Summary:
The integration of metabolomics with other omics (“multi-omics”) offers complementary insights into disease biology. However, this integration remains challenging due to the fragmented landscape of current methodologies, which often require programming experience or bioinformatics expertise. Moreover, existing approaches are limited in their ability to accommodate unidentified metabolites, resulting in the exclusion of a significant portion of data from untargeted metabolomics experiments. Here, we introduce iModMix, a novel approach that uses a graphical lasso to construct network modules for integration and analysis of multi-omics data. iModMix uses a horizontal integration strategy, allowing metabolomics data to be analyzed alongside proteomics or transcriptomics to explore complex molecular associations within biological systems. Importantly, it can incorporate both annotated and unidentified metabolites, addressing a key limitation of existing methodologies. iModMix is available as a user-friendly R Shiny application that requires no programming experience (https://imodmix.moffitt.org), and it includes example data from several publicly available multi-omic studies for exploration. An R package is available for advanced users (https://github.com/biodatalab/iModMix).
Availability and implementation:
Shiny application: https://imodmix.moffitt.org. The R package and source code: https://github.com/biodatalab/iModMix.
Introduction
Integration of metabolomics with other omics modalities, such as proteomics and transcriptomics, is essential for a comprehensive understanding of complex biological systems and disease mechanisms. The scale and complexity of these so-called “multi-omics” data necessitate computational approaches for effective integration and analysis. However, multi-omics analysis presents several challenges. These include the need for computational tools that are adaptable to diverse experimental setups and data types, as well as user-friendly enough to be used without extensive programming experience (Jendoubi 2021). Moreover, many approaches are unable to incorporate unidentified metabolites, resulting in the exclusion of large portions of data from untargeted metabolomics experiments, thereby limiting the potential for novel discoveries. Current pathway tools often restrict the inclusion of metabolite identifications due to naming mismatches and inconsistent use of standard identifiers like KEGG or HMDB IDs. This results in exclusion of some identified metabolites which cannot be fully utilized for analysis and further narrowing and hindering comprehensive insights (Jendoubi 2021).
In general, approaches for integrating metabolomics with other omics can be categorized into vertical or horizontal integration. Vertical integration takes multiple omics datasets as input and produces a single output, such as clustering assignments or a merged dataset, which no longer reflects the individual features of the original data. iClusterPlus (Q et al. 2013) is an example of vertical integration: it takes multiple omics datasets from the same samples as input, extracts latent factors across omics datasets, clusters samples in latent space, and outputs the clustering assignments by sample. In contrast, horizontal integration utilizes multiple omics datasets in parallel while maintaining the context of the original inputs. PIUmet (Benedetti et al. 2023) is an example of horizontal integration: it takes metabolomics and proteomics data as input, and the resulting network output contains both metabolites and proteins.
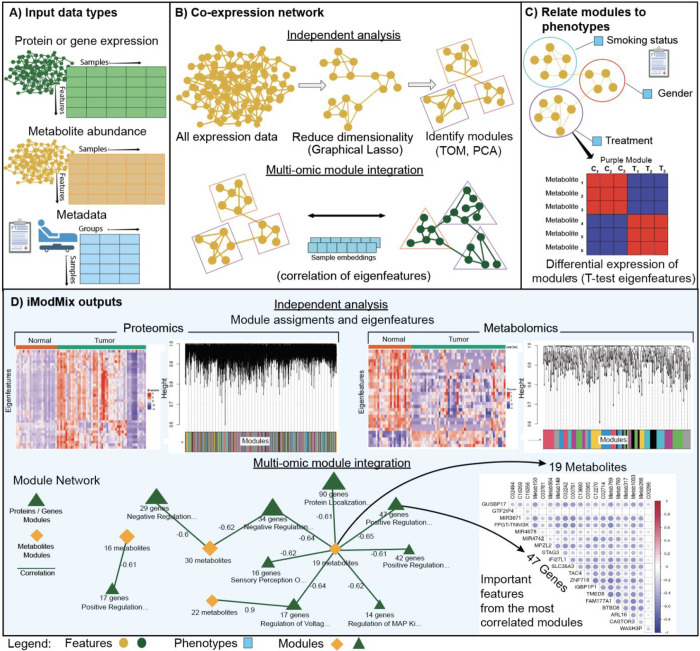
iModMix is a horizontal integration framework that constructs network modules from two input omics datasets (e.g., metabolomics and proteomics, metabolomics and transcriptomics). It first uses graphical lasso to estimate a sparse Gaussian Graphical Model (GGM) (Friedman, Hastie, and Tibshirani 2008) for each input omics dataset. GGMs capture direct associations within the input omics datasets, which is an improvement over Weighted Gene Correlation Network Analysis (WGCNA) (Langfelder et al. 2008) module creation that includes both direct and indirect associations. Similar to WGCNA, a Topological Overlap Matrix (TOM) is next calculated (Yip et al. 2007), which quantifies the extent to