Abstract
Comparative analysis of genomic sequences is a powerful approach to discover functional sites in these sequences. Herein, we present a WWW-based software system for multiple alignment of genomic sequences. We use the local alignment tool CHAOS to rapidly identify chains of pairwise similarities. These similarities are used as anchor points to speed up the DIALIGN multiple-alignment program. Finally, the visualization tool ABC is used for interactive graphical representation of the resulting multiple alignments. Our software is available at Göttingen Bioinformatics Compute Server (GOBICS) at http://dialign.gobics.de/chaos-dialign-submission
INTRODUCTION
During the last few years, cross-species sequence comparison has become a widely used approach to genome sequence analysis. The underlying idea is that functional regions of genomic sequences tend to be more conserved during evolution than non-functional parts. Thus, islands of local sequence similarity among two or several genomic sequences usually indicate biological functionality. This phylogenetic footprinting principle has been used by many researchers to detect novel functional elements in genomic sequences. Genomic sequence comparison has been used for gene prediction (1–5), to discover regulatory elements (6,7) and to study genomic duplications (8,9). Recently, multiple sequence comparison has been used to identifiy signature sequences of bacteria and viruses for rapid detection of pathogene microorganisms as part of the US biodefense program (10) and to detect non-coding functional RNA (11).
All these studies rely on pair-wise or multiple alignments of genomic sequences; their accuracy is therefore limited by the accuracy of the underlying alignment tools. Consequently, development of algorithms for genomic sequence alignment has become a high priority in Bioinformatics research, see (12,13) for a survey. A systematic evaluation of the currently used software tools for multiple alignment of genomic sequences has been carried out by Pollard et al. (14).
THE CHAOS/DIALIGN APPROACH
DIALIGN is a general-purpose alignment program that combines global and local alignment features (15,16). Such an approach is particularly appropriate when genomic sequences are to be aligned where locally conserved regions may be separated by non-related parts of the sequences. As a stand-alone tool, however, DIALIGN is too slow to align long genomic sequences as the program running time grows quadratically with the average sequence length. Therefore, an anchoring option has been implemented. Here, user-specified anchor points can be used to reduce the alignment search space, thereby improving the program running time (17). To find suitable anchor points, we use the local alignment program CHAOS (18).
In a first step, our system applies CHAOS to identify chains of local similarities among all pairs of input sequences in a multiple sequence set. In a second step, DIALIGN is used to accurately align the regions between the similarities identified by CHAOS. Our anchored-alignment approach can be applied for pair-wise as well as multiple alignment. For multiple alignment, CHAOS is run on all possible pairs of input sequences. The resulting local pair-wise similarities are then checked for consistency by DIALIGN and non-consistent ones are eliminated. This procedure is similar to the greedy approach that DIALIGN uses to construct multiple alignments, see (16).
ALIGNMENT VISUALIZATION WITH ABC
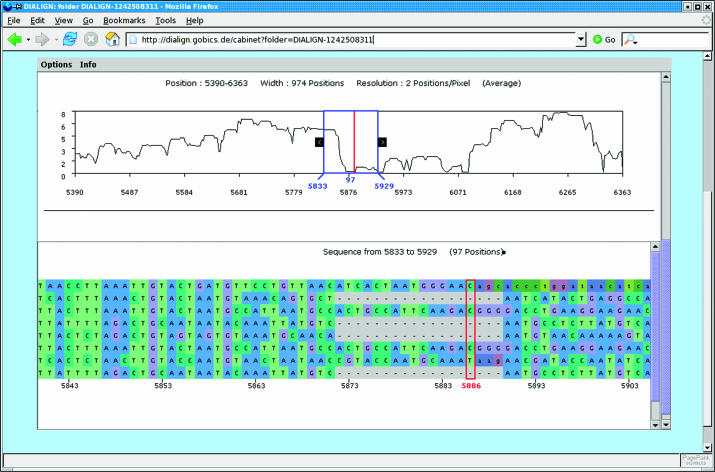
Alignments of large genomic sequences are hard to interpret without specialized visualisation tools. ABC (Application for Browsing Constraints) is an interactive Java tool that has recently been developed by Cooper et al. (19) for intuitive and efficient exploration of multiple alignments of genomic sequences. It can be used to move quickly from a summary view of the entire alignment via arbitrary levels of resolution down to the level of individual nucleotides. ABC can graphically represent additional information, such as the degree of local sequence conservation or annotation data, such as the locations of genes, etc. (Figure 1).
