Abstract
Brain age is a powerful marker of general brain health. Furthermore, brain age models are trained on large datasets, thus giving them a potential advantage in predicting specific outcomes – much like the success of finetuning large language models for specific applications. However, it is also well-accepted in machine learning that models trained to directly predict specific outcomes (i.e., direct models) often perform better than those trained on surrogate outcomes. Therefore, despite their much larger training data, it is unclear whether brain age models outperform direct models in predicting specific brain health outcomes. Here, we compare large-scale brain age models and direct models for predicting specific health outcomes in the context of Alzheimer’s Disease (AD) dementia. Using anatomical T1 scans from three continents (N = 1,848), we find that direct models outperform brain age models without finetuning. Finetuned brain age models yielded similar performance as direct models, but importantly, did not outperform direct models although the brain age models were pretrained on 1000 times more data than the direct models: N = 53,542 vs N = 50. Overall, our results do not discount brain age as a useful marker of general brain health. However, in this era of large-scale brain age models, our results suggest that small-scale, targeted approaches for extracting specific brain health markers still hold significant value.
1. INTRODUCTION
There is significant interest in using biological age as a marker of disease risk and mortality (Belsky et al., 2015; Chen et al., 2016; Tian et al., 2023). In the case of brain age, this involves training a machine learning model to predict chronological age from brain imaging data of healthy individuals (Cole et al., 2017; Dosenbach et al., 2010; Franke et al., 2010). The brain age gap (BAG) – the difference between predicted and chronological age – serves as a marker of accelerated aging and development. A positive BAG is associated with worse cognitive performance in older adults (Cumplido-Mayoral et al., 2024; Wrigglesworth et al., 2022), better cognitive performance in healthy children (Cheng et al., 2024; Erus et al., 2014), brain disorders (Constantinides et al., 2023; Han et al., 2021; Kaufmann et al., 2019), poor physical health (Cole, 2020; Franke et al., 2013; Ronan et al., 2016) and mortality (Cole et al., 2018; Paixao et al., 2020). Overall, these studies suggest the utility of BAG as a marker of general brain health.
In addition to associations at the group-level, BAG has been used to directly predict individual-level mortality (Cole et al., 2018), predict progression of mild cognitive impairment (MCI) to AD dementia (Choi et al., 2023; Gaser et al., 2013; Löwe et al., 2016) and classify psychiatric disorders (Koutsouleris et al., 2013; Leonardsen et al., 2022). However, summarizing a person’s brain health with a single number (BAG) might lose too much information. Therefore, some studies extract intermediate-level representations from pretrained brain age models, which are then used as input features for training new models to predict MCI progression (Gao et al., 2020), and classify neurological disorders (Leonardsen et al., 2022; Zheng, Pfahringer, & Mayo, 2022). Finally, when deep neural networks (DNNs) are used for brain age prediction, the resulting models can be finetuned to diagnose brain disorders (Bashyam et al., 2020; Lu et al., 2022). The fine-tuning process can improve prediction by enabling the pretrained model to adapt to the unique characteristics of the new dataset – such as demographics or MRI scanner specifications – which may differ significantly from the data used to train the brain age model. Overall, these studies have demonstrated the utility of brain age models to predict specific health outcomes.
However, it remains unclear whether brain age derived models are better than models directly trained to predict specific health outcomes, which we refer to as “direct models”. On the one hand, there are orders of magnitude more brain imaging data with age-only information, compared with brain imaging data with target outcomes. Therefore, similar to the success of finetuning large language models for specific tasks (Tinn et al., 2023; Yang et al., 2022), a brain age model trained on tens of thousands of participants might yield better target prediction than direct models trained on hundreds of participants. On the other hand, a well-accepted machine learning principle is that training a model to directly predict a target variable of interest yields better prediction performance than training the model to predict a surrogate variable (that is only correlated with the target variable). Therefore, direct models might perform better than brain age derived models.
Motivated by this question, here we compare brain age derived models and direct models in two classification tasks. The first task is to predict whether a participant is cognitively normal (CN) or has AD dementia. We refer to this task as AD classification. The second task is to predict whether a participant with MCI would progress to AD dementia within 3 years. We refer to this task as MCI progression prediction. We chose these tasks because previous studies have suggested that brain age derived models can perform well in these tasks (Bashyam et al., 2020; Gaser et al., 2013; Lu et al., 2022). Furthermore, age is the largest risk factor for AD dementia (Daviglus et al., 2010; van der Flier & Scheltens, 2005), in contrast to other target outcomes of interest, such as psychiatric disorders, where other risk factors might be more prominent. Therefore, if brain age derived models cannot outperform direct models in these tasks, then it would seem unlikely that brain age derived models can outperform direct models in other tasks.
In this study, we consider the brain age model trained on one of the largest and most diverse datasets assembled (N = 53,542; Leonardsen et al., 2022). The brain age model utilizes the same convolutional neural network architecture as the winner of the Predictive Analysis Challenge for brain age prediction in 2019 (Gong et al., 2021; Peng et al., 2021). At the time of publication, the brain age model achieved state-of-the-art performance on data from unseen MRI scanners (Leonardsen et al., 2022). Intermediate representations from the pretrained model could also be used to classify various brain disorders via a transfer learning procedure (Leonardsen et al., 2022). As such, we believe the pretrained brain age model remains one of the best in the field. Evaluation was performed using anatomical T1 scans from three datasets (N = 1,848). We note that the evaluation datasets were not used to train the pretrained brain age model, so are truly out of sample.
Consistent with previous work (Leonardsen et al., 2022), we found that classifiers trained from intermediate representations of the pretrained brain age model (brainage64D) perform a lot better than BAG, suggesting that too much information is lost when summarizing a person’s biological age with a single number (i.e., BAG). Yet, direct models perform significantly better than brainage64D. Finetuning the brainage64D classifiers yields similar prediction performance to the direct models, but do not outperform direct models even when sample size is very small (N = 50). Overall, our results do not dispute transfer learning as a general strategy to improve prediction or that brain age is a powerful marker of general brain health. However, given the computational demands of training and then finetuning large models, our results suggest that targeted approaches for extracting specific markers of brain health from small datasets continue to hold significant value.
2. RESULTS
2.1. Overview
Here we considered data from three datasets: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset (Jack et al., 2010; Jack et al., 2008; Mueller et al., 2005), the Australian Imaging, Biomarkers and Lifestyle (AIBL) study (Ellis et al., 2009; Ellis et al., 2010; Fowler et al., 2021) and the Singapore Memory Aging and Cognition center (MACC) Harmonization cohort (Chong et al., 2017; Hilal et al., 2015; Hilal et al., 2020; Xu et al., 2015). More details about the datasets and preprocessing can be found in Methods (Sections 5.1 and 5.4).
Following Leonardsen and colleagues (Leonardsen et al., 2022), we considered age-matched and sex-matched cognitively normal (CN) participants and participants diagnosed with AD dementia (N = 1272) for the AD classification task. We also considered age-matched and sex-matched MCI participants who progressed to AD dementia within three years (i.e., progressive MCI or pMCI) and remained as MCI (i.e., stable MCI or sMCI) for the MCI progression task (N = 576). For more details, see Methods (Section 5.2).
For both classification tasks, we utilized a nested (inner-loop) cross-validation procedure, in which participants were assigned to a development set and a test set with an 80:20 ratio. The development set was in turn divided into a training set and a validation set with an 80:20 ratio. In general, models were trained on the training set and hyperparameters were tuned on the validation set. The final model was evaluated in the test set. This training-validation-test procedure was repeated 50 times for robustness. For more details, see Methods (Section 5.3).
2.2. Leveraging a pretrained brain age model does not improve AD classification over training a model directly
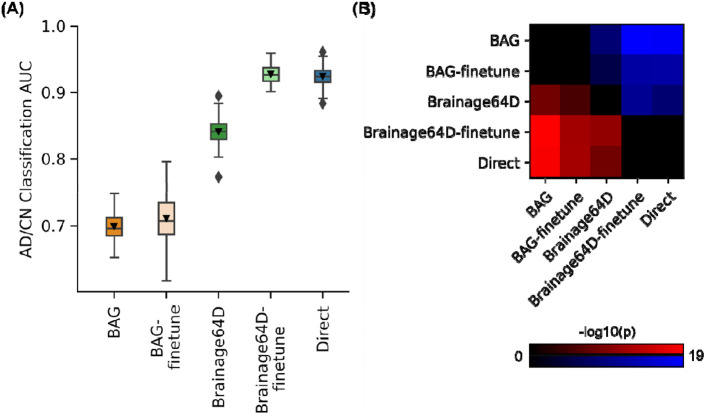
For AD classification, we compared five approaches (Figure 1) - the direct model and four brain age models (BAG, BAG-finetune, Brainage64D and Brainage64D-finetune) derived from a state-of-the-art pretrained brain age model (Leonardsen et al., 2022). The brain age model was previously trained on 53,542 participants across diverse datasets (Leonardsen et al., 2022). For more details, see Methods (Sections 5.5 and 5.6). Figure 2A shows the AD classification AUC (across 50 training-validation-test splits) for all five approaches. Figure 2B illustrates the p values from comparing pairs of approaches using the corrected resampled t-test (Nadeau & Bengio, 2003). Table 1 reports the actual p values.
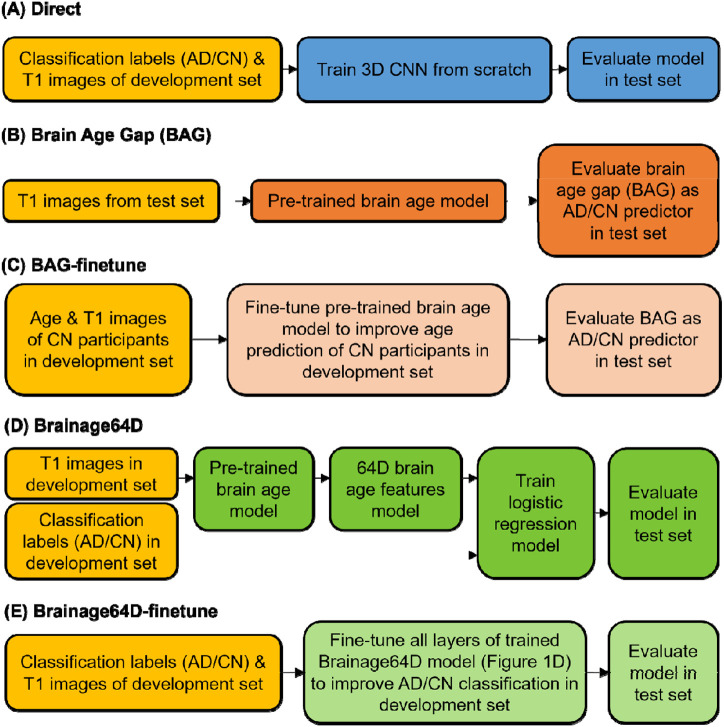
Figure 1.
