Abstract
Formalin-fixed, paraffin-embedded (FFPE) patient tissues are a valuable resource for proteomic studies with the potential to associate the derived molecular insights with clinical outcomes. Here we present an optimized, partially automated workflow for FFPE proteomics combining pathology-guided macro-dissection with Adaptive Focused Acoustics (AFA) sonication for lysis and decrosslinking, S-Trap digestion to peptides, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using Orbitrap, Astral or timsTOF HT instrumentation. The workflow enables analysis of up to 96 dissected FFPE tissue samples or whole 10 μM scrolls, identifying 8,000-10,000 unique proteins with <20% median CVs. Key optimizations include improved tissue lysis, protein quantification for normalization, and peptide cleanup prior to LC-MS/MS analysis. Application to lung adenocarcinoma (LUAD) FFPE blocks confirms the platform’s effectiveness in processing complex samples, achieving deep proteome coverage and quantitative robustness comparable to TMT-based methods. Using the newly released Orbitrap Astral with short, 24-minute gradients, the workflow identifies up to ~10,000 unique proteins and ~11,000 localized phosphosites in LUAD FFPE tissue. This high-throughput, scalable workflow advances biomarker discovery and proteomic research in archival tissue samples.
Introduction:
Formalin-fixed, paraffin-embedded (FFPE) tissue samples with clinical annotation represent an invaluable resource. Patient tissue profiling by global proteomics using liquid chromatography-tandem mass spectrometry (LC-MS/MS) provides a molecular foundation for exploration of disease mechanisms and prediction of clinical outcomes that goes beyond what can be achieved by traditional pathology or immunohistochemistry (1–5). The extensive collections of FFPE samples in biobanks and pathology archives facilitate large-scale studies, aiding in the identification of molecular markers, biomarkers, and genetic alterations associated with diverse pathologies (6).
Mass spectrometry-based proteomic analyses of FFPE tissue present a number of challenges. Many FFPE proteomics workflows have relied on laborious xylene and ethanol washes for deparaffinization (7, 8), although alternative approaches have started to emerge such as the use of the Beatbox (Preomics), which leverages bead-based tissue lysis (9); substitution of SafeClear (Fisher Scientific) for xylene (10); and lysis using the Covaris Adaptive Focused Acoustics (AFA) technology (3, 11, 12). Despite these innovations, extraction of proteins and peptides from FFPE tissues has proven difficult, constraining depth of coverage and quantitative characterization of post-translational modifications (PTMs) (5, 7, 13). Tissue preservation techniques can also impact quantitative reproducibility relative to fresh frozen tissue (14) and contaminants such as wax in FFPE can be problematic for downstream nanoflow LC-MS/MS applications. Process improvements are needed to achieve deep-scale analysis of FFPE tissues and take full advantage of the invaluable resource afforded by repositories of well-annotated FFPE specimens.
In this study, we describe a highly optimized and partially automated workflow that enables reproducible deepscale proteomic analysis of FFPE tissue yielding ~8,000-10,000 unique proteins and up to ~14,000 localized phosphosites per sample. The plate-based workflow employs the Covaris AFA sonicator for FFPE sample processing and decrosslinking, an Opentrons OT-2 robot for complex liquid handling steps, positive pressure manifold-assisted S-Trap-based digestion and purification, and LC-MS/MS analysis on Orbitrap or timsTOF instrumentation. The set-up allows us to process and analyze up-to 96 samples in less than 4.5 days.
Result:
Workflow and method optimization
The schematic representation of the final, optimized workflow is illustrated in Figure 1A. Individual 10 μm tissue scrolls are transferred into wells of a Covaris TPX plate, prefilled with Covaris Tissue Lysis Buffer (TLB). Ultrasonication by AFA is applied for tissue deparaffinization, followed by heating the plate to 90°C for protein decrosslinking. A second AFA ultrasonication step ensures thorough tissue homogenization, effectively bypassing both xylene-based deparaffinization and traditional probe-based sonication. This streamlined approach minimizes sample loss and contamination while enhancing processing efficiency and reproducibility.
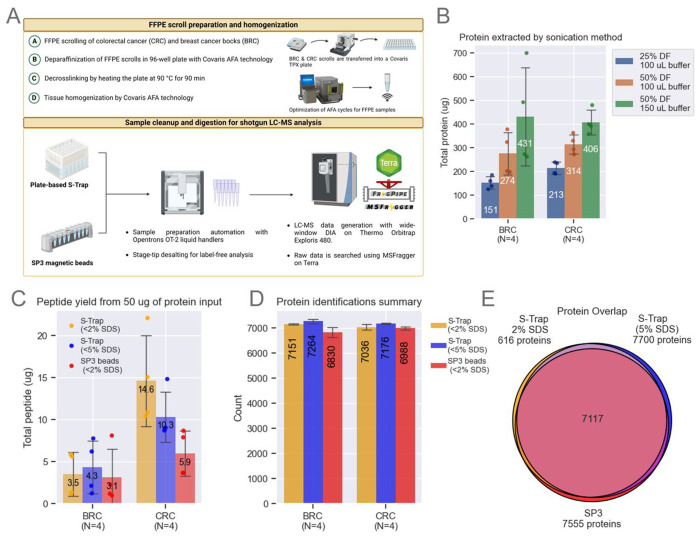
Figure 1: Method Optimization.
A. The FFPE workflow processes entire scrolls using a Covaris TPX plate. Breast cancer (BRC) and colorectal cancer (CRC) blocks were scrolled and transferred into a Covaris plate containing tissue lysis buffer. Sonication conditions were optimized to maximize protein extraction. SP3 beads and S-Trap plates were evaluated, starting with 50 μg of protein. Samples were analyzed on a Neo Vanquish LC coupled to an Exploris 480 (Thermo).
B. Optimization of protein extraction efficiency by adjusting sonication duty factors (DF) and buffer volume. Bar plots represent data from n=4 scrolls per block, showing means and standard deviation (SD) error bars.
C. Comparison of SP3 and S-Trap methods for digestion and cleanup using a fluorometric peptide assay. Bar plots represent data from n=4 scrolls per block, with means and SD error bars.
D. Unique protein identifications using MSFragger-DIA across the three tested workflows. Bar plots represent data from n=4 scrolls per block, with means and SD error bars.
E. Venn diagram illustrating protein overlap, with each circle representing the total number of unique proteins identified by each tested workflow.
Following FFPE sample lysis, the protein content of each sample was quantified colorimetrically using the Micro BCA Protein Assay Kit (Thermo Fisher) in a 96-well plate. The lysates were reduced in 5 mM tris(2-carboxyethyl)phosphine (TCEP) and alkylated by 50 mM 2-chloroacetamide (CAA). To remove detergents, the lysates were then suspension-trapped and processed using the plate-based S-Trap (Protifi) workflow. An additional chloroform/methanol wash was implemented to eliminate any residual paraffin wax from the S-Trap wells. Proteins were then digested overnight with LysC and trypsin, followed by stage-tip desalting with styrene-divinylbenzene–reverse-phase sulfonate (SDB-RPS) (15, 16) for label-free LC-MS analysis. An Opentrons OT-2 robot was used to normalize peptide input amounts for stage-tip desalting, reducing sample processing time and enhancing reproducibility. LC-MS data was acquired using Data-Independent Acquisition (DIA) methods on the Exploris 480, timsTOF HT, or Orbitrap Astral, achieving a depth of 8,000 - 10,000 unique proteins from each scroll with a turnaround time around of 4.5 days for a full 96-well plate of FFPE material. Data was uploaded to Terra (https://app.terra.bio/), a cloud-based environment running on Linux, designed to support Spectronaut (17), FragPipe (18), and DIA-NN (19), enabling fast and scalable data processing to meet the intensive computational demands of DIA pipelines in large-scale experiments.
This workflow was the product of systematic optimization of critical sample processing parameters that impact protein/peptide yields, sample cleanup, and the reproducibility of proteomic identifications as detailed below. Our initial experiments focused on refining the recommended Covaris workflow for clinical proteomics (20) using 10 μm scrolls from colorectal cancer (CRC) and breast cancer (BRC) blocks that had been stored ~20 years (reflecting real-world FFPE repositories). We found that modifications to the tissue lysis buffer (TLB) volume and sonication duty factor (DF) on the Covaris LE220-plus focused-ultrasonicator led to significant improvements in protein yield from the FFPE scrolls. Increasing DF to 50% and TLB volume to 150 μL increased protein yields by more than 100% compared to the Covaris-recommended protocol of 25% DF in 100 μL of TLB (Figure 1B). A small pellet remained in most wells after processing; initial optimization studies utilized the clear lysate (see below).