which pairs of features share common neighbors. Hierarchical clustering is then performed on TOM dissimilarity (1 – TOM), using a dynamic tree cutoff to group related features into modules. iModMix next takes the first principal component of the abundances of the features in the module, called an eigenfeature (e.g., eigenmetabolites for metabolomics, eigenproteins for proteomics). These eigenfeatures are representative of the contents of the module, and they can be used in place of normal metabolite abundance or protein expression for testing for differential expression or association with experimental conditions. Finally, integration between omics types is achieved by correlating the eigenfeatures from different omics experiments.
A major advantage of iModMix is its ability to generate network modules and their associated eigenfeatures independently of feature annotation. This allows iModMix to incorporate annotated and unidentified metabolites into its results. Since this method does not rely on existing pathway databases like KEGG, it can uncover new associations among features. iModMix is available as an R Shiny application (https://imodmix.moffitt.org) that provides an easy to use graphical interface and detailed documentation for users without programming expertise, and it is available as an R package (https://github.com/biodatalab/iModMix). iModMix can run on standard desktop computers with at least 8GB of RAM and a multi-core processor. The running time depends on the size and complexity of the dataset, but typical data sizes of 20,000 variables, the analysis can be completed in under an hour.
Implementation
Data Upload
The iModMix workflow accepts two omics datasets at a time (Fig. 1A). If available, users can provide feature-level annotation corresponding to the input omics dataset. A separate metadata file containing sample IDs and at least one sample grouping column (e.g., treatment, control) are necessary for conducting PCA, heatmaps, and boxplots. Once uploaded, the tool scales the data by centering each feature to have a mean of zero and a standard deviation of one. Missing values are imputed using the k-nearest neighbors’ algorithm, or users can also provide their own imputed data if a different method is preferred. We provide ten datasets encompassing gene expression and metabolomics data to illustrate iModMix’s capabilities. These datasets, from (Benedetti et al. 2023), have been formatted to meet iModMix specifications, with variable columns labeled as “Feature_ID” and sample columns labeled as “Samples”. These datasets are designed to facilitate user engagement with iModMix and are readily accessible for download on Zenodo (https://doi.org/10.5281/zenodo.13988161). We also provide an in-house dataset of 20 lung adenocarcinoma mouse samples (10 wild type, 10 knockout), with 7353 metabolomics features and 7928 protein groups, to highlight iModMix’s functionality in handling unidentified metabolites.
Figure 1:
Overview of iModMix. (A) iModMix integrates transcriptomics, proteomics, metabolomics, and clinical data to understand biological systems comprehensively. (B) Co-expression network: The tool performs independent analyses of each omics dataset to identify biologically relevant modules, and it then integrates the datasets by correlating eigenfeatures. (C) iModMix evaluates associations between phenotype-related modules across different data types. (D) Results of the integrated analysis of transcriptomics and metabolomics data from normal and tumor samples using iModMix. The network interaction and most correlated modules are also visualized.
Module Construction
iModMix first estimates partial correlations using the glassoFast R package (Friedman, Hastie, and Tibshirani 2008) (Fig. 1B) to build a GGM. Next, the TOM is computed and TOM dissimilarity (1 – TOM) is used for hierarchical clustering (Yip et al. 2007). Modules are created using the dynamic tree cut method, specifically the ‘hybrid’ method, which refines assignments derived from a static cut height by analyzing the shape of each branch (Langfelder, Zhang, and Horvath 2008). Finally, iModMix calculates eigenfeatures for each module using the moduleEigengenes function from the WGCNA R package (Langfelder et al. 2008) (Fig. 1B) that are used for downstream omics analyses.