Workflow of five AD classification models. Given a T1 MRI scan, each model predicted a classification label (AD or CN). “Direct” is a model trained from scratch, while the other four models involved a brain age model that was previously trained from 53,542 participants across diverse datasets (Leonardsen et al., 2022). (A) “Direct” used the same 3D CNN architecture as the brain age model, except for the final binary classification layer. Classification labels (AD or CN) and T1 images of the development set were used to train the 3D CNN from scratch. (B) BAG model subtracted the chronological age of each test participant from the predicted age of the pretrained brain age model. The resulting brain age gap (BAG) was used as the AD/CN predictor. (C) Age & T1 images of CN participants in the development set were used to finetune the pretrained brain age model to improve age prediction of CN participants in development set. The finetuned brain age model was then used to compute BAG of each test participant, and the resulting BAG was used as the AD/CN predictor. (D) “Brainage64D” extracted 64-dimensional (64D) brain age features from the output of the global averaging pooling layer of the pretrained brain age model. The 64D features and classification labels (AD/CN) of the development set were used to train a logistic regression model. The final model consisted of the concatenation of the pretrained brain age model up to (and including) the global average pooling layer and the trained logistic regression model. (E) “Brainage64D-finetune” involved finetuning all layers of the trained Brainage64D model (from panel D) to improve AD classification in the development set.
Figure 2.
AD classification AUC. (A) Box plots show the test AUC across 50 random training-validation-test splits. For each box plot, the horizontal line indicates the median across 50 test AUC values. Triangle indicates the mean. The bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Outliers are defined as data points beyond 1.5 times the interquartile range. The whiskers extend to the most extreme data points not considered outliers. (B) Minus log10 p values between all pairs of AD classification approaches based on the corrected resampled t-test (Nadeau & Bengio, 2003). Larger −log10(p) indicates greater statistical significance. Non-black color indicates significant differences after false discovery rate (FDR) correction (q < 0.05). For each pair of approaches, red (or blue) color indicates that the approach on the row outperformed (or underperformed) the approach on the column. For example, if we focus on the “Direct” row, the red color indicates that “Direct” statistically outperformed BAG, BAG-finetune and Brainage64D.
Table 1.
AD classification AUC (mean ± std) and uncorrected p values between pairs of approaches. AUC was averaged across 50 random training-validation-test splits. P values were computed using the corrected resampled t-test (Nadeau & Bengio, 2003). Bolded p values indicate statistical significance after FDR correction (q < 0.05).
| AD/NC Classification AUC (mean ± std) |
P values | ||||
|---|---|---|---|---|---|
| BAG | BAG- finetune |
Brainage64D | Brainage64D- finetune |
Direct | |
| BAG (0.70 ± 0.02) | 0.438 | 5.62e-9 | 4.20e-19 | 1.47e-18 | |
| BAG-finetune (0.71 ± 0.04) | 6.64e-6 | 2.23e-12 | 1.42e-12 | ||
| Brainage64D (0.84 ± 0.02) | 2.73e-11 | 7.44e-9 | |||
| Brainage64D-finetune (0.93 ± 0.01) | 0.655 | ||||
| Direct (0.92 ± 0.01) | |||||
Direct and Brainage64D-finetune performed the best, with no statistical difference between the two approaches. BAG and BAG-finetune performed the worst with no statistical difference between the two approaches. Brainage64D (without finetuning) achieved an intermediate level of performance. Importantly, the performance of Brainage64D (mean AUC=0.84) was comparable to the results reported by Leonardsen and colleagues (2022) (mean AUC=0.83).
Overall, this suggests that with the largest training set size of 997, leveraging a pretrained brain age model did not improve AD classification performance, compared with simply training a model from scratch.
2.3. Even when sample size is small, leveraging a pretrained brain age model does not improve AD classification over training a model directly
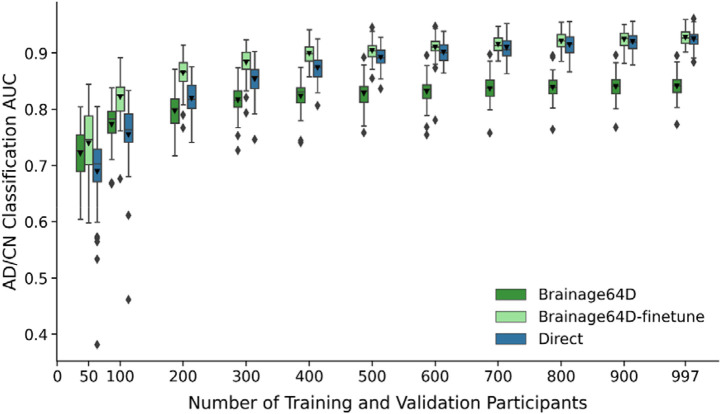
Adapting a pretrained model (trained from large datasets) for a new classification task might be more advantageous when the sample size available for the new task is small. Figure 3 shows the AD classification AUC (across 50 training-validation-test splits) for Direct, Brainage64D and Brainage64D-finetune across different development set sizes. Given their poor performance (Figure 2), we did not consider BAG and BAG-finetune in this analysis. Table 2 reports the actual AUC, while Table 3 reports the p values obtained from comparing the Direct approach with Brainage64D and Brainage64D-finetune using the corrected resampled t-test.
Figure 3.
AD classification AUC of Brainage64D, Brainage64D-finetune, and Direct across different development set sizes. Boxplots showing test AUC across different development set sizes (number of training and validation participants). Test sets were identical across development set sizes, so AUCs were comparable across sample sizes. For each boxplot, the horizontal line indicates the median across 50 test AUC values. Triangle indicates the mean. The bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Outliers are defined as data points beyond 1.5 times the interquartile range. The whiskers extend to the most extreme data points not considered outliers.
Table 2.
AD classification AUC (mean ± std) of different approaches for AD classification task across different development set sizes (i.e., number of training and validation participants). Given their poor performance (Figure 2), we did not consider BAG and BAG-finetune in this analysis.
| Number of training and validation participants |
Brainage64D | Brainage64D-finetune | Direct |
|---|---|---|---|
| 50 | 0.7214 ± 0.0465 | 0.7393 ± 0.0595 | 0.6893 ± 0.0742 |
| 100 | 0.7729 ± 0.0384 | 0.8216 ± 0.0368 | 0.7547 ± 0.0596 |
| 200 | 0.7968 ± 0.0319 | 0.8643 ± 0.0284 | 0.8195 ± 0.0307 |
| 300 | 0.8160 ± 0.0269 | 0.8833 ± 0.0251 | 0.8533 ± 0.0288 |
| 400 | 0.8231 ± 0.0277 | 0.8996 ± 0.0185 | 0.8736 ± 0.0221 |
| 500 | 0.8279 ± 0.0255 | 0.9042 ± 0.0163 | 0.8919 ± 0.0185 |
| 600 | 0.8315 ± 0.0246 | 0.9101 ± 0.0235 | 0.9013 ± 0.0195 |
| 700 | 0.8362 ± 0.0234 | 0.9157 ± 0.0144 | 0.9094 ± 0.0191 |
| 800 | 0.8389 ± 0.0226 | 0.9211 ± 0.0152 | 0.9148 ± 0.0182 |
| 900 | 0.8403 ± 0.0213 | 0.9241 ± 0.0152 | 0.9200 ± 0.0172 |
| 997 | 0.8411 ± 0.0213 | 0.9276 ± 0.0144 | 0.9246 ± 0.0146 |
Table 3.
Uncorrected p values between AUC of Direct and Brainage64D, as well as between Direct and Brainage64D-finetune across different development set sizes (i.e., number of training and validation participants). Bolded p values indicate statistical significance after FDR correction (q < 0.05). Given their poor performance (Figure 2), we did not consider BAG and BAG-finetune in this analysis.
| Number of training and validation participants |
Direct vs Brainage64D | Direct vs Brainage64D-finetune |
|---|---|---|
| 50 | 0.8816 | 0.8314 |
| 100 | 0.8696 | 0.5990 |
| 200 | 0.6743 | 0.2287 |
| 300 | 0.2941 | 0.2888 |
| 400 | 0.0368 | 0.1035 |
| 500 | 0.0010 | 0.3727 |
| 600 | 0.0001 | 0.5728 |
| 700 | 2.7106e-5 | 0.5171 |
| 800 | 2.0102e-6 | 0.3845 |
| 900 | 4.0756e-8 | 0.5238 |
| 997 | 7.4415e-9 | 0.6551 |
Brainage64D was numerically better than the Direct approach for development set sizes of 50 and 100, but the improvement was not statistically significant. The Direct approach was numerically better than Brainage64D from a development set size of 200 onwards, which became statistically significant when development set size was at least 500.
On the other hand, the Brainage64D-finetune was numerically better than the direct approach for all sample sizes, but differences were not significant even when sample size was very small (N = 50). Overall, this suggests that leveraging a pretrained brain age model did not improve AD classification over training a model directly.
2.4. Leveraging a pretrained brain age model does not improve MCI progression prediction over training a model directly
For the MCI progression prediction, we compared two direct models (Direct and Direct-AD2prog) with the best brain age derived model (Brainage64D-finetune-AD2prog; Figure 4). Both Direct-AD2prog and Brainage64D-finetune-AD2prog utilized intermediate representations (features) from the best AD classification models (Direct and Brainage64D-finetune) as inputs to train a new classifier for predicting MCI progression (Figure 4). For more details, see Methods (Section 5.7).
Figure 4.
Workflow of three MCI progression prediction models. Given a T1 MRI scan, each model predicted a classification label (sMCI or pMCI). (A) “Direct” used the same 3D CNN architecture as the pretrained brain age model (Leonardsen et al., 2022), except for the final binary classification layer. Classification labels (sMCI or pMCI) and T1 images of the development set were used to train the 3D CNN from scratch. (B) “Direct-AD2prog” extracted 64-dimensional (64D) features from the output of the global averaging pooling layer of the previously trained Direct AD classification model (Figure 1A). The 64D features and classification labels (sMCI/pMCI) of the development set were used to train a logistic regression model. (C) “Brainage64D-finetune-AD2prog” extracted 64-dimensional (64D) features from the output of the global averaging pooling layer of the previously trained brainage64D-finetune AD classification model (Figure 1E). The 64D features and classification labels (sMCI/pMCI) of the development set were used to train a logistic regression model.
