Abstract
PRofile ALIgNEment (PRALINE) is a fully customizable multiple sequence alignment application. In addition to a number of available alignment strategies, PRALINE can integrate information from database homology searches to generate a homology-extended multiple alignment. PRALINE also provides a choice of seven different secondary structure prediction programs that can be used individually or in combination as a consensus for integrating structural information into the alignment process. The program can be used through two separate interfaces: one has been designed to cater to more advanced needs of researchers in the field, and the other for standard construction of high confidence alignments. The web-based output is designed to facilitate the comprehensive visualization of the generated alignments by means of five default colour schemes based on: residue type, position conservation, position reliability, residue hydrophobicity and secondary structure, depending on the options set. A user can also define a custom colour scheme by selecting which colour will represent one or more amino acids in the alignment. All generated alignments are also made available in the PDF format for easy figure generation for publications. The grouping of sequences, on which the alignment is based, can also be visualized as a dendrogram. PRALINE is available at http://ibivu.cs.vu.nl/programs/pralinewww/.
INTRODUCTION
The alignment of two or more sequences has become an essential sequence analysis technique in biological research. State-of-the-art multiple sequence alignment (MSA) methods, such as T-COFFEE (1) and MUSCLE (2), as well as other MSA methods available to date, perform alignments by only using the sequences in the given set. Although they use profile technology to match distant sequence sets, they do not use further homology information for the sequences that are available in current sequence databases. The benefit of using homologous information to align distant sequences has been shown in a number of studies (3), while the use of profiles to represent the additional homologous information has been shown to have many advantages (4,5). For this reason, the PRALINE toolbox (6,7) has been recently re-designed to include homology-extended multiple alignment (8), where as an initial step a profile for each sequence in a given set is built by using PSI-BLAST (9,10) and the progressive alignment then proceeds using the PSI-BLAST profiles instead of the given sequences. This approach has been previously applied with success to local pairwise alignment methods for homology modelling (11–15) and is extended in PRALINE for global MSA. The recently updated MAFFT alignment tool (3,16) also uses homologous sequences to improve the alignment quality of distant sequences. However, in the MAFFT approach, the additional information is not incorporated in profiles for each of the query sequences, but homologous sequences are added to the original set and then aligned together using the various MAFFT alignment strategies. In the end, the homologous sequences are removed, leaving the aligned original sequences to form the final alignment.
In this paper we present the new web server for the PRALINE toolbox (6,7), where we have added two new alignment features: homology-extended multiple alignment (8) and the integration of predicted secondary structure information with iteration capabilities (V. A. Simossis and J. Heringa, submitted for publication). We show results for the cytochrome P450 HOMSTRAD (17) sequence set as an example to demonstrate how the homology-extended strategy and integrating secondary structure information, in combination with the visualization possibilities of the server output can lead to meaningful interpretations. Details about the PRALINE strategies and optimizations have been described previously (6–8,18).
HOMOLOGY-EXTENDED MULTIPLE ALIGNMENT
The homology-extended MSA strategy enriches the information for each of the sequences in a given set by collecting putative homologous sequences. Each sequence is submitted as a query to PSI-BLAST over a database of choice [default: non-redundant (NR)]. The resulting PSI-BLAST alignments are then filtered for redundancy (100% sequence identity). In the event that no hits or only redundant hits are detected, the PSI-BLAST E-value threshold is automatically adjusted to a 10-fold less stringent setting (e.g. from 10 × 10−6 to 10 × 10−5) and the query is re-submitted. Once all the sequences to be aligned have at least one additional putative homologue, each PSI-BLAST alignment is converted into a profile and progressively aligned. A more detailed account of the PRALINE homology-extended multiple alignment algorithm and its performance is available in Ref. (8).
The advantage of this strategy is that it uses a much larger amount of position-specific information in the homology-extended profiles to score the alignment of two or more positions. As a result, the cases that benefit the most are those that evolution has changed so extensively (<30% identity) that the homology (common ancestry) between them is almost undetectable when compared directly (8).
In Table 1, the performance of the homology-extended alignment strategy on 254 HOMSTRAD (17) multiple alignment cases has been compared with the state-of-the-art methods T-COFFEEv2.03 and MUSCLEv3.51. The results show that for the strictest quality measure, column scoring, the overall improvement of the PRALINEPSI strategy is >3.5% relative to T-COFFEE and MUSCLE. Moreover, the improvement is >5% for the most distant and difficult test cases with sequences <30% sequence identity. In addition, PRALINEPSI has also been compared with the PRALINE standard global progressive alignment strategy (PRALINEBASIC) (6) and the PRALINEBASIC and PRALINEPSI strategies with integrated predicted [PSIPRED (19) and YASPIN (20)] secondary structure information, respectively, named as PRALINEBASIC-PSIPRED, PRALINEBASIC-YASPIN, PRALINEPSI-PSIPRED and PRALINEPSI-YASPIN. The latter secondary structure-guided alignment strategies of PRALINE are discussed in the next section. As shown in Table 1, the improvement in alignment quality achieved by homology-extended alignment (PRALINEPSI) as compared with other methods is significant in the more difficult alignment cases with average sequence identity percentages <60%. As would be expected, in the easier alignment cases that share >60% sequence identity, all the alignments are of comparable high quality.