Next, two sample digestion methods, S-Trap and SP3, the former at two SDS concentrations, were compared with respect to peptide yield starting with 50 μg protein from BRC and CRC FFPE replicate sections (Figure 1C). Yields from CRC scrolls were ~3X those from BRC scrolls, possibly due to the higher fat content of the latter, which may interfere with the protein BCA assay, explaining the difference in observed yields across different tissues based on their heterogeneity. BRC peptide yields were roughly comparable across the three tested conditions: S-Trap with tissue lysis buffer (TLB) (~2% SDS), S-Trap with TLB spiked with additional SDS (~5% SDS), and SP3 beads with TLB (~2% SDS). In contrast, peptide yields for CRC scrolls were more than twice as high for S-Trap than for SP3 methods using 2% SDS, and somewhat reduced yield when S-Trap SDS was increased to 5% (Figure 1C). The possible effects of variations in the protocol on the numbers of proteins identified were then evaluated using BRC and CRC FFPE tissue. Comparable numbers of proteins were identified for CRC and BRC samples by analyzing the equivalent of one μg of peptide (normalized based on the peptide yields shown in Figure 1C) (Figure 1D, see methods) with over 90% overlap in identified proteins irrespective of the processing method used (Figure 1E). The S-Trap method, however, identified ~10% more peptides than the SP3 method (Figure S1A). Furthermore, total ion chromatograms (TICs) were both higher and more reproducible for samples processed by S-Trap than SP3 (Figure S1B), despite slightly better digestion efficiency with SP3 (Figure S1C). In addition, cleaner samples are obtained using the S-Trap workflow due to the chloroform/methanol wash step that enabled the processing of whole tissue rather than clear lysate only, enhancing peptide recovery almost two-fold. Together, the improved peptide identifications, better TIC reproducibility and cleaner samples – which reduced the need for frequent LC column changes and instrument cleaning – demonstrated the comparative advantage of S-trap sample digestion in our protocol.
Application of the optimized method to macro-dissected tissue
The challenges to tissue homogenization and emulsification posed by the wax encapsulated FFPE tissue samples are ameliorated but not eliminated by powerful sonication devices like the Covaris. Laser microdissection (LMD) of tumor-rich areas is the established strategy for addressing those challenges and pathologist-guided manual macro-dissection (MMD) provides a viable alternative (21). Moreover, enrichment for tumor epithelial cells within heterogenous cancer tissue has utility to inferring cancer-specific proteins, post-translational modifications (PTMs), and pathways.
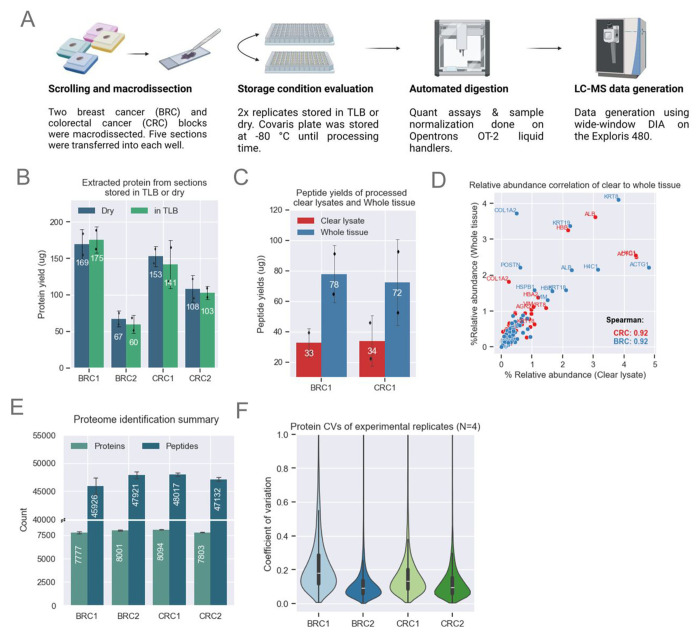
For a subset of CRC and BRC samples, we optimized parameters for pathology-guided macrodissection of tumor-rich areas from three to five 10 μm thick scrolls per sample (Figure 2A, Table S1). Transferring fragments from MMD samples to empty wells was impeded by electrostatic interaction; however, transfer was made easier and more consistent when wells were pre-filled with TLB. To assess the downstream impact of wet vs. dry stabilization, samples transferred to both dry wells and pre-filled wells were frozen at −80°C and processed after several weeks. Protein yields from the clear lysates were nearly identical between MMD samples stored in dry versus pre-filled wells (Figure 2B), as were peptide yields obtained by processing 50 μg of protein (data not shown). A single dry-stored BRC2 sample exhibited an unexpectedly high peptide yield (Figure S2A). As noted above, despite utilizing a 50% duty factor (DF) and 150 μl TLB, we consistently observed a small pellet in the processed wells. Post-homogenization lysate clarity was inconsistent, as centrifugation never fully removed insoluble pellet particulates. We hypothesized that the unexpectedly higher yield of peptides of this sample might have resulted from our inadvertently loading more of this pellet material into the S-Trap. To examine this possibility, we processed additional BRC1 and CRC1 lysates, this time including the undissolved tissue that we refer to here as “whole tissue” (Figure 2C). The S-Trap mesh trapped insoluble tissue material, enabling further protein digestion and extraction of additional peptides. After minimizing excess FFPE matrix by MMD processing and incorporating an additional methanol/chloroform S-Trap wash step (see Methods), proteolytic digestion of whole tissue resulted in a greater than two-fold increase in peptide yields (Figure 2C), while maintaining equivalent depth and protein abundance profile (Figure S2B–C). To investigate whether specific proteins were being extracted more efficiently using the whole tissue approach, we calculated relative abundance for each protein within a sample, where the abundance of each protein was divided by the protein abundance sum of the sample. The calculated relative abundances of certain cytoskeletal proteins, such as COL1A2 and KRT19, were increased (Figure 2D). Notably, the relative abundance across all identified proteins was still highly correlated (spearman correlation = 0.92), indicating strong global agreement between clear lysate and whole tissue processing (Figure 2D). Overall, we identified up to 8,000 unique proteins in each of the MMD samples. This increase in protein counts resulted from analyzing 1 μg of peptides, compared to the 500 ng used in Figure 1 (Figure 2E). Median protein abundance coefficient of variation (CV) was <20% across 16 MMD samples representing four replicates each from two CRC blocks and two BRC blocks, demonstrating a high level of reproducibility (Figure 2F). Initial column and MS performance, however, deteriorated over a relatively short period. Despite desalting peptides using standard tC18 stage-tips, we consistently observed the deposition of white material on the column emitter, likely contributing to performance degradation. To address this, we tested peptide cleanup with styrene-divinylbenzene–reverse-phase sulfonate (SDB-RPS) as the stage-tip material, which is known for its potential to improve peptide recovery (15, 16). SDB-RPS cleanup resulted in the near-complete disappearance of these troublesome emitter deposits, enabling LC-MS/MS analysis to be carried out over extended periods without interruption. While no improvements in peptide recovery were observed with SDB-RPS, there was a slight reduction in miscleavage rates. The number of identifications was close to that seen with tC18 (Figure S2D–E), with consistency in protein abundance distributions (Figure S2F).
Figure 2: Method Optimization for MMD Samples.
A. Schematic showing MMD sample collection and processing. Proteomics data for this figure was generated on an Exploris 480.
B. Bar plots showing comparable protein yields (n=2 scrolls per condition) across both storage conditions. The height of each bar represents the mean, with error bars indicating the standard deviation.
C. Bar plot showing enhanced peptide yields from whole tissue loaded on the S-Trap compared to clear lysates only. Demonstrated with BRC1 (n=2) and CRC1 (n=2) samples. The height of each bar represents the mean, with error bars indicating the standard deviation.
D. Comparison of relative abundance across shared proteins identified from clear lysate and whole tissue. Relative abundance is calculated by dividing each protein’s abundance by the total protein abundance for each sample. Both methods show strong Spearman correlation.
E. Unique protein and peptide identifications from MMD tissue with a median area of ~57 μm2 for CRC and BRC samples (n=4). The height of each bar represents the mean, with error bars indicating the standard deviation.
F. Protein abundance CVs among experimental replicates (n=4) for each block. The violin plot illustrates data distribution based on the kernel density estimate (KDE). The inner lines represent the 25th, 50th, and 75th percentiles of the IQR range.