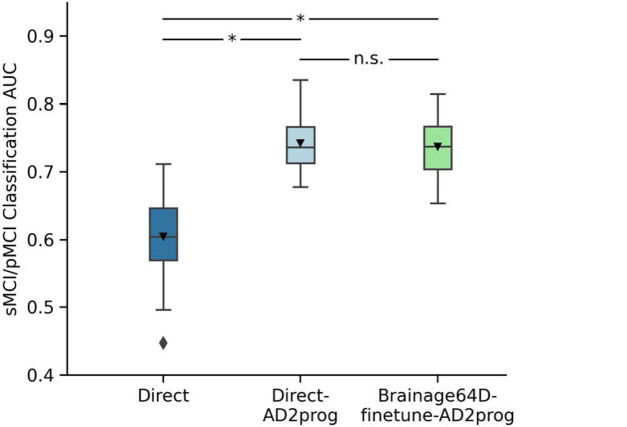
Figure 5 shows the MCI progression prediction AUC for all three approaches. Table 4 reports the p values from comparing pairs of approaches using the corrected resampled t-test (Nadeau & Bengio, 2003). Both Direct-AD2prog and Brainage64D-finetune-AD2prog performed better than the direct approach. There was no statistical difference between Direct-AD2prog and Brainage64D-finetune-AD2prog.
Figure 5.
MCI progression prediction AUC. Box plots show the test AUC across 50 random training-validation-test splits. For each box plot, the horizontal line indicates the median across 50 test AUC values. Triangle indicates the mean. The bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Outliers are defined as data points beyond 1.5 times the interquartile range. The whiskers extend to the most extreme data points not considered outliers. P values are computed using the corrected resampled t-test. “*” indicates statistical significance after FDR correction (q < 0.05) and “n.s.” indicates not significant after FDR correction.
Table 4.
MCI progression prediction AUC (mean ± std) and uncorrected p values between pairs of approaches. AUC was averaged across 50 random training-validation-test splits. P values were computed using the corrected resampled t-test (Nadeau & Bengio, 2003). Bolded p values indicate statistical significance after FDR correction (q < 0.05).
| sMCI/pMCI classification AUC (mean ± std) |
P values | ||
|---|---|---|---|
| Direct | Direct-AD2prog | Brainage64D-finetune- AD2prog |
|
| Direct (0.6047 ± 0.0587) | 2.90e-5 | 3.00e-4 | |
| Direct-AD2prog (0.7426 ± 0.0382) | 0.756 | ||
| Brainage64D-finetune-AD2prog (0.7375 ± 0.0440) | |||
Consistent with previous studies (Lian et al., 2020; Oh et al., 2019; Wen et al., 2020), our results suggest that MCI progression prediction can be improved by transferring features from previously trained AD classification models. However, we did not observe any additional benefit from leveraging features of a pretrained brain age model.
2.5. Even when sample size is small, leveraging a pretrained brain age model does not improve MCI prediction over training a model directly
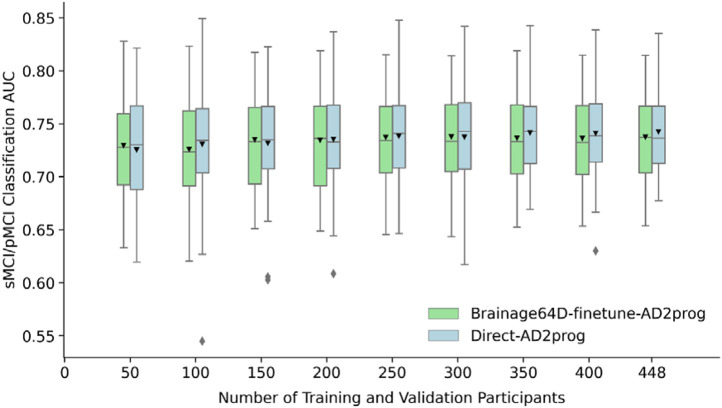
Adapting a pretrained model (trained from large datasets) for a new classification task might be more advantageous when the sample size available for the new task is small. Figure 6 shows the MCI prediction AUC (across 50 training-validation-test splits) for Direct-AD2prog and Brainage64D-finetune-AD2prog across different development set sizes. We did not consider the “direct” approach for this analysis, given its poor performance (Figure 5). Table 5 shows the actual AUC values and p values from comparing Direct-AD2prog and Brainage64D-finetune-AD2prog. Across all sample sizes, there was no statistical difference between Direct-AD2prog and Brainage64D-finetune-AD2prog. Overall, this suggests that even when sample size is small, there was not a significant advantage in leveraging features from a pretrained brain age model.
Figure 6.
MCI progression prediction AUC of Brainage64D-finetune-AD2prog and Direct-AD2prog across different development set sizes. Boxplots showing test AUC across different development set sizes (number of training and validation participants). Test sets were identical across development set sizes, so AUCs were comparable across sample sizes. For each boxplot, the horizontal line indicates the median across 50 test AUC values. Triangle indicates the mean. The bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Outliers are defined as data points beyond 1.5 times the interquartile range. The whiskers extend to the most extreme data points not considered outliers.
Table 5.
MCI prediction AUC (mean ± std) and p values comparing Direct-AD2prog and Brainage64D-finetune-AD2prog across different development set sizes (i.e., number of training and validation participants). Given its poor performance (Figure 5), we did not include the “Direct” approach in this analysis. Across all sample sizes, there was no statistically significant difference.
| Number of training and validation participants |
Direct-AD2prog | Brainage64D- finetune-AD2prog |
P values |
|---|---|---|---|
| 50 | 0.7253 ± 0.0482 | 0.7294 ± 0.0450 | 0.9587 |
| 100 | 0.7307 ± 0.0508 | 0.7257 ± 0.0484 | 0.9240 |
| 150 | 0.7316 ± 0.0475 | 0.7350 ± 0.0452 | 0.9296 |
| 200 | 0.7352 ± 0.0451 | 0.7345 ± 0.0458 | 0.9839 |
| 250 | 0.7385 ± 0.0418 | 0.7373 ± 0.0457 | 0.9624 |
| 300 | 0.7376 ± 0.0463 | 0.7377 ± 0.0450 | 0.9965 |
| 350 | 0.7415 ± 0.0402 | 0.7365 ± 0.0441 | 0.7939 |
| 400 | 0.7411 ± 0.0417 | 0.7362 ± 0.0441 | 0.7905 |
| 448 | 0.7426 ± 0.0382 | 0.7375 ± 0.0440 | 0.7558 |
3. DISCUSSION
In this study, we evaluated the utility of brain age models in generating specific markers of brain health in the domain of AD dementia. We found that models directly trained to predict AD-related outcomes (i.e., direct models) performed as well as, or even better than, brain age derived models. Even when training data was scarce (N = 50), brain age derived models did not statistically outperform direct models. Overall, given the widespread availability of brain data with age-only information, we believe that brain age can still be useful as a marker of general brain health. However, our results suggest that current large-scale brain age models do not offer a strong advantage for predicting specific health outcomes.
To interpret our results, it is useful to think of brain age derived models as a form of transfer learning (Weiss, Khoshgoftaar, & Wang, 2016; Zhuang et al., 2021), which can be broadly defined as using past experience from one or more source tasks to improve learning on a target task (Hospedales et al., 2022). In the case of brain age derived models, we first train a model to predict age in a large dataset, followed by model transfer to predict another phenotype in a new dataset. There are a few factors that might influence the success of transfer learning in the current study.
The first factor to consider is sample size. Given that the brain age model is trained from a very large dataset (N > 50,000), the hope is that the features will be more robust than those learned directly from a small dataset with target outcomes of interest (N < 1000), thus improving the performance of the target outcomes. We note that in the scenario with very small sample sizes (N = 50), there were >1000 times more data for training the brain age model than the target task, yet brain age models did not yield a significant advantage over direct models.
The second factor to consider is the similarity between source and target tasks. Transfer learning is easier if source and target tasks are more similar. Conversely, transfer learning is harder if source and target tasks are more different. However, given that age is the strongest risk factor for AD dementia, we believe the source and target tasks in our study are relatively well-aligned.
The final factor is the impact of well-documented MRI site differences (An et al., 2024; Fortin et al., 2018; Pomponio et al., 2020) which may degrade model transfer between the original large-scale data to the new data. Previous studies have suggested that training models on large diverse datasets could overcome site differences without having to explicitly perform harmonization (Abraham et al., 2017; Chen et al., 2024). Since the pretrained brain age model was trained on a wide variety of scanners and populations, we did not think that this would be a serious issue for the brain age derived models.
Overall, when considering all three factors, we note that it was not a foreordained result that brain age derived models and direct models would have similar performance. Indeed, an interesting finding is that BAG exhibited the worst performance, and finetuning did not improve the performance of BAG. This suggests that while BAG might be a powerful marker of general brain health, too much information is lost by reducing a person’s brain health to a single number, thus diminishing its utility for predicting specific brain health outcomes.
Furthermore, Brainage64D exhibited worse performance than Brainage64D-finetune and Direct models. On the other hand, Brainage64D-finetune and Direct models performed similarly well in the AD classification task. Brainage64-finetune-AD2prog and Direct-AD2prog models also performed similarly well in the MCI progression task. Overall, this suggests the importance of finetuning brain age models to new tasks potentially due to site differences or due to task misalignment between predicting chronological age and AD-related health outcomes.
Finally, it is important to note that brain age derived models are just one approach of transfer learning. There is a plethora of transfer learning approaches in the brain imaging (Deepak & Ameer, 2019; Malik & Bzdok, 2022; Mei et al., 2022) and machine learning (Bengio, 2011; Palatucci et al., 2009; Shin et al., 2016) literature. We believe that the idea of translating models trained from large-scale datasets to predict new phenotypes in small datasets remains a promising one. However, similarity between the source and target tasks is an important factor that needs to be taken care of to maximize transfer learning performance (Chen et al., 2024; He et al., 2022; Wulan et al., 2024).