Module Exploration
The phenotype analysis section provides a framework for classifying phenotypes based on eigenfeatures. Users can upload metadata, select phenotypes of interest, and set significance thresholds for statistical tests. The module performs classification using Student’s t-test, comparing selected phenotypes and generating results that include t-statistics and p-values. Significant eigenfeatures are visualized through boxplots. Additionally, users can explore specific modules and view associated features, PCA loading, and heatmap plots to view feature behavior across different phenotypes (Fig. 1C). iModMix supports pathway enrichment analysis of genes or proteins by utilizing all available libraries in Enrichr (Kuleshov et al. 2016). This allows users to choose their preferred library, such as KEGG, GO, Human Gene Atlas, or WikiPathways.
Multi-omics analysis
iModMix integrates modules across omics datasets by calculating the Spearman correlation between eigenfeatures. iModMix then constructs an interactive network, where protein/gene modules are represented as green triangles and metabolite modules as yellow diamonds, with correlation coefficients shown on connecting arrows. Users can select the number of top multi-omics module correlations to visualize and explore detailed correlations within these modules through corrplots and tables. Lists of metabolites and proteins/genes within highly correlated modules are provided, facilitating further pathway analysis and offering insights into the relationships between different omics layers. Additionally, classification between features of each layer and phenotypes using t-tests and boxplots is provided, enabling users to visualize and identify significant differences. (Fig. 1B).
Case-Study: ccRCC Dataset with Identified Metabolites
We used 24 normal and 52 tumor clear cell renal cell carcinoma (ccRCC) samples (Golkaram et al. 2022; Tang et al. 2023; Benedetti et al. 2023) as a case study. It contained 23001 genes from RNA-seq and 904 identified metabolites from untargeted metabolomics. Applying iModMix identified 401 gene modules and 24 metabolite modules. Differential expression analysis of the modules through t-test confirmed changes in metabolite abundance between groups recapitulated those from prior work (Benedetti et al. 2023) highlighting reduced levels of adenosine/C00212 (module ME#443A83FF, p-value = 0.0000), gamma-glutamyltyrosine (module ME#472D7BFF, p-value = 0.0001), creatinine/C00791 and 1-methyladenosine/C02494 (module ME#1F9A8AFF p-value = 0.0000) in tumors compared to normal tissue. Conversely, metabolites with increased abundance in tumors compared to normal tissues included fructose/C00095 (module ME#472D7BFF, p-value = 0.0001), proline/C00148 and glutamine/C00064 (module ME#35B779FF, p-value = 0. 0023) (Benedetti et al. 2023). The top 5 correlated metabolomic modules to transcriptomic modules included pathways such as mitochondria fission and regulation of MAPK activity. Mitochondria fission results in lowered respiratory rates and emphasizes tumor reliance on glucose resulting in higher glycolysis rates common in renal cell carcinoma (Linehan et al. 2019).
Case-Study: Lung Adenocarcinoma Dataset with Unidentified Metabolites
Matched proteomics and metabolomics dataset was generated using two mouse models for lung adenocarcinoma (10 wild type, 10 knockout), with 7353 metabolomics features (annotated and unidentified) and 7928 protein groups. Applying iModMix generated 267 gene modules and 191 metabolite modules. High correlations were found between these modules, starting from a correlation threshold of 0.95. The second most correlated modules (protein module: ME#3ABA76FF with metabolite module: ME#238A8DFF), with a correlation of −0.94, included 14 genes (Gng11, Cnbp, Pkm, Aprt, Pgk1, Rbx1, Baz1b, Pgam1, Mif, Lgals3, Sp110, Map2, Mmrn2, Aldoc) and 21 metabolites, three of which are identified metabolites (Cortisol 21-acetate, Thymine, Deoxyuridine). Pathway analysis of these 14 genes and 3 metabolites using MetaboAnalyst (Pang et al. 2024) revealed significant enrichment in Glycolysis or Gluconeogenesis (P-value: 7.94E-7) and Pyrimidine metabolism (P-value: 0.0043961). Glycolysis and gluconeogenesis are known to be activated in non-small cell lung cancer (NSCLC) in a manner modulated by tumor size and oxygenation, and they differentially correlate with patient outcomes. The frequent co-activation of these pathways in NSCLC suggests their potential as targets for future therapeutic strategies aimed at cancer cell metabolism (Smolle et al. 2020). Additionally, high expression levels of pyrimidine metabolic rate-limiting enzymes have been identified as adverse prognostic factors in lung adenocarcinoma (LUAD). This reprogramming of metabolism is associated with clinical outcomes, indicating that the pyrimidine metabolism signaling pathway plays a significant role in LUAD prognosis (Wang et al. 2020).