Formalin tissue fixation results in a range of poorly defined chemical modifications of proteins and peptides (5, 22, 23). The decrosslinking step aims to reverse these formalin-induced chemical modifications. To investigate the decrosslinking efficiency of our workflow, we used FragPipe’s open-search tools to identify fixation-related mass shifts from DDA runs of pooled a BRC/CRC sample by peptide-spectrum matches (PSM) (Figure S3A, see methods, (24, 25). The distribution of the observed mass shifts spanned ~400 Da, with a peak at zero, representing 70% of the PSM library (Figure S3B). Approximately 10% of all PSMs exhibited mass shifts, including common formalin modifications such as methylation, formylation, formaldehyde adducts, and methylol groups (Figure S3C). These experimental FFPE modifications were highly localized to lysine and arginine residues (Figure S3D), consistent with previously reported studies (5). Globally, we identified approximately 74,000 peptidoforms, with 13% of peptides only identified in their chemically modified form, and 25% of peptides showing both modified and unmodified forms (Figure S3E). The percentage of chemically modified peptides found in our study is somewhat smaller than previously reported (5).
Evaluation of quantitative accuracy using multiple instrument platforms
Quantitative accuracy and reproducibility of DIA-generated data can be compromised as a result of fragment ion interference especially as gradients are shortened. To assess this, we tested instruments that offer high-throughput capabilities, including the Bruker timsTOF HT and the ThermoFisher Orbitrap Astral. The detailed timsTOF HT and Astral data acquisition methods are described in Methods and Figure S4A.
To assess quantification, we utilized FFPE blocks obtained from wild-type (WT) mice and mice over-expressing (OE) Ncoa4 (26). As illustrated in Figure 3A–B, we conducted three independent experiments using scrolls obtained from the two WT and two OE blocks. Each FFPE block contained multiple organs from a single WT or NCOA4 expressing mouse, with a few specific exceptions highlighted in Figures S4C–D. Aliquots of each sample were analyzed on the Exploris 480 (110 min gradient), timsTOF HT (35 min gradient) and the Orbitrap Astral (24 min gradient). DDA data was also generated by TMT labeling ~50 ug peptide aliquots of four replicates each of WT and OE, in an 18-plex. Following offline fractionation into 24 fractions, data was acquired on the Exploris 480 using MS2-based peptide quantification. DIA data were searched using DIA-NN on Terra (see Methods), while DDA data was processed using FragPipe.
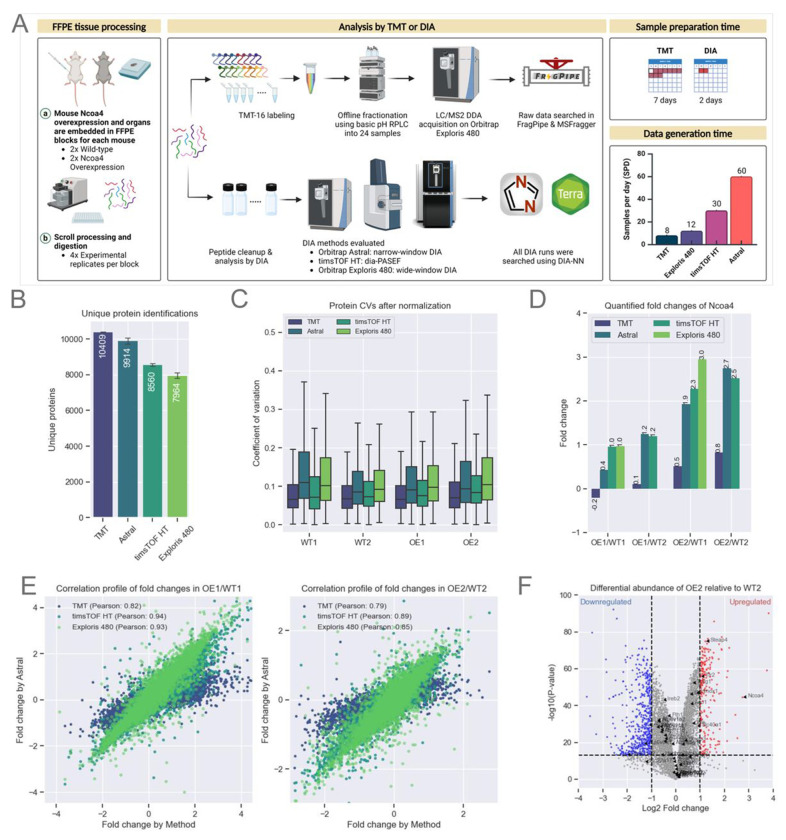
Figure 3: Evaluation of Quantitative Accuracy Using Multiple Instrument Platforms.
A. Schematic representation of mouse Ncoa4 WT and OE FFPE blocks. Peptides were either TMT-labeled or analyzed label-free using various DIA methods and instruments, as highlighted in the figure. TMT data was searched using FragPipe while all DIA analysis were performed using DIA-NN as that supported both Thermo and Bruker raw files.
B. Protein identifications for each experiment (n=16 per experiment). Bars represent the average, with error bars indicating the standard deviation.
C. Normalized protein abundance CVs from experimental replicates (n=4 per sample). The box represents the interquartile range (IQR), with the top and bottom edges indicating the 75th and 25th percentiles, respectively. The line within the box denotes the median, and whiskers extend to include data within 1.5x the IQR.
E. Quantified Ncoa4 fold-change between WT and OE mouse models across each TMT and DIA method (X-axis) compared to DIA on Astral (Y-axis). Scatter plots show protein fold-change for two WT and OE pairs, labeled WT1, WT2, OE1, and OE2.
F. Volcano plot illustrating differential protein abundance between WT2 and OE2.
As anticipated, the highly fractionated TMT DDA data provided the deepest coverage, encompassing over 10,000 unique proteins. However, the need to analyze each of the 24 fractions using a 110 min gradient resulted in a sequencing speed of just 8 samples per day (SPD) (Figure 3B). In contrast, DIA on the Astral (4 m/z windows) yielded ~9,900 unique proteins per sample using a 24 min gradient, corresponding to 60 SPD. diaPASEF on the timsTOF HT yielded ~8,500 unique proteins per sample using a 35 min gradient, corresponding to 30SPD. Wide-window (~15 m/z) DIA on the Exploris 480 identified ~8,000 unique proteins using a 110 min gradient corresponding to 12 SPD (Figure 3B). DDA analysis of the highly fractionated TMT-labeled sample quantified ~108,000 unique peptides, while Astral DIA achieved a depth of ~90,000 unique peptides per sample and dia-PASEF ~74,000 unique peptides per sample (Figure S3B). Given the slower speed of the Exploris 480, DIA acquisition on this instrument used wider windows and shorter mass range compared to the Astral (25 Da windows, m/z: 400-850 vs. 4 Da, m/z: 380-980) and quantified about ~56,000 unique peptides, 40% less than the other methods we tested. The median protein abundance CVs for each of the four experimental replicates were all below 20%, calculated using the DIA-NN quantification algorithm in FragPipe without any sample-wise normalization (Figure 3C).
Differential expression between OE and WT was evaluated to assess quantitative reliability and reproducibility of the TMT and DIA datasets. Ncoa4 was prominently up-regulated in the OE FFPE samples in each of the DIA datasets (Figure 3D), with high correlation of protein fold-change between OE and WT across the different platforms (Pearson’s correlation > 0.8) using the DIA dataset generated on the Astral platform as a comparator (Figure 4E). As expected, the TMT data were compressed despite the use of 24 off-line fractions.
Figure 4: Application to Clinical LUAD FFPE Samples.
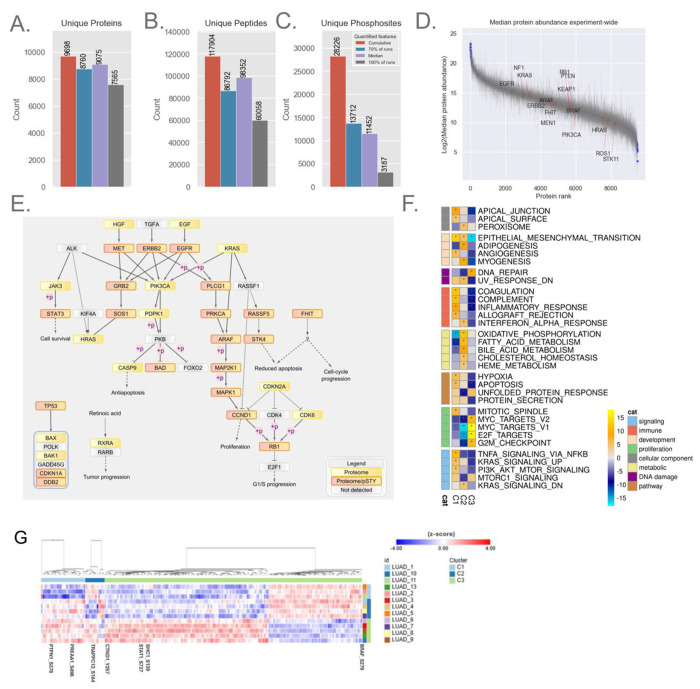
A-B. Bar plot showing unique protein and peptide depth from lung adenocarcinoma (LUAD) FFPE blocks processed with MMD (n=13).