Indeed, a recent study suggests that age prediction based on neuropsychological measures leads to better MCI progression prediction than BAG from brain imaging data (Garcia Condado, Cortes, & Initiative, 2023). Furthermore, there are also studies that have developed biological age models based on mortality (Levine et al., 2018). It is possible that brain age models trained on mortality, rather than chronological age, could yield better markers of specific brain health. We leave this to future work.
A limitation of the current study is that we only considered one pretrained brain age model (Leonardsen et al., 2022). We believe that this model remains the best (or one of the best) in the field, but we do not preclude the possibility that other brain age models trained on even larger and more diverse datasets might yield better transfer learning results. However, new brain age models are typically trained to improve chronological age prediction (Dartora et al., 2024; Kalc et al., 2024). In doing so, the models might learn better features for predicting age, but not necessarily for predicting other phenotypes.
Another limitation is that for fair comparison, the neural network architecture and training procedure were constrained to be the same between brain age derived models and direct models. For example, given the relatively small sample sizes available to the direct models, it might make sense to use a less complex neural network architecture (e.g., less layers). Consequently, our AD classification and MCI progression prediction results are not the best in the field.
Studies in the past five years have reported AD classification AUC ranging from 0.82 to 0.99 (Ashtari-Majlan, Seifi, & Dehshibi, 2022; Bashyam et al., 2020; Kang et al., 2023; Leonardsen et al., 2022; Lu et al., 2022; Ocasio & Duong, 2021; Yin et al., 2024; Zarei et al., 2024; Zheng, Pfahringer, & Mayo, 2022), and MCI progression prediction AUC ranging from 0.62 to 0.94 (Ashtari-Majlan, Seifi, & Dehshibi, 2022; Bron et al., 2021; Gao et al., 2020; Kang et al., 2023; Li et al., 2021; Luo et al., 2024; Nanni et al., 2020; Ocasio & Duong, 2021; Zhou et al., 2024). Therefore, while our AD classification AUC (0.92) and MCI progression prediction AUC (0.74) are not the best, they are still in line with recent studies, especially given our architecture limitations.
Furthermore, it is important to emphasize that prediction performance is not directly comparable across studies because of different patient selection criteria, problem set up, dataset differences, and so on. However, a meaningful comparison can be made with Leonardsen and colleagues (Leonardsen et al., 2022) because we have strived to align our setup with theirs as closely as possible. The highly similar AD classification AUCs between our Brainage64 model (0.84) and theirs (0.83) suggest that our implementation is not biased against brain age derived models.
4. CONCLUSION
Brain age is a powerful marker of general brain health. However, it remains unclear whether brain age derived models are better than models directly trained to predict the specific brain health outcomes. Surprisingly, despite three orders of magnitude more training data (50,000 vs 50), brain age models did not outperform direct models for predicting AD-related health outcomes. Overall, our results suggest that if we are interested in specific markers of brain health, then currently, it might be more advantageous to directly train models from datasets with target outcomes of interest.
5. METHODS
5.1. Datasets
In this study, we considered three datasets: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset (Jack et al., 2010; Jack et al., 2008; Mueller et al., 2005), the Australian Imaging, Biomarkers and Lifestyle (AIBL) study (Ellis et al., 2009; Ellis et al., 2010; Fowler et al., 2021) and the Singapore Memory Aging and Cognition center (MACC) Harmonization cohort (Chong et al., 2017; Hilal et al., 2015; Hilal et al., 2020; Xu et al., 2015). Each dataset included both MRI data and clinical data collected at multiple timepoints.
In the first classification task, our goal was to predict whether an individual was cognitively normal (CN) or diagnosed with AD dementia at baseline using the baseline anatomical T1 scan. We refer to this task as AD classification. In the second classification task, our goal was to predict whether an individual with mild cognitive impairment (MCI) at baseline progressed to AD dementia within 36 months (i.e., progressive MCI or pMCI) or remained mild cognitively impaired (i.e., stable MCI or sMCI) based on the baseline anatomical T1 scan. We refer to this task as MCI progression prediction. Individuals who (1) exhibited more than one diagnosis changes (e.g., MCI → AD → MCI), (2) reverted to CN (i.e., MCI → CN), or (3) had missing diagnoses (such that we could not determine whether the individual should be considered pMCI or sMCI) were excluded. We note that there were no overlapping participants used for AD classification and MCI progression prediction tasks
For the ADNI dataset, we considered participants from ADNI 1, ADNIGo/2, and ADNI3. T1 scans were acquired using 1.5T and 3T scanners from Siemens, Philips, and General Electric. At baseline, there were 2,039 scans, comprising 942 CN individuals, 432 individuals with AD dementia, 391 individuals who were sMCI, and 274 individuals who were pMCI. For the AIBL dataset, T1 scans were collected from Siemens 3T scanners. At baseline, there were 580 scans from 479 CN individuals, 78 individuals with AD dementia, 13 individuals who were sMCI, and 10 individuals who were pMCI. For the MACC dataset, T1 scans were collected from a Siemens 3T Tim Trio scanner, and a Siemens 3T Prisma scanner. At baseline, there were 457 scans from 132 CN individuals, 207 individuals with AD dementia, 79 individuals who were sMCI, and 39 individuals who were pMCI.
5.2. Sample stratification
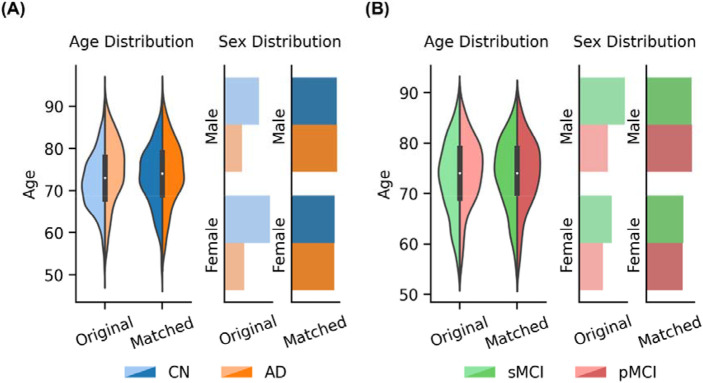
Following Leonardsen and colleagues (Leonardsen et al., 2022), for the AD classification task, age and sex matching were performed for each scanner model to ensure the same age and sex distribution between the two diagnostic groups. For instance, suppose Scanner Model X in Dataset A has more participants with AD dementia than CN participant, then for each CN participant, we found the closest matching AD participant in terms of sex and age. Once all the CN participants were successfully matched, any excess AD participants were excluded from subsequent analyses. After matching, there were 636 CN participants and 636 participants with AD dementia. Figure 7A and Table 6 show the age and sex distributions of participants before and after matching. The same matching procedure was performed for the MCI progression prediction task (Figure 7B and Table 7), yielding 288 participants with sMCI and 288 participants with pMCI.
Figure 7.
Age and sex distributions for each group before and after matching. (A) Age and sex distributions of CN participants and participants with AD dementia, before and after matching. (B) Age and sex distributions of participants with sMCI and pMCI, before and after matching.
Table 6.
Participant demographics of three datasets (AIBL, ADNI, MACC) used for AD classification before and after matching age and sex. After matching, the paired t-test p value between the age of CN participants and participants with AD dementia was 0.031. For sex, the p value for the chi-square goodness of fit test between the two diagnostic groups was 0.99.
| Participants | Age (mean ± std years) |
Sex (female) |
||
|---|---|---|---|---|
| Before matching | CN | 1553 (AIBL 479, ADNI 942, MACC 132) | 71.9 ± 6.71 | 57% |
| AD | 717 (AIBL 78, ADNI 432, MACC 207) | 75.0 ± 7.64 | 53% | |
| After matching | CN | 636 (AIBL 78, ADNI 428, MACC 130) | 73.3 ± 7.21 | 49% |
| AD | 636 (AIBL 78, ADNI 428, MACC 130) | 74.2 ± 7.60 | 48% | |
Table 7.
Participant demographics of three datasets (AIBL, ADNI, MACC) used for MCI progression prediction before and after matching age and sex. After matching, the paired t-test p value between the age of the two groups (sMCI and pMCI) was 0.87. For sex, p value for chi-square goodness of fit test between the two groups was 0.99.
| Participants | Age (mean years ± SD) |
Sex (female) |
||
|---|---|---|---|---|
| Before matching | sMCI | 483 (AIBL: 13, ADNI 391, MACC: 79) | 73.0 ± 7.38 | 42% |
| pMCI | 323 (AIBL: 10, ADNI 274, MACC: 39) | 74.2 ± 6.83 | 45% | |
| After matching | sMCI | 288 (AIBL 10, ADNI 239, MACC 39) | 73.9 ± 6.93 | 45% |
| pMCI | 288 (AIBL 10, ADNI 239, MACC 39) | 74.0 ± 6.87 | 44% | |
5.3. Train, validation, and test split
For the AD classification task, we split the matched participants into a development set and test set, maintaining an 80:20 ratio. The development set was further divided into a training set and a validation set, also maintaining an 80:20 ratio. This results in training-validation-test split ratio of 64:16:20. The split into training, validation and test sets was performed separately for each scanner model of each dataset.
In general, the training set was used to train the parameters of each classification model. The validation set was used for early stopping (see details in Section 5.6). Finally, the test set was used to evaluate the performance of the model. This procedure was repeated 50 times with a different split of the participants into training, validation and test sets. The same procedure was repeated for the MCI progression prediction task.
5.4. T1 preprocessing
We performed the same T1 preprocessing as Leonardsen et al., 2022. Briefly, the T1 scans underwent skull stripping using FreeSurfer recon-all (Reuter, Rosas, & Fischl, 2010; Ségonne et al., 2004), which generated a brain mask to remove non-brain areas and the skull. Subsequently, the brain was aligned to the standard FSL orientation via fslreorient2std (Jenkinson et al., 2012). Afterward, the images were linearly registered to MNI152 space using FLIRT (Greve & Fischl, 2009; Jenkinson et al., 2002; Jenkinson & Smith, 2001) with linear interpolation and a rigid body transformation with 6 degrees of freedom. The registration process used the FSL MNI152 1mm template. After registration, the images were cropped along the borders at [6:173, 2:214, 0:160] (using python indexing convention), resulting in 3D volumes of dimensions 167 × 212 × 160. This cropping procedure resulted in a compact cuboid that preserved almost all brain-related information. Finally, the voxel intensity values of all images were normalized to a range of [0, 1] by dividing all voxel intensities by 255.