Benchmarking
We benchmark IModMix to identify the optimal parameters for gene network construction using the iModMix algorithm. The process considers three input parameters: num_genes, num_fold, and lambda. Expression data from RC20 and lung adenocarcinoma mouse samples were used, and genes with the highest variance were selected, constrained by num_genes. To identify the best sparsity parameter (lambda), we applied five-fold cross-validation using the Graphical Lasso (Glasso) algorithm, which calculates a sparse Gaussian Graphical Model (GGM) for each input omics dataset. The Glasso model was fitted using the glassoFast function with lambda, and metrics such as the number of non-zero partial correlations, log-likelihood, and execution time were evaluated. Hierarchical clustering analysis was then used to identify gene modules. Stability was achieved in both datasets with an alpha value of 0.25 (Fig. S1A, Fig. S1B), allowing for efficient and accurate model evaluation across various configurations. This benchmarking process demonstrates that IModMix offers a refined approach to network construction by focusing on direct associations, making it a robust alternative to WGCNA for complex omics data.
Summary:
The iModMix pipeline captures interactions that could not be observable with metabolomics or transcriptomics alone. iModMix progresses the field of multi-omic analysis with advantages such as de novo network generation, independent of existing pathway databases and the ability to integrate annotated and unidentified metabolites for multi-omic analysis.
Supplementary Material
Funding:
This work was supported in part by:
• The Biostatistics and Bioinformatics Shared Resource and the Proteomics and Metabolomics Shared Resource at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292)
• The Cancer Research Institute (Stewart)
• P01CA250984, ‘Identifying Metabolic Vulnerabilities in Lung Cancer,’
• T32 CA233399/CA/NCI NIH HHS/United States
• The Anna D. Valentine and Charles L. Oehler Award (Li/Fridley/Chen) funded prior work.
References
- Benedetti E., Liu EM., Tang C., Kuo F., Buyukozkan M., Park T., Park J., Correa F., Hakimi AA., Intlekofer AM., Krumsiek J., and Reznik E.. 2023. ‘A multimodal atlas of tumour metabolism reveals the architecture of gene-metabolite covariation’, Nature metabolism, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman Jerome, Hastie Trevor, and Tibshirani Robert. 2008. ‘Sparse inverse covariance estimation with the graphical lasso’, Biostatistics, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golkaram Mahdi, Kuo Fengshen, Gupta Sounak, Carlo Maria I., Salmans Michael L., Vijayaraghavan Raakhee, Tang Cerise, Makarov Vlad, Rappold Phillip, Blum Kyle A., Zhao Chen, Mehio Rami, Zhang Shile, Godsey Jim, Pawlowski Traci, DiNatale Renzo G., Morris Luc G. T., Durack Jeremy, Russo Paul, Kotecha Ritesh R., Coleman Jonathan, Chen Ying-Bei, Reuter Victor E., Motzer Robert J., Voss Martin H., Liu Li, Reznik Ed, Chan Timothy A., Hakimi A. Ari, Golkaram Mahdi, Kuo Fengshen, Gupta Sounak, Carlo Maria I., Salmans Michael L., Vijayaraghavan Raakhee, Tang Cerise, Makarov Vlad, Rappold Phillip, Blum Kyle A., Zhao Chen, Mehio Rami, Zhang Shile, Godsey Jim, Pawlowski Traci, DiNatale Renzo G., Morris Luc G. T., Durack Jeremy, Russo Paul, Kotecha Ritesh R., Coleman Jonathan, Chen Ying-Bei, Reuter Victor E., Motzer Robert J., Voss Martin H., Liu Li, Reznik Ed, Chan Timothy A., and Hakimi A. Ari. 2022. ‘Spatiotemporal evolution of the clear cell renal cell carcinoma microenvironment links intra-tumoral heterogeneity to immune escape’, Genome Medicine 2022 14:1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendoubi Takoua. 