C. Phosphoproteome depth from LUAD FFPE blocks (n=12). The bar plot represents confidently localized (localization probability > 0.75) and unique phosphosites from 12 samples, each with 50 μg of peptide available for enrichment.
D. Rank plot displaying the dynamic range of quantified proteins across all blocks; each point represents median expression, and error bars indicate the quantified range for the experiment.
E. Pathway diagram highlighting non-small cell lung cancer pathway-relevant proteins and phosphosites quantified by the workflow. Nodes detected by proteome alone or by both proteome and phosphoproteome are marked with yellow and pink-yellow boxes, respectively.
F. NMF clustering of the proteome dataset, resulting in three clusters (C1-3). The heatmap shows pathway terms from Gene Set Enrichment Analysis (GSEA) using the MSigDB hallmark gene sets and pathways with FDR < 0.05 is indicated with an asterisk.
G. Heatmap showing the significant phosphosites driving NMF clustering, with a handful of sites highlighted.
Ncoa4 selectively regulates autophagic degradation of ferritin, the key cytosolic iron storage complex (27). In addition to observing the expected upregulation of Ncoa4 protein, we also observed increased levels of proteins involved in iron metabolism such as Steap4, a metalloreductase that reduces Fe3+ to Fe2+ (28) and Tfr2 (transferrin receptor 2) that allows iron transportation inside cells, which likely are feedback mechanism in response to Ncoa4 overexpression (Figure 3F).
Proteome and phosphoproteome analysis of LUAD FFPE blocks using optimized protocol
Having developed a processing pipeline capable of deep-scale proteome profiling of FFPE BRC and CRC tissue, we benchmarked the workflow’s performance in a cohort of lung-adenocarcinoma (LUAD) FFPE blocks of resection tissues from 13 patients. The blocks were macrodissected to enrich tumor-rich sections and digested for global proteome analysis. Twelve of the 13 samples yielded enough peptide to allow 50 μg phosphoproteome enrichment using Fe(III)-NTA AssayMap cartridges on the automated Agilent Bravo liquid handling robot (see methods).
Based on the robust performance of the Orbitrap Astral in prior experiments, this instrument was selected to evaluate the LUAD FFPE samples (see Methods) using the 60 SPD (24 min) Astral DIA (4 m/z) method. This approach cumulatively quantified approximately 9,700 unique proteins and 120,000 peptides, with a median of 9,075 unique proteins and 98,000 peptides per sample (Figure 4A–B). Anticipating that Astral’s enhanced performance would similarly benefit phosphoproteome analysis, we applied the same Astral DIA method (4 m/z) to these samples. In total, we identified ~28,000 fully localized unique phosphosites, with a median of 11,000 fully localized phosphosites per sample. The abundance of quantified proteins spanned a range of over 20-fold in magnitude (Figure 4D).
To verify the biological relevance of quantified proteins, we mapped these proteins back to the non-small cell lung cancer (NSCLC) pathways in the KEGG database. We captured 36 proteins that are actively implicated in the NSCLC cancer pathways reported by KEGG (29), of which 21 of these proteins also exhibited phosphorylation events (Figure 4E).
Given the limited clinical metadata attached to these samples, we further explored the intrinsic structure of the cohort using non-negative matrix factorization (NMF)-based clustering of proteome or phosphoproteome datasets (30). NMF clustering using proteome and phosphoproteome resulted in three clusters. The choice of three clusters was supported by multiple metrics including the dispersion and cophenetic correlation coefficients as well as membership consensus (Figure S5A–C). Both proteome- and phosphoproteome-only NMF clustering separated samples into three groups. Some samples clustered differently in the phosphoproteome-only clusters, although overall pathway patterns between the individual clusters were retained (Proteome: Figure 4F., Phosphoproteome: Figure S5E). However, we show that phosphoproteome-only clustering may capture subtle heterogeneity in tumors, although the small sample size hinders us from deeper exploration. The first proteomics cluster (C1) showed upregulation of gene sets related to immune signaling such as coagulation, interferon alpha response and proteins related to EMT and apical junction. The second cluster (C2) demonstrated upregulation of metabolic pathways such as oxidative phosphorylation, fatty acid metabolism, and downregulation of Myc and E2F targets. The third cluster (C3) was characterized by downregulation of immune signaling pathways, and upregulation of Myc targets, and DNA repair (Figure 4F). Overall, GSEA results illustrate the ability for FFPE proteomics to undergo standard data analysis approaches to yield biologically relevant differences despite this small cohort size. At the level of phosphoproteome, we identified phosphorylation of several key proteins with an established role in LUAD (Figure S5F) and explored the driver phosphosites that are signatures to each phosphoproteome NMF cluster (Figure 4G). In C1, phosphorylation events such as S496 in PRKAA1 and S378 in PTPN1 are relatively downregulated, potentially reflecting a shift in cellular processes such as energy metabolism (31) and phosphatase activity regulation (32). In C2, S184 in TRAPPC12 was relatively downregulated, with dephosphorylation events in TRAPPC12 known to be necessary for cell cycle progression (33). In cluster C3, several notable phosphorylation events were differentially regulated. Phosphorylation of BRAF at S729, a site known to act as a negative feedback regulator, was lower compared to other clusters. Downregulation of the inhibitory site may lead to BRAF mediated MAPK/ERK signaling, leading to enhanced cellular proliferation. These findings suggest that the oncogenic MAPK/ERK pathway may be more active in C3 (34, 35). Additionally, SHC1 phosphorylation at S139 was upregulated, a key site in receptor tyrosine kinase (RTK) signaling. It has been reported to be upregulated in renal clear cell (RCC) carcinoma and downregulated in colon cancer (38), highlighting a diverse role in oncogenic processes. Another interesting observation was the increased phosphorylation of CTND1 at Y257, a critical modulator of the Ras-MAPK pathway, influencing cellular proliferation, survival, and migration (36). Lastly, STAT1 phosphorylation at S727 was higher in C3 relative to other clusters, a site known to promote AML cell proliferation and differentiation in JAK-STAT pathway-driven neoplasms (37).
In summary, using this small sample set, we demonstrate the overall feasibility of identifying over ~9,000 unique proteins from our LUAD subset, which are mapped back to relevant biological processes. The identification of ~28,000 localized unique phosphosites, with a median of 11,000 phosphosites per sample demonstrates the feasibility of phosphopeptide analysis of FFPE tissue samples using our workflow.
Discussion:
We have developed a method of high-throughput FFPE tissue processing workflows and optimized data acquisition methods for proteomic and phosphoproteomic applications with increased data depth and completeness as compared to previous DIA-based FFPE analyses. By utilizing ultra-sonication and adaptive focused acoustics, we eliminate the need for xylene-based deparaffinization. Tissue digestion using plate-based S-Trap with an additional chloroform/methanol wash and SDB-RPS stage-tip desalting allowed for cleaner samples. Label-free DIA analysis shortens the time needed for sample preparation as it avoids chemical labeling, achieving a turnaround time of less than a week for proteome-level analysis of a 96-well plate of FFPE material, with an additional 3 days required for phosphoproteome analysis. The use of DIA in this platform achieves a proteome depth similar to that obtained with TMT, identifying up to 10,000 unique proteins from each sample, but with much faster sample processing and data generation speed. These same approaches are applicable to tissue microarray samples (manuscript in preparation).
While PTM enrichment in FFPE samples has traditionally been challenging due to sample preservation and processing complexities, our study demonstrates its feasibility in a label-free setting. Using an automated workflow, over 11,000 localized phosphosites were enriched from macrodissected LUAD FFPE tissue samples and data collected on the Astral. Bioinformatic analysis of the proteome and phosphoproteome data generated on the small set of LUAD samples from archived blocks expands proteomics utility in FFPE characterization, with the potential of adding critical insights for investigating driver genes across multiple omes. The application of proteogenomic data analysis techniques to this small cohort of samples demonstrates the utility of high-throughput, plate-based FFPE proteomics and phosphoproteomics for derivation of meaningful biology from a larger clinical cohort of samples. Future studies are aimed at obtaining a deeper understanding of the biological relevance of these and other post-translationally modified peptides and their stability in FFPE samples compared to flash-frozen tissue. In conclusion, our experiments improve upon the use of DIA for proteomic and phosphoproteomic data collection, allowing for broader applications in proteomics and biomarker discovery of samples.