5.5. Neural network backbone
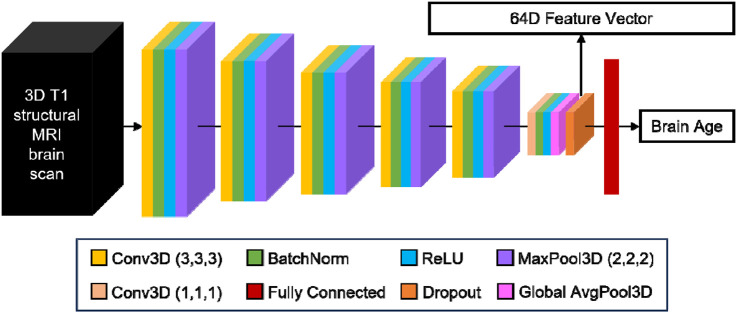
The pretrained brain age model (Leonardsen et al., 2022) utilized the Simple Fully Convolutional Network (SFCN) backbone (Leonardsen et al., 2022; Peng et al., 2021). Therefore, we used the SFCN architecture for all tested models (Figure 2). The SFCN backbone comprised 5 repeated convolutional blocks and an appended 6th convolutional block. Each of the 5 repeated convolutional blocks consisted of a 3D convolutional layer with a filter size of (3, 3, 3), zero padding with a padding size of 1, and a stride of 1. This results in convolutional layers of the same size as the input image. Each convolutional layer is followed by a batch normalization layer, rectified linear activation function (ReLU) activation, and a max pooling layer with a pooling size of (2, 2, 2). This results in the output size of each convolutional block being reduced by half across the height, width, and depth, compared to the input size. The appended 6th convolutional block incorporates a channel-wise convolutional layer, a final batch normalization layer, and a global average pooling layer. The number of filters used in the convolutional layers are [32, 64, 128, 256, 256, 64], so the output of the global average pooling layer is of length 64. In the SFCN-regression model (Leonardsen et al., 2022), the 64 features were entered into a fully connected layer (with one output node) to directly predict chronological age. Because the pretrained SFCN-regression model achieved the best brain age prediction performance on external data (Leonardsen et al., 2022), we used the SFCN-regression model for the above analyses.
5.6. AD classification models
We compared five approaches for AD classification (Figure 1): Direct approach (Figure 1A), brain age gap (BAG; Figure 1B), BAG-finetune (Figure 1C), Brainage64D (Figure 1D) and Brainage64D-finetune (Figure 1E). All computations involving SFCN utilized NVIDIA RTX 3090 GPUs with 24GB memory and CUDA 11.0.
5.6.1. Direct approach
In the “Direct” approach, we trained a SFCN model from scratch to predict the classification label. More specifically, we replaced the fully connected layer (with one output node) of the SFCN-regression model with a fully connected layer with two output nodes for CN and AD classification labels respectively (Figure 1A). Stochastic gradient descent (SGD) was used to minimize the cross-entropy loss. Because of the computational cost, no hyperparameter was tuned. Instead, we used a fixed set of hyperparameters: weight decay = 1e-4, dropout rate = 0.5, and initial learning rate = 0.1. The learning rate was decreased by a factor of 10 every 30 epochs. The training batch size was set to 6 due to GPU memory constraints. For each training-validation-test split, the Direct approach was trained from scratch (using randomly initialized weights) for 150 epochs in the training set. Model parameters with the highest area under the receiver operating characteristic curve (AUC) in the validation set was used for evaluation in the test set. The evaluation metric was also AUC. For each training-validation-test split, the training duration was approximately 5 hours.
5.6.2. Brain age gap (BAG) approach
The remaining four approaches involved the pretrained SFCN-regression model (Leonardsen et al., 2022). The first brain-age approach was simply the brain age gap (BAG) generated by the pretrained SFCN-regression model (Figure 1B). More specifically, the preprocessed T1 image of a test participant was fed into the pretrained brain age model. The BAG was then defined as the predicted age minus the actual chronological age of the participant. If the BAG was above a certain BAG threshold, we would classify the participant as having AD dementia. Otherwise, we classified the participant as CN. By varying the BAG threshold, we could compute the area under the receiver operating characteristic curve (AUC) for this approach. We note that the training and validation sets were not used at all for the BAG approach.
5.6.3. Brain age gap finetune (BAG-finetune) approach
As seen in Section 2.2, the BAG approach did not result in a good classification accuracy. One potential reason is that the new datasets (ADNI, AIBL and MACC) were too different from the original multi-site datasets used to train the brain age model (Leonardsen et al., 2022). Therefore, we considered the BAG-finetune approach (Figure 1C), in which the pretrained brain age model was finetuned to predict chronological age in the CN participants. More specifically, for each training-validation-test split, the weights of the pretrained SFCN-regression model were finetuned to predict the chronological age of the CN participants in the training set using the mean absolute error (MAE) loss. The same hyperparameters were used as the Direct approach, except that the initial learning rate was set to a low value of 0.01 to avoid significant deviation from the original pretrained weights. The validation set was used to select the epoch with the hyperparameters that yielded the best chronological age prediction based on MAE. The finetuned brain age model was then used to generate brain age gap in each test participant, which was in turn used to compute AUC in the test set.
5.6.4. Brain age 64D (Brainage64D) approach
As will be seen, the BAG-finetune approach also did not perform well, so another possible hypothesis is that summarizing a participant with just a single scalar (brain age gap) might be losing too much information. Therefore, we considered the brain age 64D (Brainage64D) approach, in which a logistic ridge regression model was trained on the 64-dimensional output of the global averaging pooling layer in the pretrained SFCN-regression model (Figure 1D). The scikit-learn package (Pedregosa et al., 2011) was used. The logistic ridge regression model included an inverse regularization parameter λ (larger value indicated lower regularization). Model fitting was performed on the training set, and the inverse regularization parameter was determined based on AUC in the validation set.
The optimal hyperparameter was selected from 0.001, 0.01, 0.1, 1, 10, 100 or 1000. The best hyperparameter was then used to retrain the logistic regression model on the full development (training and validation) set. The final trained Brainage64D model was the concatenation of the pretrained SFCN-regression model up to (and including) the global average pooling layer and the trained logistic regression model. This final model was then evaluated in the test set.
5.6.5. Brain age 64D finetune (Brainage64D-finetune) approach
As seen in Section 2.2, the Brainage64D approach performed better than BAG (Section 5.6.2) and BAG-finetune (Section 5.6.3), but was still worse than the Direct approach. One potential reason is that the new datasets (ADNI, AIBL and MACC) were too different from the original multi-site datasets used to train the brain age model (Leonardsen et al., 2022). Therefore, we considered a Brainage64D-finetune approach (Figure 1E), in which we finetuned the previously trained Brainage64D model (Section 5.6.4). All layers of the trained Brainage64D model were finetuned, using the same cost function and hyperparameters as the Direct approach (in Section 5.6.1), except that the initial learning rate was set to a low value of 0.01 to avoid significant deviation from the original pretrained weights. Model parameters with the highest AUC in the validation set was then used for evaluation in the test set.
5.7. MCI progression prediction models
We compared three approaches for MCI progression prediction (Figure 4): Direct approach (Figure 4A), Direct-AD2prog (Figure 4B) and Brainage64D-AD2prog (Figure 4C). All computations involving SFCN utilized NVIDIA RTX 3090 GPUs with 24GB memory and CUDA 11.0.
5.7.1. Direct approach
The “Direct” approach for predicting MCI progression is the same as “Direct” approach for AD classification. In other words, we trained a SFCN model from scratch to predict whether a participant was pMCI or sMCI. Similar to Section 5.6.1, we replaced the fully connected layer (with one output node) of the SFCN-regression model with a fully connected layer with two output nodes for sMCI and pMCI classification labels respectively (Figure 4A). Stochastic gradient descent (SGD) was used to minimize the cross-entropy loss. Because of the computational cost, no hyperparameter was tuned. Instead, we used a fixed set of hyperparameters: weight decay = 1e-4, dropout rate = 0.5, and initial learning rate = 0.1. The learning rate was decreased by a factor of 10 every 30 epochs. The training batch size was set to 6 due to GPU memory constraints. For each training-validation-test split, the Direct approach was trained from scratch (using randomly initialized weights) for 150 epochs in the training set. Model parameters with the highest area under the receiver operating characteristic curve (AUC) in the validation set was used for evaluation in the test set.
5.7.2. Direct-AD2prog
As seen in Section 2.4, the prediction performance of the “Direct” approach was not good. Previous studies have suggested that adapting an AD classification task to predict MCI progression can improve prediction performance, compared with training a model from scratch (Lian et al., 2020; Oh et al., 2019; Wen et al., 2020). Therefore, we extracted the 64-dimensional output of the global averaging pooling layer of the previously trained Direct AD classification models (Section 5.6.1; Figure 1A). We then trained a logistic ridge regression model using the 64-dimensional features to predict whether a participant progressed to AD dementia. We refer to this approach as Direct-AD2prog (Figure 4B), where “AD2prog” refers to the fact that we transferred features from the AD classification model to build a new model to predict disease progression.
Consistent with Section 5.6.4, the logistic ridge regression model included an inverse regularization parameter λ (larger value indicated lower regularization), which was determined based on AUC in the validation set. The optimal hyperparameter was selected from 0.001, 0.01, 0.1, 1, 10, 100 or 1000. The best hyperparameter was then used to retrain the logistic regression model on the development (training and validation) set. The trained model was then evaluated on the test set.
5.7.3. Brainage64D-finetune-AD2prog
Similar to Direct-AD2prog, we also extracted the 64-dimensional output of the global averaging pooling layer of the previously trained Brainage64D-finetune AD classification models (Section 5.6.5). We then trained a logistic ridge regression model using the 64-dimensional features to predict whether a participant progressed to AD dementia. We refer to this approach as Brainage64D-finetune-AD2prog (Figure 4C). Consistent with previous sections, we again select the optimal regularization hyperparameter (from 0.001, 0.01, 0.1, 1, 10, 100 or 1000) based on the validation set. The best hyperparameter was then used to retrain the logistic regression model on the full development (training and validation) set. The trained model was then evaluated on the test set.
5.8. Vary training and validation set sizes
Adapting a pretrained model (trained from large datasets) for a new classification task might be more advantageous when the sample size available for the new task is small. Therefore, we repeated the previous AD classification and MCI progression prediction tasks by varying the size of the development (training and validation) set.