Figure 1.
Visualization of multiple alignments using ABC (19). The user can interactively switch between different levels from a global view of the output alignment down to the level of individual residues.
At our server, we offer ABC to visualize multiple alignments produced by CHAOS and DIALIGN. The degree of local similarity among the input sequences is graphically represented based on the weight scores used by DIALIGN to assess the local degree of similarity among the sequences to be analyzed. The standard DIALIGN output file represents the degree of local similarity in a pair-wise or multiple alignment, using stars or numbers below the alignment. For each alignment column, the weight scores of all fragments connecting residues at this column are summed up and normalized, see (16) for a precise definition of fragment weights.
We use the same measure of local sequence similarity for graphical representation by ABC. Note that this is only a rough measure of sequence conservation. It is possible that columns with identical nucleotide composition receive different similarity values if they are connected by fragments with different weight scores. It is also important to keep in mind, that our similarity values are not absolute values but are normalized such that in every alignment the column of maximum local similarity obtains a certain fixed score. Nevertheless, our graphical representation gives a good overview of the local degree of conservation among a sequence set.
THE CHAOS/DIALIGN/ABC WWW SERVER
The input data for our web server is a single text file containing two or several genomic sequences in FASTA format. The maximum total length of the input sequences is currently 3 MB. The server runs CHAOS and DIALIGN on the input sequences. Visualization of the results with ABC can be chosen as an additional option. This requires that the user has Java installed on his computer. For small input data, the resulting alignment is immediately shown on the computer screen—either in standard DIALIGN format or using ABC if this option has been chosen. For larger sequence sets, the program output is stored at our server; the corresponding web addresses are sent to the user by email. Different output files are created: (i) the output alignment in DIALIGN format, (ii) the same alignment in FASTA format, (iii) a list of fragments, i.e. local segment pairs, that are used as building blocks for the DIALIGN alignment, and (iv) a list of anchor points identified by CHAOS. These files are provided as plain text files. In addition the optional ABC output is stored at the server together with these standard output files.
Alignments in DIALIGN format contain additional information about the degree of local sequence similarity in the multiple alignment. Also, the program distinguishes between nucleotides that could be aligned and nucleotides with no statistically significant matches to the compared sequences. Upper-case and lower-case letters are used to indicate which nucleotides are considered to be aligned. This output format and the ABC output are designed for visual inspection of the returned alignments. The output in FASTA format contains essentially the same information but is more appropriate for further automatic analysis as most sequence analysis programs accept FASTA-formatted files as input data.
The list of returned fragments is annotated with some additional information that may be useful for more detailed analyses. This includes quality scores (so called weights) of the fragments indicating the degree of local sequence similarity. In addition, calculated overlap weights are returned. Overlap weights reflect not only the similarity between two segments but also the degree of overlap with other segment pairs involving different pairs of sequences as described in (15). Finally, the fragment list states for each fragment if it was consistent with other fragments and could be included into the multiple alignment or if it had to be rejected because of non-consistency. The fragment list is also designed for automatized post-processing. It is easy to parse and contains more information than the resulting alignment alone. In addition to the fragment list, a list of anchor points created by CHAOS is returned. Our WWW server provides detailed online help regarding input and output formats.
AVAILABILITY
Our software is available through Göttingen Bioinformatics Compute Server (GOBICS): http://dialign.gobics.de/chaos-dialign-submission.