Methods:
Deparaffinization of FFPE Tissue using Covaris Ultrasonicator
Breast (BRC) and colorectal cancer (CRC) FFPE blocks (~2 x 3 cm), provided by the Van Andel Research Institute (VARI), and sectioned into serial 10 μm scrolls using the HistoCore MULTICUT (Leica). Scrolls were transferred immediately into a Covaris 96 AFA-Tube TPX Plate (Covaris, Part No. 520291) and fully submerged in wells prefilled with Covaris tissue lysis buffer (TLB) (Covaris, Part No. 52-284). FFPE tissue was deparaffinized by ultra-sonication in the Covaris LE220Rsc Focused-ultrasonicator at 3 mm Y-dither at 10 mm/s, followed by heating to 90°C in a thermocycler (Eppendorf) with a heated lid at 105°C for 90 minutes to decrosslink proteins. The tissue was then sonicated again for homogenization at 1 mm Z-dither at 20 mm/s. For the deparaffinization and homogenization steps, the ultrasonicator was maintained at 20°C, operated with a peak power of 350 W, and set to 200 cycles per burst, for 5 min for each lane of the Covaris TPX plate. The parameters evaluated included increasing the Covaris default sonication duty factor (DF) from 25% to 50%, and using either 100 μL or 150 μL of TLB. Optimal sonication conditions of 50% in 150 μL of TLB were used for subsequent experiments. Total protein amount following sonication was quantified using a BCA protein assay kit (Thermo Fisher, Cat No. 23225), with standards diluted 5X in TLB. The Covaris plate was spun down briefly at 1500 g in a centrifuge (Beckman Coulter) and 2 μL of the semi-clarified lysate was diluted 5X in water for quantification.
Sample cleanup and digestion optimization
For protein digestion and cleanup, suspension trapping (S-Trap) and solvent precipitation bead (SP3) workflows were evaluated. All lysates were initially treated with 2 mM magnesium chloride and 0.8 U/μL of benzonuclease (Sigma Aldrich, Cat No. E8263-5KU). BRC and CRC samples were normalized to 50 μg of protein lysate, followed by reduction and alkylation in 5 mM tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher, Cat No. 77720) and 50 mM 2-Chloroacetamide (CAA) (VWR, Cat No. TCC2536-25G) for one hour while mixing in darkness at room temperature (RT).
For the SP3 workflow, the beads (Cytiva Sera-Mag Hydrophilic and Sera-Mag Hydrophobic Magnetic Particles: Cat No. 44152105050250 and 24152105050250) were equilibrated at RT for 30 minutes, mixed at 1:1 ratio, washed three times with 80% acetonitrile (MeCN), and resuspended at 50 mg/mL. The bead mix was spiked into each sample at a bead to protein ratio of 10 : 1. Protein binding to the beads was initiated by adding MeCN to a final concentration of 80%. The beads were mixed during binding for 5 minutes, pelleted using a magnetic rack, and the supernatant removed by manual aspiration. The beads were then washed three times with 500 μL of 80% MeCN. On-bead protein digestion was performed overnight in 50 mM of triethylammonium bicarbonate (TEAB) pH 9 with mass spectrometry-grade trypsin (Promega) and LysC (FUJIFILM) at a ratio of 1:50 (weight/weight). Following overnight digestion, the beads were centrifuged at 3000 g, and the supernatant was collected. The beads were then washed with 50 μL of water, followed by 1% FA, where in each wash step, the samples were centrifuged and transferred to a magnetic rack, where the supernatant was collected and combined.
For suspension trapping, 1.2% phosphoric acid was added into each sample following reduction and alkylation, followed by six times the volume of 100 mM TEAB in 90% methanol (MeOH) (pH 7.1). Samples were mixed thoroughly via repeated pipette aspirations and loaded on the S-trap™ 96-well Mini Plate (Protifi) using a vacuum manifold. After loading, the wells were washed three times with 300 μL of a 50/50 (volume/volume) chloroform/methanol solution and three times with 300 μL of 100 mM TEAB in 90% MeOH pH 7.1. Samples were digested overnight in 125 μL of 50 mM TEAB pH 9, with trypsin and LysC at a ratio of 1:50 (weight/weight). Peptides were eluted into a deep-well plate (Cytiva, Part No. 7701-5200) sequentially with 80 μL of 50 mM TEAB pH 9, 0.2% formic acid (FA), and 0.2% FA in 50% MeCN.
Stage-tip desalting optimization
Peptides following digestion were quantified using a quantitative fluorometric peptide assay (Thermo Fisher, Cat No. 23290), acidified to 1% FA, and a specified amount was stage-tip desalted with Empore C18 disks (CDS Analytical, Part No. 98-0604-0217-3). The stage-tips were conditioned with 100% MeOH, 0.1% FA in 50% MeCN, and equilibrated with 1% FA. The peptides were loaded, washed three times with 0.1% FA, and eluted with 0.1% FA in 50% MeCN. The samples were dried down in a speedvac vacuum concentrator and stored at −80°C until LC-MS analysis.
SDB-RPS stage tips (Empore, Cat No. ST7600196) were implemented to reduce build-up on the column emitter. Peptides were acidified with 1% FA and stage-tips conditioned with 100% MeCN before loading. The peptides were washed twice with 1% FA in isopropyl alcohol (IPA), followed by 1% FA. Peptides were eluted with 5% ammonium hydroxide (NH3OH) in 80% MeCN. The samples were dried down and stored at −80°C until LC-MS analysis.
SDB-XC stage-tips were prepared by packing SDB-XC (Empore, Cat. No 98060402231EA) punches at a ratio of 1 punch for each 1 ug of peptides. The SDB-XC stage-tip was conditioned with 100% of MeOH, 0.1% FA in 50% MeCN, and equilibrated with 1% FA. Peptides were acidified to 1% FA and then loaded on the conditioned SDB-XC stage-tip for fractionation. The stage-tip was washed twice with 1% FA in water. Peptides were eluted from the stage-tip in five steps with 0.1% NH3OH in 5%, 10%, 15%, 20%, and 80% MeCN. Samples were dried and stored at −80°C until LC-MS analysis.
Semi-automated 96-well plate workflow
Automated aspects were implemented to enable efficient 96-well plate processing of FFPE tissue. Following plate-based Covaris sonication and S-trap loading of full pellets, the amount of trypsin and LysC added was determined based on the protein yield in each sample. The Opentrons OT-2 Robot was used to prepare the variable trypsin and lys-C input for each sample. This was implemented by using a custom Python script that reads a CSV file with the desired volume of protease for each sample in a 96-well plate. The opentrons then normalized each protease solution to a final volume of 125 μL of 50 mM TEAB. The script and CSV file were uploaded into the Opentrons application programming interface (API) for variable input steps. A multi-channel pipette was then used to transfer the digestion buffers on top of the S-trap wells. A similar custom protocol on the Opentrons OT-2 Robot was implemented to normalize peptide input amounts for stage-tip desalting. This involved transferring specified volumes from the S-trap deep-well elution plate to a secondary plate (Bio-Rad, Cat. No HSP9601), followed by normalization to a standard volume. The stage-tip desalting was carried in a 96-well plate format by removing the base from a S-trap plate and inserting the stage-tips into the wells, allowing for processing using a benchtop centrifuge (Beckman Coulter) and elution into a 96-well plate (Bio-Rad, Cat. No HSP9601) for LC-MS injection.
BRC & CRC FFPE tissue macrodissection
For BRC and CRC blocks, 10 μm sections were stained with hematoxylin and eosin (H&E) to identify and macrodissect tumor rich areas and transferred to Covaris 96-well plate in quadruplicates (performed at VARI). Each replicate consisted of tumor rich areas obtained from 5 scrolls equivalent to a total surface area of 30 mm2. To evaluate optimal tissue storage conditions, two of the four experimental replicates from each block were stored with or without TLB. The samples went through the S-trap digestion and SDB-RPS stage-tip desalting workflows described above. A pooled sample consisting of all BRC & CRC peptide samples was fractionated with SDB-XC stage-tips as described above.