Recall that we have previously repeated the training-validation-test procedure 50 times (Section 5.3). In the current analysis, for each of these 50 repetitions, we in turn randomly sub-sample the development (training and validation) set. In the case of AD classification, we varied the development set size as follows: 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, and the maximum development set size of 997. In the case of MCI progression prediction, we varied the development set size as follows: 50, 100, 150, 200, 250, 300, 350, 400, and the maximum development set size of 448.
Consistent with previous analyses, the development set was in turn divided into training and validation sets with an 80:20 ratio. The test set was also maintained to be the same across all development set sample sizes, so that the prediction accuracies were comparable across development sample sizes.
In the case of AD classification, care was taken so that when the subsampled development set had the same numbers of CN participants and participants with AD dementia. Since BAG and BAG-finetune performed poorly in the main analysis (Figure 2), for this analysis we only considered Brainage64D, Brainage64D-finetune and Direct approach.
In the case of MCI progression prediction, care was taken so that when the subsampled development set had the same numbers of sMCI and pMCI participants. Since Direct performed poorly in the main analysis (Figure 5), for this analysis we only considered Direct-AD2prog and Brainage64D-finetune-AD2prog. Furthermore, while we varied the development set size, the input features for both approaches were extracted from AD classification models trained from the full sample size (Section 5.7).
Figure 8.
Schematic of Simply Fully Convolutional Network (SFCN) architecture (Leonardsen et al., 2022; Peng et al., 2021). The number of filters used in the convolutional layers were [32, 64, 128, 256, 256, 64]. The output of the global average pooling layer was of length 64. In the pretrained SFCN-regression model (Leonardsen et al., 2022), the 64-dimensional output of the global average pooling layer was fed into a fully connected layer (with one output node) to directly predict chronological age.
ACKNOWLEDGEMENTS
This research is supported by the NUS Yong Loo Lin School of Medicine (NUHSRO/2020/124/TMR/LOA), the Singapore National Medical Research Council (NMRC) LCG (OFLCG19May-0035), NMRC CTG-IIT (CTGIIT23jan-0001), NMRC OF-IRG (OFIRG24jan-0030), NMRC STaR (STaR20nov-0003), Singapore Ministry of Health (MOH) Centre Grant (CG21APR1009), the Temasek Foundation (TF2223-IMH-01), and the United States National Institutes of Health (R01MH133334). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not reflect the views of the funders. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
REFERENCES
- Abraham A., Milham M. P., Di Martino A., Craddock R. C., Samaras D., Thirion B., & Varoquaux G. (2017). Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. Neuroimage, 147, 736–745. https://doi.org/ 10.1016/j.neuroimage.2016.10.045 [DOI] [PubMed] [Google Scholar]
- An L., Zhang C., Wulan N., Zhang S., Chen P., Ji F., Ng K. K., Chen C., Zhou J. H., & Yeo B. T. T. (2024). DeepResBat: deep residual batch harmonization accounting for covariate distribution differences. Medical Image Analysis, 103354. https://doi.org/ 10.1016/j.media.2024.103354 [DOI] [PubMed] [Google Scholar]
- Ashtari-Majlan M., Seifi A., & Dehshibi M. M. (2022). A Multi-Stream Convolutional Neural Network for Classification of Progressive MCI in Alzheimer’s Disease Using Structural MRI Images. IEEE Journal of Biomedical and Health Informatics, 26(8), 3918–3926. 10.1109/JBHI.2022.3155705 [DOI] [PubMed] [Google Scholar]
- Bashyam V. M., Erus G., Doshi J., Habes M., Nasrallah I., Truelove-Hill M., Srinivasan D., Mamourian L., Pomponio R., Fan Y., Launer L. J., Masters C. L., Maruff P., Zhuo C., Volzke H., Johnson S. C., Fripp J., Koutsouleris N., Satterthwaite T. D., … Davatzikos C. (2020). MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain, 143(7), 2312–2324. 10.1093/brain/awaa160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky D. W., Caspi A., Houts R., Cohen H. J., Corcoran D. L., Danese A., Harrington H., Israel S., Levine M. E., Schaefer J. D., Sugden K., Williams B., Yashin A. I., Poulton R., & Moffitt T. E. (2015). Quantification of biological aging in young adults. Proceedings of the National Academy of Sciences, 112(30), E4104–E4110. https://doi.org/doi: 10.1073/pnas.1506264112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengio Y. (2011). Deep learning of representations for unsupervised and transfer learning Proceedings of the 2011 International Conference on Unsupervised and Transfer Learning workshop - Volume 27, Washington, USA. [Google Scholar]
- Bron E. E., Klein S., Papma J. M., Jiskoot L. C., Venkatraghavan V., Linders J., Aalten P., De Deyn P. P., Biessels G. J., Claassen J. A. H. R., Middelkoop H. A. M., Smits M., Niessen W. J., van Swieten J. C., van der Flier W. M., Ramakers I. H. G. B., & van der Lugt A. (2021). Cross-cohort generalizability of deep and conventional machine learning for MRI-based diagnosis and prediction of Alzheimer’s disease. NeuroImage: Clinical, 31, 102712. https://doi.org/ 10.1016/j.nicl.2021.102712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. H., Marioni R. E., Colicino E., Peters M. J., Ward-Caviness C. K., Tsai P. C., Roetker N. S., Just A. C., Demerath E. W., Guan W., Bressler J., Fornage M., Studenski S., Vandiver A. R., Moore A. Z., Tanaka T., Kiel D. P., Liang L., Vokonas P., Horvath S. (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY), 8(9), 1844–1865. 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., An L., Wulan N., Zhang C., Zhang S., Ooi L. Q. R., Kong R., Chen J., Wu J., Chopra S., Bzdok D., Eickhoff S. B., Holmes A. J., & Yeo B. T. T. (2024). Multilayer meta-matching: Translating phenotypic prediction models from multiple datasets to small data. Imaging Neuroscience, 2, 1–22. 10.1162/imag_a_00233 [DOI] [Google Scholar]
- Cheng S. F., Yue W. L., Ng K. K., Qian X., Liu S., Tan T. W. K., Nguyen K.-N., Leong R. L. F., Hilal S., Cheng C.-Y., Tan A. P., Law E. C., Gluckman P. D., Chen C. L.-H., Chong Y. S., Meaney M. J., Chee M. W. L., Yeo B. T. T., & Zhou J. H. (2024). Rate of brain aging associates with future executive function in Asian children and older adults. In: eLife Sciences Publications, Ltd. [Google Scholar]
- Choi U.-S., Park J. Y., Lee J. J., Choi K. Y., Won S., & Lee K. H. (2023). Predicting mild cognitive impairments from cognitively normal brains using a novel brain age estimation model based on structural magnetic resonance imaging. Cerebral Cortex, 33(21), 10858–10866. 10.1093/cercor/bhad331 [DOI] [PubMed] [Google Scholar]
- Chong J. S. X., Liu S., Loke Y. M., Hilal S., Ikram M. K., Xu X., Tan B. Y., Venketasubramanian N., Chen C. L., & Zhou J. (2017). Influence of cerebrovascular disease on brain networks in prodromal and clinical Alzheimer's disease. Brain, 140(11), 3012–3022. 10.1093/brain/awx224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. H. (2020). Multimodality neuroimaging brain-age in UK biobank: relationship to biomedical, lifestyle, and cognitive factors. Neurobiology of Aging, 92, 34–42. https://doi.org/ 10.1016/j.neurobiolaging.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. H., Poudel R. P. K., Tsagkrasoulis D., Caan M. W. A., Steves C., Spector T. D., & Montana G. (2017). Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage, 163, 115–124. https://doi.org/ 10.1016/j.neuroimage.2017.07.059 [DOI] [PubMed] [Google Scholar]
- Cole J. H., Ritchie S. J., Bastin M. E., Valdés Hernández M. C., Muñoz Maniega S., Royle N., Corley J., Pattie A., Harris S. E., Zhang Q., Wray N. R., Redmond P., Marioni R. E., Starr J. M., Cox S. R., Wardlaw J. M., Sharp D. J., & Deary I. J. (2018). Brain age predicts mortality. Molecular Psychiatry, 23(5), 1385–1392. 10.1038/mp.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides C., Han L. K. M., Alloza C., Antonucci L. A., Arango C., Ayesa-Arriola R., Banaj N., Bertolino A., Borgwardt S., Bruggemann J., Bustillo J., Bykhovski O., Calhoun V., Carr V., Catts S., Chung Y.-C., Crespo-Facorro B., Díaz-Caneja C. M., Donohoe G., Consortium E. S. (2023). Brain ageing in schizophrenia: evidence from 26 international cohorts via the ENIGMA Schizophrenia consortium. Molecular Psychiatry, 28(3), 1201–1209. 10.1038/s41380-022-01897-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumplido-Mayoral I., Brugulat-Serrat A., Sánchez-Benavides G., González-Escalante A., Anastasi F., Milà-Alomà M., López-Martos D., Akinci M., Falcón C., Shekari M., Cacciaglia R., Arenaza-Urquijo E. M., Minguillón C., Fauria K., Molinuevo J. L., Suárez-Calvet M., Grau-Rivera O., Vilaplana V., Gispert J. D., Vilor Tejedor N. (2024). The mediating role of neuroimaging-derived biological brain age in the association between risk factors for dementia and cognitive decline in middle-aged and older individuals without cognitive impairment: a cohort study. The Lancet Healthy Longevity, 5(4), e276–e286. 10.1016/S2666-7568(24)00025-4 [DOI] [PubMed] [Google Scholar]
- Dartora C., Marseglia A., Mårtensson G., Rukh G., Dang J., Muehlboeck J.-S., Wahlund L.-O., Moreno R., Barroso J., Ferreira D., Schiöth H. B., Westman E., , f. t. A. s. D. N. I., , t. A. I. B., Ageing, L. F. S. o., , t. J. A. s. D. N. I., & , t. A. C. (2024). A deep learning model for brain age prediction using minimally preprocessed T1w images as input [Original Research]. Frontiers in Aging Neuroscience, 15. 10.3389/fnagi.2023.1303036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviglus M. L., Bell C. C., Berrettini W., Bowen P. E., Connolly E. S., Cox N. J., Dunbar-Jacob J. M., Granieri E. C., Hunt G., McGarry K., Patel D., Potosky A. L., Sanders-Bush E., Silberberg D., & Trevisan M. (2010). National Institutes of Health State-of-the-Science Conference Statement: Preventing Alzheimer Disease and Cognitive Decline. Annals of Internal Medicine, 153(3), 176–181. 10.7326/0003-4819-153-3-201008030-00260 [DOI] [PubMed] [Google Scholar]