2021. ‘Approaches to Integrating Metabolomics and Multi-Omics Data: A Primer’, Metabolites 2021, Vol. 11, Page 184, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov Maxim V., Jones Matthew R., Rouillard Andrew D., Fernandez Nicolas F., Duan Qiaonan, Wang Zichen, Koplev Simon, Jenkins Sherry L., Jagodnik Kathleen M., Lachmann Alexander, McDermott Michael G., Monteiro Caroline D., Gundersen Gregory W., and Ma’ayan Avi. 2016. ‘Enrichr: a comprehensive gene set enrichment analysis web server 2016 update’, Nucleic Acids Research, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder Peter, Horvath Steve, Langfelder Peter, and Horvath Steve. 2008. ‘WGCNA: an R package for weighted correlation network analysis’, BMC Bioinformatics 2008 9:1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder Peter, Zhang Bin, and Horvath Steve. 2008. ‘Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R’, Bioinformatics, 24. [DOI] [PubMed] [Google Scholar]
- Linehan W. Marston, Ricketts Christopher J., Linehan W. Marston, and Ricketts Christopher J.. 2019. ‘The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications’, Nature Reviews Urology 2019 16:9, 16. [DOI] [PubMed] [Google Scholar]
- Pang Zhiqiang, Xu Lei, Viau Charles, Lu Yao, Salavati Reza, Basu Niladri, Xia Jianguo, Pang Zhiqiang, Xu Lei, Viau Charles, Lu Yao, Salavati Reza, Basu Niladri, and Xia Jianguo. 2024. ‘MetaboAnalystR 4.0: a unified LC-MS workflow for global metabolomics’, Nature Communications 2024 15:1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Q Mo, Wang S, Seshan VE, Olshen AB, Schultz N, Sander C, Powers RS, Ladanyi M, and Shen R. 2013. ‘Pattern discovery and cancer gene identification in integrated cancer genomic data - PubMed’, Proceedings of the National Academy of Sciences of the United States of America, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle Elisabeth, Leko Petra, Stacher-Priehse Elvira, Brcic Luka, El-Heliebi Amin, Hofmann Lilli, Quehenberger Franz, Hrzenjak Andelko, Popper Helmut H., Olschewski Horst, and Leithner Katharina. 2020. ‘Distribution and prognostic significance of gluconeogenesis and glycolysis in lung cancer’, Molecular Oncology, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Cerise, Xie Amy X, Liu Minwei, Kuo Fengshen, Kim Minsoo, Renzo Di Natale Mahdi Golkaram, Chen Yingbei, Gupta Sounak, Motzer Robert, Russo Paul, Coleman Jonathan, Carlo Maria I, Voss Martin H, Kotecha Ritesh R, Lee Chung Han, Tansey Wesley, Schultz Nikolaus, Hakimi A Ari, and Reznik Ed. 2023. ‘Immunometabolic coevolution defines unique microenvironmental niches in ccRCC’, Cell metabolism, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Haiwei, Wang Xinrui, Xu Liangpu, Zhang Ji, Cao Hua, Wang Haiwei, Wang Xinrui, Xu Liangpu, Zhang Ji, and Cao Hua. 2020. ‘High expression levels of pyrimidine metabolic rate–limiting enzymes are adverse prognostic factors in lung adenocarcinoma: a study based on The Cancer Genome Atlas and Gene Expression Omnibus datasets’, Purinergic Signalling, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip Andy M, Horvath Steve, Yip Andy M, and Horvath Steve. 2007. ‘Gene network interconnectedness and the generalized topological overlap measure’, BMC Bioinformatics 2007 8:1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.