LC-MS analysis of BRC & CRC FFPE samples
For all BRC & CRC FFPE samples, a total of 1 μg peptides (prior to stage-tip desalting) were loaded on a Vanquish Neo™ UHPLC system (Thermo Fisher) coupled to a Orbitrap Exploris™ 480 Mass Spectrometer (Thermo Fisher) operating in data independent acquisition (DIA) mode. Peptides were separated on a 25 cm New Objective PicoFrit™ column that was packed in-house with 1.5 μm C18 beads (Reprosil-Pur, Dr. Maisch). The separation occurred over a 94-minute active gradient and a 200 nL/min flow rate: 1.6% B - 4.8% B in 1 min, 4.8% - 24% B in 83 min, and 24% B - 48% B in 10 min. MS1 spectra covered a mass range of 350-1800 m/z with a resolution of 120,000 at 200 m/z. MS1 automatic gain control (AGC) target was set to 3E6 with a maximum injection time of 25 ms. For DIA analysis, 30 variable windows were used covering the mass range of 500-740 Th as shown in the Table S2. The scans were performed at an AGC target of 5E5 with a maximum injection time of 54 ms. For fragmentation, higher energy collisional dissociation (HCD) was set to 27% normalized collision energy (NCE). The DIA method had an average cycle time of 2.2 seconds equating to ~5 data points per peak (DPPP). MS2 spectra were HCD collision energy was set to 27% NCE with charge scaling of +3.
The pooled and fractionated BRC/CRC samples were run on the same LC-MS setup described above, with the instrument operating in data dependent acquisition (DDA) mode. Data was acquired at a resolution of 60,000 with an AGC target of 3E6 for MS1. MS2 spectra were acquired using a Top-20 cycle with a resolution of 120,000 and an AGC target of 3E5. Precursors of charge 2-6 were fragmented using HCD at 30% NCE.
Evaluation of Ncoa4 overexpression by TMT or label-free with DIA
Four mouse FFPE blocks were obtained from Dr. Joseph D. Mancias at the Dana-Farber Cancer Institute (26). The blocks embedded tissues from different organs with two blocks being wild-type and two with induced overexpression of Ncoa4. The blocks were sectioned into four experimental replicates and processed with the optimized Covaris, digestion, and stage-tip parameters outlined above.
For DIA analysis, 1 μg of peptides (amount prior to SDB-RPS stage-tip) were injected on various LC-MS systems. This analysis encompassed variable-window DIA method on the Orbitrap Exploris 480 (outlined above), Orbitrap Astral™ DIA with narrow isolation windows (4 m/z, Thermo Fisher), and ion mobility-based DIA (dia-PASEF) on the timsTOF-HT™ (Bruker Daltonics) with a variable window scheme as shown in Table S3. The wide-window DIA method on the Exploris 480 was consistent with the method described in the previous section, with the exception of a wider mass range (400-850 Th) to enhance peptide depth as shown in Table S4.
For dia-PASEF, peptides were separated on a nanoElute-2™ (Bruker Daltonics), using PepSep™ ULTRA C18 column (Bruker Daltonics, 25 cm length/75 μm inner diameter/1.5 μm particle size) and a 30 minute active gradient at a flow rate of 500 nL/min: 3% B - 28% B in 28 min, 28% B - 32% B in 2 min. The dia-PASEF method on the timsTOF HT covered an m/z range of 300-1400 optimized with py-diAID using two ion mobility windows per m/z area (38). A total of 20 dia-PASEF windows were used, and an additional MS1 scan was collected. The IM scan range was set to 0.7 to 1.45 V cm−2 with 50 ms ramp and accumulation time, equating a cycle time of 1.2 s and ~6 DPPP. Collision energy was synchronized with the IM range, 59 eV at 1/K0 = 1.6 Vs cm−2 decreased to 20 eV at 1/K0 = 0.6 Vs cm−2.
For Astral DIA (4 m/z), peptides were separated using a Vanquish Neo™ UHPLC system (Thermo Fisher) coupled to a Orbitrap Astral™ (Thermo Fisher) with a Aurora Elite™ TS C18 UHPLC column (IonOpticks, 15 cm length/75 μm inner diameter/1.7 μm particle size) at a flow rate of 800 nL/min. The 21 minute active gradient was as follows: 2 %B - 4 % B in 0.5 min, 4%B - 8 %B in 0.6 min, 8%B - 22.5 %B in 12.8 min, 22.5%B - 35%B in 6.9 min, and 35%B - 55%B in 0.4 min. MS1 spectra covered a mass range of 390-980 m/z with a resolution of 240,000 at 200 m/z using the Orbitrap mass analyzer. MS1 AGC target was set to 5E6 with a maximum injection time of 3 ms, and MS1 spectra were acquired with a timed loop control of 0.6 s. 4Th isolation windows spanned the range of m/z: 390-900. Maximum injection time was set to 5 ms for MS2 scans, with an AGC target of 2E5. Normalized collision energy was set to 25%.
For TMT analysis, peptides (~50 ug) were dried down and reconstituted to ~1 mg/mL in 100 mM TEAB. Peptides were labeled in a 1:5 ratio with TMTpro 18-plex (Thermo Fisher, Cat no. A52045). All channels were fully labeled, and initially, two channels were designated to represent a mixed pool of peptides from all experimental replicates, which would later be excluded for bioinformatic analysis. Following labeling, the 18 channels were quenched with hydroxylamine to a final concentration of 0.3%. The TMT-labeled samples were then pooled and dried down before desalting with SepPak tC18 (Waters, Cat. No. WATO36820). Offline-fractionation of TMT-labeled peptides was performed using a high-pH reversed-phase gradient with a ZORBAX™ C18 column (Agilent, 250 mm length/4.6 mM inner diameter/3.5 μm particle size) as described previously (39). The resulting fractions were combined into 24 samples for LC-MS analysis in data-dependent acquisition (DDA) mode. A total of 1 μg of each fraction was analyzed on the Thermo Scientific Vanquish Neo UHPLC coupled to a Thermo Scientific Orbitrap Exploris 480 with a 25 cm New Objective PicFrit™ column that was packed in-house with 1.5 μm C18 beads. A 99-minute active gradient at a flow rate of 200 nL/min was used: 1.8% B - 5.4% B in 1 min, 5.4%B - 27% B in 86 min, and 27% B - 54% B in 12 min. MS1 spectra covered a mass range of 350-1800 m/z with a resolution of 60,000. MS1 AGC target was set to 3E6 with a maximum injection time of 25 ms. Top 20 precursors were isolated, and monoisotopic peak selection (MIPS) filter was set to peptides with relaxed restrictions when too few precursors were found. The precursor intensity threshold for selection was 5E3 with a fit threshold of 50% and a window of 1.2 m/z. Precursors of charge states 2-6 were selected for fragmentation with an isolation width of 0.7 Th, the dynamic exclusion was set to 20 seconds, and NCE was set to 34%. MS2 scans were acquired with a resolution of 45,000, with an AGC target of 3E5, and a maximum injection time of 105 ms.
Proteome and phosphoproteome analysis of LUAD FFPE blocks
The LUAD FFPE samples were macrodissected by VARI as described above and directly placed into a Covaris 96 AFA-Tube TPX Plate pre-filled with TLB buffer. The samples had varying total tissue area (9 - 63 mm2 ). Covaris plates with dissected tissue were kept at −80C until time of processing with optimal Covaris, digestion, and stage-tip parameters described above. The proteome and phosphoproteome data was collected on the Orbitrap Astral using the same LC setup and 4 m/z isolation windows DIA method described above.
For phosphopeptide enrichment, 50 μg of peptides following S-Trap digestion were desalted using a Sep-Pak™ tC18 96-well plate (Waters, Cat No. 196002320) and eluted in 200 μL of 80% MeCN/0.1% trifluoroacetic acid (TFA). An additional 1% TFA was added prior to phosphopeptide enrichment on the AssayMAP Bravo liquid handling system (Agilent). Fe(III)-NTA cartridges (Agilent, 5 μL) were initially primed with 100 μL of 50% MeCN/0.1%TFA and equilibrated with 50 μL of 80% MeCN/0.1% TFA, followed by sample loading (195 μL, 5 μL/min). The cartridges were then washed with 50 μL of 80% MeCN/0.1%TFA and the phosphorylated peptides eluted with 20 μL of 1% NH4OH directly into a 96-well plate (Bio-Rad, Cat. no HSP9601) containing 2.5 μL of 100% formic acid. Following elution, 20 μL of 100% ACN was added to each sample using a multi-channel pipette. The samples were dried down and stored at −80°C until LC-MS analysis.
Data analysis
For all experiments, data was searched against the reviewed human proteome database obtained from UniProt, without isoforms (20,412 entries). Contaminants were appended to the FASTA file, and decoys were generated using the reverse sequence approach.
DIA data from the FFPE workflow optimization and macrodissections were searched in library free mode on FragPipe V20.0. Default parameters of DIA_Speclib_Quant workflow were loaded.
Peak matching: precursor mass tolerance 20 PPM, fragment mass tolerance: 20 PPM, calibration and optimization: mass calibration and parameter optimization, Isotope error: 0/1/2. Require precursor was enabled to discard for PSMs without any identified parent peaks.
