange R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 8.Hofacker I.L., Priwitzer B., Stadler P.F. Prediction of locally stable RNA secondary structures for genome-wide surveys. Bioinformatics. 2004;20:186–190. doi: 10.1093/bioinformatics/btg388. [DOI] [PubMed] [Google Scholar]
- 9.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coventry A., Kleitman D.J., Berger B. MSARI: multiple sequence alignments for statistical detection of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:12102–12107. doi: 10.1073/pnas.0404193101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews D.H., Turner D.H. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- 15.Major F., Turcotte M., Gautheret D., Lapalme G., Fillion E., Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991;253:1225–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- 16.Clote P. An efficient algorithm to compute the landscape of locally optimal RNA secondary structures with respect to the Nussinov–Jacobson energy model. J. Comput. Biol. 2005;1:83–101. doi: 10.1089/cmb.2005.12.83. [DOI] [PubMed] [Google Scholar]
- 17.Šali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 18.Cupal J., Hofacker I., Stadler P. Dynamic programming algorithm for the density of states of RNA secondary structures. In: Hofstädt R., Lengauer T., Löffler M., Schomburg D., editors. Proceedings of the German Conference on Bioinformatics (Computer Science and Biology) Germany: Universität Leipzig; 1996. pp. 184–186. [Google Scholar]
- 19.Flamm C., Fontana W., Hofacker I.L., Schuster P. RNA folding at elementary step resolution. RNA. 2000;6:325–338. doi: 10.1017/s1355838200992161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flamm C., Hofacker I.L., Stadler P.F., Wolfinger M. Barrier trees of degenerate landscapes. Z. Phys. Chem. 2002;216:155–173. [Google Scholar]
- 21.Evers D.J., Giegerich R. Reducing the conformation space in RNA structure prediction. Proceedings of the German Conference on Bioinformatics; 2001. pp. 118–124. [Google Scholar]
- 22.Nussinov R., Jacobson A.B. Fast algorithm for predicting the secondary structure of single stranded RNA. Proc. Natl Acad. Sci. USA. 1980;77:6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clote P., Backofen R. Computational Molecular Biology: An Introduction. NY: John Wiley & Sons; 2000. [Google Scholar]
- 24.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clote P., Ferrè F., Kranakis E., Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005 doi: 10.1261/rna.7220505. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]