- Deepak S., & Ameer P. M. (2019). Brain tumor classification using deep CNN features via transfer learning. Computers in Biology and Medicine, 111, 103345. https://doi.org/ 10.1016/j.compbiomed.2019.103345 [DOI] [PubMed] [Google Scholar]
- Dosenbach N. U. F., Nardos B., Cohen A. L., Fair D. A., Power J. D., Church J. A., Nelson S. M., Wig G. S., Vogel A. C., Lessov-Schlaggar C. N., Barnes K. A., Dubis J. W., Feczko E., Coalson R. S., Pruett J. R., Barch D. M., Petersen S. E., & Schlaggar B. L. (2010). Prediction of Individual Brain Maturity Using fMRI. Science, 329(5997), 1358–1361. https://doi.org/doi: 10.1126/science.1194144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K. A., Bush A. I., Darby D., De Fazio D., Foster J., Hudson P., Lautenschlager N. T., Lenzo N., Martins R. N., Maruff P., Masters C., Milner A., Pike K., Rowe C., Savage G., Szoeke C., Taddei K., Villemagne V., Woodward M., … Group A. R. (2009). The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. Int Psychogeriatr, 21(4), 672–687. 10.1017/S1041610209009405 [DOI] [PubMed] [Google Scholar]
- Ellis K. A., Rowe C. C., Villemagne V. L., Martins R. N., Masters C. L., Salvado O., Szoeke C., Ames D., & group A. r. (2010). Addressing population aging and Alzheimer's disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer's Disease Neuroimaging Initiative. Alzheimers Dement, 6(3), 291–296. 10.1016/j.jalz.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Erus G., Battapady H., Satterthwaite T. D., Hakonarson H., Gur R. E., Davatzikos C., & Gur R. C. (2014). Imaging Patterns of Brain Development and their Relationship to Cognition. Cerebral Cortex, 25(6), 1676–1684. 10.1093/cercor/bht425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., Cullen N., Sheline Y. I., Taylor W. D., Aselcioglu I., Cook P. A., Adams P., Cooper C., Fava M., McGrath P. J., McInnis M., Phillips M. L., Trivedi M. H., Weissman M. M., & Shinohara R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. Neuroimage, 167, 104–120. https://doi.org/ 10.1016/j.neuroimage.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C., Rainey-Smith S. R., Bird S., Bomke J., Bourgeat P., Brown B. M., Burnham S. C., Bush A. I., Chadunow C., Collins S., Doecke J., Dore V., Ellis K. A., Evered L., Fazlollahi A., Fripp J., Gardener S. L., Gibson S., Grenfell R., … the, A. i. (2021). Fifteen Years of the Australian Imaging, Biomarkers and Lifestyle (AIBL) Study: Progress and Observations from 2,359 Older Adults Spanning the Spectrum from Cognitive Normality to Alzheimer's Disease. J Alzheimers Dis Rep, 5(1), 443–468. 10.3233/ADR-210005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Gaser C., Manor B., & Novak V. (2013). Advanced BrainAGE in older adults with type 2 diabetes mellitus [Original Research]. Frontiers in Aging Neuroscience, 5. 10.3389/fnagi.2013.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Ziegler G., Klöppel S., & Gaser C. (2010). Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage, 50(3), 883–892. 10.1016/j.neuroimage.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Gao F., Yoon H., Xu Y., Goradia D., Luo J., Wu T., & Su Y. (2020). AD-NET: Age-adjust neural network for improved MCI to AD conversion prediction. NeuroImage: Clinical, 27, 102290. https://doi.org/ 10.1016/j.nicl.2020.102290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Condado J., Cortes J. M., & Initiative, f. t. A. s. D. N. (2023). NeuropsychBrainAge: A biomarker for conversion from mild cognitive impairment to Alzheimer's disease. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, 15(4), e12493. https://doi.org/ 10.1002/dad2.12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaser C., Franke K., Kloppel S., Koutsouleris N., Sauer H., & Alzheimer's Disease Neuroimaging I. (2013). BrainAGE in Mild Cognitive Impaired Patients: Predicting the Conversion to Alzheimer's Disease. PLoS One, 8(6), e67346. 10.1371/journal.pone.0067346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Beckmann C. F., Vedaldi A., Smith S. M., & Peng H. (2021). Optimising a Simple Fully Convolutional Network for Accurate Brain Age Prediction in the PAC 2019 Challenge [Original Research]. Frontiers in Psychiatry, 12. 10.3389/fpsyt.2021.627996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve D. N., & Fischl B. (2009). Accurate and robust brain image alignment using boundary-based registration. Neuroimage, 48(1), 63–72. https://doi.org/ 10.1016/j.neuroimage.2009.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L. K. M., Dinga R., Hahn T., Ching C. R. K., Eyler L. T., Aftanas L., Aghajani M., Aleman A., Baune B. T., Berger K., Brak I., Filho G. B., Carballedo A., Connolly C. G., Couvy-Duchesne B., Cullen K. R., Dannlowski U., Davey C. G., Dima D., … Schmaal L. (2021). Brain aging in major depressive disorder: results from the ENIGMA major depressive disorder working group. Molecular Psychiatry, 26(9), 5124–5139. 10.1038/s41380-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T., An L., Chen P., Chen J., Feng J., Bzdok D., Holmes A. J., Eickhoff S. B., & Yeo B. T. T. (2022). Meta-matching as a simple framework to translate phenotypic predictive models from big to small data. Nature Neuroscience, 25(6), 795–804. 10.1038/s41593-022-01059-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilal S., Chai Y. L., Ikram M. K., Elangovan S., Yeow T. B., Xin X., Chong J. Y., Venketasubramanian N., Richards A. M., Chong J. P. C., Lai M. K. P., & Chen C. (2015). Markers of cardiac dysfunction in cognitive impairment and dementia. Medicine (Baltimore), 94(1), e297. 10.1097/MD.0000000000000297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilal S., Tan C. S., van Veluw S. J., Xu X., Vrooman H., Tan B. Y., Venketasubramanian N., Biessels G. J., & Chen C. (2020). Cortical cerebral microinfarcts predict cognitive decline in memory clinic patients. J Cereb Blood Flow Metab, 40(1), 44–53. 10.1177/0271678X19835565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospedales T., Antoniou A., Micaelli P., & Storkey A. (2022). Meta-Learning in Neural Networks: A Survey. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(9), 5149–5169. 10.1109/TPAMI.2021.3079209 [DOI] [PubMed] [Google Scholar]
- Jack C. R. Jr., Bernstein M. A., Borowski B. J., Gunter J. L., Fox N. C., Thompson P. M., Schuff N., Krueger G., Killiany R. J., Decarli C. S., Dale A. M., Carmichael O. W., Tosun D., Weiner M. W., & Alzheimer's Disease Neuroimaging, I. (2010). Update on the magnetic resonance imaging core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement, 6(3), 212–220. 10.1016/j.jalz.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C. R. Jr., Bernstein M. A., Fox N. C., Thompson P., Alexander G., Harvey D., Borowski B., Britson P. J., J L. W., Ward C., Dale A. M., Felmlee J. P., Gunter J. L., Hill D. L., Killiany R., Schuff N., Fox-Bosetti S., Lin C., Studholme C., … Weiner M. W., (2008). The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging, 27(4), 685–691. 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., & Smith S. (2002). Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. Neuroimage, 17(2), 825–841. https://doi.org/ 10.1006/nimg.2002.1132 [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C. F., Behrens T. E. J., Woolrich M. W., & Smith S. M. (2012). FSL. Neuroimage, 62(2), 782–790. https://doi.org/ 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Jenkinson M., & Smith S. (2001). A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5(2), 143–156. https://doi.org/ 10.1016/S1361-8415(01)00036-6 [DOI] [PubMed] [Google Scholar]
- Kalc P., Dahnke R., Hoffstaedter F., Gaser C., & Initiative A. s. D. N. (2024). BrainAGE: Revisited and reframed machine learning workflow. Human Brain Mapping, 45(3), e26632. https://doi.org/ 10.1002/hbm.26632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W., Lin L., Sun S., & Wu S. (2023). Three-round learning strategy based on 3D deep convolutional GANs for Alzheimer's disease staging. Sci Rep, 13(1), 5750. 10.1038/s41598-023-33055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T., van der Meer D., Doan N. T., Schwarz E., Lund M. J., Agartz I., Alnæs D., Barch D. M., Baur-Streubel R., Bertolino A., Bettella F., Beyer M. K., Bøen E., Borgwardt S., Brandt C. L., Buitelaar J., Celius E. G., Cervenka S., Conzelmann A., … Karolinska Schizophrenia P. (2019). Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nature Neuroscience, 22(10), 1617–1623. 10.1038/s41593-019-0471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouleris N., Davatzikos C., Borgwardt S., Gaser C., Bottlender R., Frodl T., Falkai P., Riecher-Rössler A., Möller H.-J., Reiser M., Pantelis C., & Meisenzahl E. (2013). Accelerated Brain Aging in Schizophrenia and Beyond: A Neuroanatomical Marker of Psychiatric Disorders. Schizophrenia Bulletin, 40(5), 1140–1153. 10.1093/schbul/sbt142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsen E. H., Peng H., Kaufmann T., Agartz I., Andreassen O. A., Celius E. G., Espeseth T., Harbo H. F., Hogestol E. A., Lange A. M., Marquand A. F., Vidal-Pineiro D., Roe J. M., Selbaek G., Sorensen O., Smith S. M., Westlye L. T., Wolfers T., & Wang Y. (2022). Deep neural networks learn general and clinically relevant representations of the ageing brain. Neuroimage, 256, 119210. 10.1016/j.neuroimage.2022.119210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Li F., Elahifasaee F., Liu M., Zhang L., & the Alzheimer’s Disease Neuroimaging, I. (2021). Hippocampal shape and asymmetry analysis by cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Brain Imaging and Behavior, 15(5), 2330–2339. 10.1007/s11682-020-00427-y [DOI] [PubMed] [Google Scholar]