Protein digestion was set to strict trypsin, with KR cuts at the C-Terminus and max cleavages set to 2. Peptide length was set to a minimum of 7 and a max of 50. Peptide mass range was kept at 500 to 5000 Da. N-terminal methionine excision was enabled.
Methionine oxidation and N-terminal acetylation were set as variable modifications, while carbimidomethylation of cysteine was selected as fixed modification.
An additional 1% run-specific protein FDR filtering through the DIA-NN quantification step was enabled.
The DDA data from the pooled and fractioned BRC/CRC samples were searched in FragPipe with the localization-aware open-search parameters in MSFragger and PTM-Shepherd. The default parameters were used (see workflow parameters in Table S5).
For all Ncoa4 overexpression data a DIA-NN search was performed because of its compatibility with all acquired raw files and to control for any database searching variability. An in-silico digest spectral library was generated in DIA-NN against mouse FASTA, that was downloaded from UniProt, with unreviewed sequences to include NCOA4 sequences. The fragment ion range was m/z: 100-2000 m/z, and the precursor ion range was set to m/z: 200-1600, with charge states 2-4. The peptide length was 7-30 with a max number of variable mods of 3 and 1 missed cleavage. Carbamidomethylation was selected as a fixed mod. N-terminal methionine excision, methionine oxidation and N-terminal acetylation were chosen as variable mods. TMT data was searched in FragPipe V20 based on the parameters from the TMT16 workflow. The precursor mass tolerance was set to +/− 20 ppm. Peptide length was set to 7-50 with 2 missed cleavages. Carbamidomethylation of cysteines and TMT-tag of lysine and N-terminal were chosen as static modifications. N-terminal acetylation, methionine oxidation, pyroglutamic acid, pyrocarbamidomethylation of cysteine, proline hydroxylation and N-terminal deamidation were searched for as variable mods. The number of maximum variable mods per peptide was set to 3. Phospho data was searched in Spectronaut 18 in directDIA mode. Cysteine carbamidomethylation was set as a fixed modification, while methionine oxidation, N-term acetylation and phosphorylation of serine, threonine and tyrosine were set as variable mods, with a maximum variable modification count of 5. A PTM probability threshold of 0 was used to allow for phosphosites of Class II and above, with a cutoff filter of ≥ 0.75 being applied post-analysis to determine localized Class I phosphosites (40).
MSFragger-DIA and Spectronaut instantiation on Terra platform
DIA data was searched using MSFragger-DIA workflow, which was added in FragPipe v20.0(18). MSFragger-DIA was implemented as a Google Cloud-based pipeline on the Terra platform (https://app.terra.bio) using a headless configuration to pass commands to a Fragpipe docker. To enable searching of DIA files collected from ThermoFisher instruments, MSConvert, (41) was adapted to Terra for parallelized Fragpipe-compatible raw-to-mzML conversion. The default conversion settings of MSConvert are reflected in the standard Terra configuration: MS1 peak picking (centroiding) and removal of consecutive extra zeros in MS1. The Terra-hosted Fragpipe pipeline requires users to input a GUI generated workflow file, containing parameters for running relevant Fragpipe modules with full functionality. Additional documentation for the Terra MSConvert and Fragpipe workflows is available on GitHub (https://github.com/broadinstitute/PANOPLY) (42). Spectronaut version 19.3 was also implemented as a Terra pipeline with functionality for multiple report generation, customized enzyme database integration, and combination of SNE files. (41)
Supplementary Material
Acknowledgements:
This work was supported by the following awards and grants - Broad Institute SPARC (#800444) awards to S.S and S.A.C, National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium (CPTAC) grants U24CA270823 to S.S, M.A.G and S.A.C, U01CA271402 to M.A.G and S.A.C and, Dr Miriam and Sheldon G. Adelson Medical Research Foundation to S.A.C. We also thank Dr. Joseph Mancias and Matthew J Dorman (Dana Farber Cancer Institute, Harvard Medical School) for the Ncoa4 wild-type and over-expressing mice FFPE blocks. We also thank Lia Abarzua and Sameer Vasantgadkar from Covaris, Woburn for their guidance with protocols and Lilian Heil from Thermo Fischer Scientific, San Jose for her assistance with data generation in the Thermo Fischer Science demo laboratory in San Jose, California.
Footnotes
Conflict of interest:
S. A. C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, Seer, and PrognomIQ. All other authors declare no competing interests.
References:
- 1.Faktor J., Kote S., Bienkowski M., Hupp T. R., and Marek-Trzonkowska N. (2024) Novel FFPE proteomics method suggests prolactin induced protein as hormone induced cytoskeleton remodeling spatial biomarker. Commun. Biol. 7, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buczak K., Ori A., Kirkpatrick J. M., Holzer K., Dauch D., Roessler S., Endris V., Lasitschka F., Parca L., Schmidt A., Zender L., Schirmacher P., Krijgsveld J., Singer S., and Beck M. (2018) Spatial Tissue Proteomics Quantifies Inter-and Intratumor Heterogeneity in Hepatocellular Carcinoma (HCC). Mol. Cell. Proteomics 17, 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y., Weiss T., Zhang Q., Sun R., Wang B., Yi X., Wu Z., Gao H., Cai X., Ruan G., Zhu T., Xu C., Lou S., Yu X., Gillet L., Blattmann P., Saba K., Fankhauser C. D., Schmid M. B., Rutishauser D., Ljubicic J., Christiansen A., Fritz C., Rupp N. J., Poyet C., Rushing E., Weller M., Roth P., Haralambieva E., Hofer S., Chen C., Jochum W., Gao X., Teng X., Chen L., Zhong Q., Wild P. J., Aebersold R., and Guo T. (2019) High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol. Oncol. 13, 2305–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tüshaus J., Eckert S., Fraefel M., Zhou Y., Pfeiffer P., Halves C., Fusco F., Weigl J., Hönikl L., Butenschön V., Todorova R., Rauert-Wunderlich H., The M., Rosenwald A., Heinemann V., Holch J., Meyer B., Weichert W., Mogler C., Hendrik-Kuhn P., and Kuster B. (2024) Towards routine proteome profiling of FFPE tissue: Insights from a 1,200 case pan-cancer study. Pathology, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coscia F., Doll S., Bech J. M., Schweizer L., Mund A., Lengyel E., Lindebjerg J., Madsen G. I., Moreira J. M., and Mann M. (2020) A streamlined mass spectrometry-based proteomics workflow for large-scale FFPE tissue analysis. J. Pathol. 251, 100–112 [DOI] [PubMed] [Google Scholar]
- 6.Velasquez E., Szadai L., Zhou Q., Kim Y., Pla I., Sanchez A., Appelqvist R., Oskolas H., Marko-Varga M., Lee B., Kwon H. J., Malm J., Szász A. M., Gil J., Betancourt L. H., Németh I. B., and Marko-Varga G. (2021) A biobanking turning-point in the use of formalin-fixed, paraffin tumor blocks to unveil kinase signaling in melanoma. Clin Transl Med 11, e466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich C., Schallenberg S., Kirchner M., Ziehm M., Niquet S., Haji M., Beier C., Neudecker J., Klauschen F., and Mertins P. (2021) Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat. Commun. 12, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert S., Chang Y.-C., Bayer F. P., The Matthew, Kuhn P.-H., Weichert W., and Kuster B. (2021) Evaluation of Disposable Trap Column nanoLC–FAIMS–MS/MS for the Proteomic Analysis of FFPE Tissue. J. Proteome Res. 20, 5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soni R. K. (2023) Protocol for deep proteomic profiling of formalin-fixed paraffin-embedded specimens using a spectral library-free approach. STAR Protoc 4, 102381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mar D., Babenko I. M., Zhang R., Noble W. S., Denisenko O., Vaisar T., and Bomsztyk K. (2024) A High-Throughput PIXUL-Matrix-Based Toolbox to Profile Frozen and Formalin-Fixed Paraffin-Embedded Tissues Multiomes. Lab. Invest. 104, 100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pujari G. P., Mangalaparthi K. K., Madden B. J., Bhat F. A., Charlesworth M. C., French A. J., Sachdeva G., Daviso E., Thomann U., McCarthy P., Vasantgadkar S., Bhattacharyya D., and Pandey A. (2023) A High-Throughput Workflow for FFPE Tissue Proteomics. J. Am. Soc. Mass Spectrom. 34, 1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchione D. M., Ilieva I., Devins K., Sharpe D., Pappin D. J., Garcia B. A., Wilson J. P., and Wojcik J. B. (2020) HYPERsol: High-quality data from archival FFPE tissue for clinical proteomics. J. Proteome Res. 19, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakabayashi M., Yoshihara H., Masuda T., Tsukahara M., Sugiyama N., and Ishihama Y. (2014) Phosphoproteome analysis of formalin-fixed and paraffin-embedded tissue sections mounted on microscope slides. J. Proteome Res. 13, 915–924 [DOI] [PubMed] [Google Scholar]