Acknowledgments
We would like to thank Michael Brudno, Gregory Cooper and Arend Sidow for helping us with CHAOS and ABC. The work was supported by Deutsche Forschungsgemeinschaft (DFG), project MO 1048/1-1 to BM. Funding to pay the Open Access publication charges for this article was provided by the University of Göttingen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bafna V., Huson D.H. The conserved exon method for gene finding. Bioinformatics. 2000;16:190–202. [PubMed] [Google Scholar]
- 2.Batzoglou S., Pachter L., Mesirov J.P., Berger B., Lander E.S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17(Suppl. 1):S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 4.Wiehe T., Gebauer-Jung S., Mitchell-Olds T., Guigó R. SGP-1, Prediction and validation of homologous genes based on sequence alignments. Genome Res. 2001;11:1574–1583. doi: 10.1101/gr.177401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taher L., Rinner O., Gargh S., Sczyrba A., Brudno M., Batzoglou S., Morgenstern B. AGenDA: homology-based gene prediction. Bioinformatics. 2003;19:1575–1577. doi: 10.1093/bioinformatics/btg181. [DOI] [PubMed] [Google Scholar]
- 6.Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 7.Göttgens B., Barton L.M., Gilbert J.G.R., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 8.Prohaska S., Fried C., Flamm C., Wagner G.P., Stadler P.F. Surveying phylogenetic footprints in large gene clusters: applications to Hox cluster duplications. Mol. Phylogenet. Evol. 2004;31:581–604. doi: 10.1016/j.ympev.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Fried C., Prohaska S.J., Stadler P.F. Independent Hox-cluster duplications in lampreys. J. Exp. Zoolog. B Mol. Dev. Evol. 2003;299:18–25. doi: 10.1002/jez.b.37. [DOI] [PubMed] [Google Scholar]
- 10.Fitch J.P., Gardner S.N., Kuczmarski T.A., Kurtz S., Myers R., Ott L.L., Slezak T.R., Vitalis E.A., Zemla A.T., McCready P.M. Rapid development of nucleic acid diagnostics. Proceedings of the IEEE. 2002;90:1708–1721. [Google Scholar]
- 11.Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller W. Comparison of genomic DNA sequences: solved and unsolved problems. Bioinformatics. 2001;17:391–397. doi: 10.1093/bioinformatics/17.5.391. [DOI] [PubMed] [Google Scholar]
- 13.Chain P., Kurtz S., Ohlebusch E., Slezak T. An applications-focused review of comparative genomics tools: capabilities, limitations, and future challenges. Brief. Bioinform. 2003;4:105–123. doi: 10.1093/bib/4.2.105. [DOI] [PubMed] [Google Scholar]
- 14.Pollard D.A., Bergman C.M., Stoye J., Celniker S.E., Eisen M.B. Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics. 2004;5:6. doi: 10.1186/1471-2105-5-6. http://www.biomedcentral.com/1471-2105/5/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgenstern B., Dress A.W.M., Werner T. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl Acad. Sci. USA. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgenstern B. DIALIGN 2, improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 17.Morgenstern B., Rinner O., Abdeddaïm S., Haase D., Mayer K., Dress A., Mewes H.-W. Exon discovery by genomic sequence alignment. Bioinformatics. 2002;18:777–787. doi: 10.1093/bioinformatics/18.6.777. [DOI] [PubMed] [Google Scholar]
- 18.Brudno M., Chapman M., Göttgens B., Batzoglou S., Morgenstern B. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics. 2003;4:66. doi: 10.1186/1471-2105-4-66. http://www.biomedcentral.com/1471-2105/4/66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper G.M., Singaravelu S.A.G., Sidow A. ABC: software for interactive browsing of genomic multiple sequence alignment data. BMC Bioinformatics. 2004;5:192. doi: 10.1186/1471-2105-5-192. [DOI] [PMC free article] [PubMed] [Google Scholar]