- Lian C., Liu M., Zhang J., & Shen D. (2020). Hierarchical Fully Convolutional Network for Joint Atrophy Localization and Alzheimer's Disease Diagnosis Using Structural MRI. IEEE Transactions on Pattern Analysis and Machine Intelligence, 42(4), 880–893. 10.1109/TPAMI.2018.2889096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe L. C., Gaser C., Franke K., & for the Alzheimer’s Disease Neuroimaging, I. (2016). The Effect of the APOE Genotype on Individual BrainAGE in Normal Aging, Mild Cognitive Impairment, and Alzheimer’s Disease. PLoS One, 11(7), e0157514. 10.1371/journal.pone.0157514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Li H.-X., Chang Z.-K., Li L., Chen N.-X., Zhu Z.-C., Zhou H.-X., Li X.-Y., Wang Y.-W., Cui S.-X., Deng Z.-Y., Fan Z., Yang H., Chen X., Thompson P. M., Castellanos F. X., & Yan C.-G. (2022). A practical Alzheimer’s disease classifier via brain imaging-based deep learning on 85,721 samples. Journal of Big Data, 9(1), 101. 10.1186/s40537-022-00650-y [DOI] [Google Scholar]
- Luo M., He Z., Cui H., Ward P., & Chen Y.-P. P. (2024). Dual attention based fusion network for MCI Conversion Prediction. Computers in Biology and Medicine, 182, 109039. https://doi.org/ 10.1016/j.compbiomed.2024.109039 [DOI] [PubMed] [Google Scholar]
- Malik N., & Bzdok D. (2022). From YouTube to the brain: Transfer learning can improve brain-imaging predictions with deep learning. Neural Networks, 153, 325–338. https://doi.org/ 10.1016/j.neunet.2022.06.014 [DOI] [PubMed] [Google Scholar]
- Mei X., Liu Z., Robson P. M., Marinelli B., Huang M., Doshi A., Jacobi A., Cao C., Link K. E., Yang T., Wang Y., Greenspan H., Deyer T., Fayad Z. A., & Yang Y. (2022). RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning. Radiology: Artificial Intelligence, 4(5), e210315. 10.1148/ryai.210315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S. G., Weiner M. W., Thal L. J., Petersen R. C., Jack C., Jagust W., Trojanowski J. Q., Toga A. W., & Beckett L. (2005). The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am, 15(4), 869–877, xi-xii. 10.1016/j.nic.2005.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau C., & Bengio Y. (2003). Inference for the Generalization Error. Machine Learning, 52(3), 239–281. 10.1023/A:1024068626366 [DOI] [Google Scholar]
- Nanni L., Interlenghi M., Brahnam S., Salvatore C., Papa S., Nemni R., & Castiglioni I. (2020). Comparison of Transfer Learning and Conventional Machine Learning Applied to Structural Brain MRI for the Early Diagnosis and Prognosis of Alzheimer's Disease. Front Neurol, 11, 576194. 10.3389/fneur.2020.576194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocasio E., & Duong T. Q. (2021). Deep learning prediction of mild cognitive impairment conversion to Alzheimer's disease at 3 years after diagnosis using longitudinal and whole-brain 3D MRI. PeerJ Comput Sci, 7, e560. 10.7717/peerj-cs.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K., Chung Y.-C., Kim K. W., Kim W.-S., & Oh I.-S. (2019). Classification and Visualizatio n of Alzheimer’s Disease using Volumetric Convolutional Neural Network and Transfer Learning. Scientific Reports, 9(1), 18150. 10.1038/s41598-019-54548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixao L., Sikka P., Sun H., Jain A., Hogan J., Thomas R., & Westover M. B. (2020). Excess brain age in the sleep electroencephalogram predicts reduced life expectancy. Neurobiology of Aging, 88, 150–155. https://doi.org/ 10.1016/j.neurobiolaging.2019.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatucci M., Pomerleau D., Hinton G., & Mitchell T. M. (2009). Zero-shot learning with semantic output codes Proceedings of the 22nd International Conference on Neural Information Processing Systems, Vancouver, British Columbia, Canada. [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., & Duchesnay É., (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res., 12(null), 2825–2830. [Google Scholar]
- Peng H., Gong W., Beckmann C. F., Vedaldi A., & Smith S. M. (2021). Accurate brain age prediction with lightweight deep neural networks. Medical Image Analysis, 68, 101871. https://doi.org/ 10.1016/j.media.2020.101871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponio R., Erus G., Habes M., Doshi J., Srinivasan D., Mamourian E., Bashyam V., Nasrallah I. M., Satterthwaite T. D., Fan Y., Launer L. J., Masters C. L., Maruff P., Zhuo C., Völzke H., Johnson S. C., Fripp J., Koutsouleris N., Wolf D. H., … Davatzikos C. (2020). Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. Neuroimage, 208, 116450. https://doi.org/ 10.1016/j.neuroimage.2019.116450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Rosas H. D., & Fischl B. (2010). Highly accurate inverse consistent registration: A robust approach. Neuroimage, 53(4), 1181–1196. https://doi.org/ 10.1016/j.neuroimage.2010.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan L., Alexander-Bloch A. F., Wagstyl K., Farooqi S., Brayne C., Tyler L. K., & Fletcher P. C. (2016). Obesity associated with increased brain age from midlife. Neurobiology of Aging, 47, 63–70. https://doi.org/ 10.1016/j.neurobiolaging.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F., Dale A. M., Busa E., Glessner M., Salat D., Hahn H. K., & Fischl B. (2004). A hybrid approach to the skull stripping problem in MRI. Neuroimage, 22(3), 1060–1075. https://doi.org/ 10.1016/j.neuroimage.2004.03.032 [DOI] [PubMed] [Google Scholar]
- Shin H. C., Roth H. R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., & Summers R. M. (2016). Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Transactions on Medical Imaging, 35(5), 1285–1298. 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. E., Cropley V., Maier A. B., Lautenschlager N. T., Breakspear M., & Zalesky A. (2023). Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nature Medicine, 29(5), 1221–1231. 10.1038/s41591-023-02296-6 [DOI] [PubMed] [Google Scholar]
- Tinn R., Cheng H., Gu Y., Usuyama N., Liu X., Naumann T., Gao J., & Poon H. (2023). Fine-tuning large neural language models for biomedical natural language processing. Patterns, 4(4). 10.1016/j.patter.2023.100729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier W. M., & Scheltens P. (2005). Epidemiology and risk factors of dementia. Journal of Neurology, Neurosurgery & Psychiatry, 76(suppl 5) , v2. 10.1136/jnnp.2005.082867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K., Khoshgoftaar T. M., & Wang D. (2016). A survey of transfer learning. Journal of Big Data, 3(1), 9. 10.1186/s40537-016-0043-6 [DOI] [Google Scholar]
- Wen J., Thibeau-Sutre E., Diaz-Melo M., Samper-González J., Routier A., Bottani S., Dormont D., Durrleman S., Burgos N., & Colliot O. (2020). Convolutional neural networks for classification of Alzheimer's disease: Overview and reproducible evaluation. Medical Image Analysis, 63, 101694. https://doi.org/ 10.1016/j.media.2020.101694 [DOI] [PubMed] [Google Scholar]
- Wrigglesworth J., Yaacob N., Ward P., Woods R. L., McNeil J., Storey E., Egan G., Murray A., Shah R. C., Jamadar S. D., Trevaks R., Ward S., Harding I. H., & Ryan J. (2022). Brain-predicted age difference is associated with cognitive processing in later-life. Neurobiology of Aging, 109, 195–203. https://doi.org/ 10.1016/j.neurobiolaging.2021.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulan N., An L., Zhang C., Kong R., Chen P., Bzdok D., Eickhoff S. B., Holmes A. J., & Yeo B. T. T. (2024). Translating phenotypic prediction models from big to small anatomical MRI data using meta-matching. Imaging Neuroscience, 2, 1–21. 10.1162/imag_a_00251 [DOI] [Google Scholar]
- Xu X., Hilal S., Collinson S. L., Chong E. J., Ikram M. K., Venketasubramanian N., & Chen C. L. (2015). Association of Magnetic Resonance Imaging Markers of Cerebrovascular Disease Burden and Cognition. Stroke, 46(10), 2808–2814. 10.1161/STROKEAHA.115.010700 [DOI] [PubMed] [Google Scholar]
- Yang X., Chen A., PourNejatian N., Shin H. C., Smith K. E., Parisien C., Compas C., Martin C., Costa A. B., Flores M. G., Zhang Y., Magoc T., Harle C. A., Lipori G., Mitchell D. A., Hogan W. R., Shenkman E. A., Bian J., & Wu Y. (2022). A large language model for electronic health records. npj Digital Medicine, 5(1), 194. 10.1038/s41746-022-00742-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T. T., Cao M. H., Yu J. C., Shi T. Y., Mao X. H., Wei X. Y., & Jia Z. Z. (2024). T1-Weighted Imaging-Based Hippocampal Radiomics in the Diagnosis of Alzheimer's Disease. Academic Radiology. https://doi.org/ 10.1016/j.acra.2024.06.012 [DOI] [PubMed] [Google Scholar]
- Zarei A., Keshavarz A., Jafari E., Nemati R., Farhadi A., Gholamrezanezhad A., Rostami H., & Assadi M. (2024). Automated classification of Alzheimer's disease, mild cognitive impairment, and cognitively normal patients using 3D convolutional neural network and radiomic features from T1-weighted brain MRI: A comparative study on detection accuracy. Clinical Imaging, 115, 110301. https://doi.org/ 10.1016/j.clinimag.2024.110301 [DOI] [PubMed] [Google Scholar]
- Zheng C., Pfahringer B., & Mayo M. (2022, 18-23 July 2022). Alzheimer’s Disease Detection vi a a Surrogate Brain Age Prediction Task using 3D Convolutional Neural Networks. 2022 International Joint Conference on Neural Networks (IJCNN), [Google Scholar]
- Zhou X., Balachandra A. R., Romano M. F., Chin S. P., Au R., & Kolachalama V. B. (2024). Adversarial Learning for MRI Reconstruction and Classification of Cognitively Impaired Individuals. IEEE Access, 12, 83169–83182. 10.1109/ACCESS.2024.3408840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F., Qi Z., Duan K., Xi D., Zhu Y., Zhu H., Xiong H., & He Q. (2021). A Comprehensive Survey on Transfer Learning. Proceedings of the IEEE, 109(1), 43–76. 10.1109/JPROC.2020.3004555 [DOI] [Google Scholar]