- 14.Zeneyedpour L., Stingl C., Dekker L. J. M., Mustafa D. A. M., Kros J. M., and Luider T. M. (2020) Phosphorylation ratio determination in fresh-frozen and formalin-fixed paraffin-embedded tissue with targeted mass spectrometry. J. Proteome Res. 19, 4179–4190 [DOI] [PubMed] [Google Scholar]
- 15.Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 16.Myers S. A., Rhoads A., Cocco A. R., Peckner R., Haber A. L., Schweitzer L. D., Krug K., Mani D. R., Clauser K. R., Rozenblatt-Rosen O., Hacohen N., Regev A., and Carr S. A. (2019) Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated Murine immune cells. Mol. Cell. Proteomics 18, 995–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruderer R., Bernhardt O. M., Gandhi T., Miladinović S. M., Cheng L.-Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., and Reiter L. (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu F., Teo G. C., Kong A. T., Fröhlich K., Li G. X., Demichev V., and Nesvizhskii A. I. (2023) Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform. Nat. Commun. 14, 4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demichev V., Messner C. B., Vernardis S. I., Lilley K. S., and Ralser M. (2020) DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller T., Kalxdorf M., Longuespée R., Kazdal D. N., Stenzinger A., and Krijgsveld J. (2020) Automated sample preparation with SP3 for low-input clinical proteomics. Mol. Syst. Biol. 16, e9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makhmut A., Qin D., Fritzsche S., Nimo J., König J., and Coscia F. (2023) A framework for ultra-low-input spatial tissue proteomics. Cell Syst. 14, 1002–1014.e5 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Muller M., Xu B., Yoshida Y., Horlacher O., Nikitin F., Garessus S., Magdeldin S., Kinoshita N., Fujinaka H., Yaoita E., Hasegawa M., Lisacek F., and Yamamoto T. (2015) Unrestricted modification search reveals lysine methylation as major modification induced by tissue formalin fixation and paraffin embedding. Proteomics 15, 2568–2579 [DOI] [PubMed] [Google Scholar]
- 23.Lai Z. W., Weisser J., Nilse L., Costa F., Keller E., Tholen M., Kizhakkedathu J. N., Biniossek M., Bronsert P., and Schilling O. (2016) Formalin-fixed, paraffin-embedded tissues (FFPE) as a robust source for the profiling of native and protease-generated protein amino termini. Mol. Cell. Proteomics 15, 2203–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F., Teo G. C., Kong A. T., Haynes S. E., Avtonomov D. M., Geiszler D. J., and Nesvizhskii A. I. (2020) Identification of modified peptides using localization-aware open search. Nat. Commun. 11, 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiszler D. J., Kong A. T., Avtonomov D. M., Yu F., Leprevost F. da V., and Nesvizhskii A. I. (2021) PTM-Shepherd: Analysis and summarization of post-translational and chemical modifications from open search results. Mol. Cell. Proteomics 20, 100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santana-Codina N., Del Rey M. Q., Kapner K. S., Zhang H., Gikandi A., Malcolm C., Poupault C., Kuljanin M., John K. M., Biancur D. E., Chen B., Das N. K., Lowder K. E., Hennessey C. J., Huang W., Yang A., Shah Y. M., Nowak J. A., Aguirre A. J., and Mancias J. D. (2022) NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins. Cancer Discov. 12, 2180–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancias J. D., Wang X., Gygi S. P., Harper J. W., and Kimmelman A. C. (2014) Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scarl R. T., Lawrence C. M., Gordon H. M., and Nunemaker C. S. (2017) STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J. Endocrinol. 234, R123–R134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., and Sato Y. (2020) KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillette M. A., Satpathy S., Cao S., Dhanasekaran S. M., Vasaikar S. V., Krug K., Petralia F., Li Y., Liang W.-W., Reva B., Krek A., Ji J., Song X., Liu W., Hong R., Yao L., Blumenberg L., Savage S. R., Wendl M. C., Wen B., Li K., Tang L. C., MacMullan M. A., Avanessian S. C., Kane M. H., Newton C. J., Cornwell M., Kothadia R. B., Ma W., Yoo S., Mannan R., Vats P., Kumar-Sinha C., Kawaler E. A., Omelchenko T., Colaprico A., Geffen Y., Maruvka Y. E., da Veiga Leprevost F., Wiznerowicz M., Gümüş Z. H., Veluswamy R. R., Hostetter G., Heiman D. I., Wyczalkowski M. A., Hiltke T., Mesri M., Kinsinger C. R., Boja E. S., Omenn G. S., Chinnaiyan A. M., Rodriguez H., Li Q. K., Jewell S. D., Thiagarajan M., Getz G., Zhang B., Fenyö D., Ruggles K. V., Cieslik M. P., Robles A. I., Clauser K. R., Govindan R., Wang P., Nesvizhskii A. I., Ding L., Mani D. R., Carr S. A., and Clinical Proteomic Tumor Analysis Consortium (2020) Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 182, 200–225.e35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Zhou X., Cheng L., Wang X., Zhang Q., Zhang Y., and Sun S. (1AD) PRKAA1 Promotes Proliferation and Inhibits Apoptosis of Gastric Cancer Cells Through Activating JNK1 and Akt Pathways. OR 28, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson B., Bock A. J., Holth A., and Nymoen D. A. (2021) The phosphatase PTPN1/PTP1B is a candidate marker of better chemotherapy response in metastatic high-grade serous carcinoma. Cytopathology 32, 161–168 [DOI] [PubMed] [Google Scholar]
- 33.Milev M. P., Hasaj B., Saint-Dic D., Snounou S., Zhao Q., and Sacher M. (2015) TRAMM/TrappC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment. J Cell Biol 209, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen C.-H., Yuan P., Perez-Lorenzo R., Zhang Y., Lee S. X., Ou Y., Asara J. M., Cantley L. C., and Zheng B. (2013) Phosphorylation of BRAF by AMPK impairs BRAF-KSR1 association and cell proliferation. Molecular cell 52, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J., Gao G., Li L., Ding J., Chen X., Lei J., Long H., Wu L., Long X., He L., Shen Y., Yang J., Lu Y., and Sun Y. (2022) Pan-Cancer Study of SHC-Adaptor Protein 1 (SHC1) as a Diagnostic, Prognostic and Immunological Biomarker in Human Cancer. Front. Genet. 13, 817118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav L., Pietilä E., Öhman T., Liu X., Mahato A. K., Sidorova Y., Lehti K., Saarma M., and Varjosalo M. (2020) PTPRA Phosphatase Regulates GDNF-Dependent RET Signaling and Inhibits the RET Mutant MEN2A Oncogenic Potential. iScience 23, 100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitulescu I. I., Meyer S. C., Wen Q. J., Crispino J. D., Lemieux M. E., Levine R. L., Pelish H. E., and Shair M. D. (2017) Mediator Kinase Phosphorylation of STAT1 S727 Promotes Growth of Neoplasms With JAK-STAT Activation. EBioMedicine 26, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowronek P., Thielert M., Voytik E., Tanzer M. C., Hansen F. M., Willems S., Karayel O., Brunner A.-D., Meier F., and Mann M. (2022) Rapid and In-Depth Coverage of the (Phospho-)Proteome With Deep Libraries and Optimal Window Design for dia-PASEF. Mol. Cell. Proteomics 21, 100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satpathy S., Jaehnig E. J., Krug K., Kim B.-J., Saltzman A. B., Chan D. W., Holloway K. R., Anurag M., Huang C., Singh P., Gao A., Namai N., Dou Y., Wen B., Vasaikar S. V., Mutch D., Watson M. A., Ma C., Ademuyiwa F. O., Rimawi M. F., Schiff R., Hoog J., Jacobs S., Malovannaya A., Hyslop T., Clauser K. R., Mani D. R., Perou C. M., Miles G., Zhang B., Gillette M. A., Carr S. A., and Ellis M. J. (2020) Microscaled proteogenomic methods for precision oncology. Nat. Commun. 11, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bekker-Jensen D. B., Bernhardt O. M., Hogrebe A., Martinez-Val A., Verbeke L., Gandhi T., Kelstrup C. D., Reiter L., and Olsen J. V. (2020) Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 11, 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M.-Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mani D. R., Maynard M., Kothadia R., Krug K., Christianson K. E., Heiman D., Clauser K. R., Birger C., Getz G., and Carr S. A. (2021) PANOPLY: a cloud-based platform for automated and reproducible proteogenomic data analysis. Nat Methods 18, 580–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.