Table 1.
The quality assessment of 254 HOMSTRAD multiple alignment cases generated by different alignment strategies
| Alignment method | Overall (%) | 0–30 (%) | 30–60 (%) | 60–100 (%) | P (0–100) |
|---|---|---|---|---|---|
| Column score | |||||
| PRALINEBASIC | 63.8 | 38.7 | 68.5 | 95.5 | – |
| PRALINEBASIC-YASPIN | 68.0 | 45.3 | 72.2 | 96.3 | 0.106 |
| PRALINEBASIC-PSIPRED | 67.4 | 43.5 | 72.1 | 95.9 | 0.337 |
| PRALINEPSI | 70.2 | 50.2 | 73.6 | 96.7 | 0.025 |
| PRALINEPSI-YASPIN | 70.0 | 49.7 | 73.6 | 96.5 | 0.042 |
| PRALINEPSI-PSIPRED | 70.1 | 50.2 | 73.5 | 96.7 | 0.014 |
| TCOFFEEv2.03 | 67.6 | 44.0 | 72.2 | 95.8 | 0.237 |
| MUSCLEv3.51 | 67.5 | 45.0 | 71.6 | 96.3 | 0.461 |
The significance of the results (P-value from Kolmogorov–Smirnov test) is calculated with regard to the PRALINEBASIC method. The column scores are the percentage correctly aligned columns with regard to the HOMSTRAD structure alignment.
When used as an option on the server, the homology-extended alignment strategy can further be customized by manually entering the desired iteration count, starting E-value cut-off and database to be searched by PSI-BLAST for the building of the homology-extended profiles (default: 3 iterations, starting with a cut-off of 10 × 10−6 on the NR database). The default parameters have been optimized by testing different settings on the HOMSTRAD database of structural alignments (8).
INTEGRATION OF SECONDARY STRUCTURE
The rule-of-thumb that structure is more conserved than sequence is a well-documented fact (21–24). As a result, many studies have shown that its use to guide sequence alignment improves alignment quality, especially between distant sequences (6–8,11–15,25). To this end, we have devised a secondary structure scoring scheme for the alignment algorithm that combines exchange weights from four types of matrices: sequence or profile positions that have not been assigned the same secondary structure class are scored using a generic matrix (default: BLOSUM62), otherwise the positions that have matching helix, strand or coil assignments use the Lüthy (26) helix-, strand- and coil-specific matrices, respectively. The use of the secondary structure information significantly improves the PRALINEBASIC alignment quality and also boosts the PRALINEPSI alignments in the very difficult alignment cases <20% sequence identity (V. A. Simossis and J. Heringa, submitted for publication). In Table 1, it is clearly shown that the use of the secondary structure is beneficial for PRALINEBASIC (>4% improvement in cases with <60% identity), albeit not as significant as the improvements seen with PRALINEPSI.
The secondary structure integration options of PRALINE involve the use of any one of the seven prediction methods that are listed [PHDpsi (27), PROFsec (B. Rost, unpublished data), SSPRO 2.01 (28), YASPIN (20), PSIPRED (19), JNET (29) and PREDATOR (30,31)] to predict the secondary structure of the input sequences. In addition, the user can optionally select to also search the Protein Data Bank (PDB) to find 3D structure information for the input sequences and use the DSSP-derived secondary structure for the alignment. If both DSSP and a prediction method are selected, the predictions will only be integrated into the alignment for those sequences that do not have a PDB entry. Finally, in the same list as the seven prediction methods, an optimally segmented (24) or majority voting consensus can be alternatively used that currently combines the predictions of PROFsec, YASPIN and PSIPRED.
PROFILE PRE-PROCESSING AND ITERATION
PRALINE provides a number of alignment strategies, such as profile pre-processing and iterative alignment optimization (6,7). The secondary structure-guided strategies using PHD, PROFsec, JNET and SSPRO, and the profile pre-processing strategies can be set to use consistency information to drive subsequent alignment rounds (iterations), each time drawing upon the theoretically higher quality information from the previous cycle. A detailed account of these strategies can be found in previously published work (6,7,18,25,32,33).
THE NEW PRALINE SERVER
The PRALINE program is designed to use two or more input protein sequences in the FASTA format (34). The proposed maximum number of sequences that should be submitted to the server is set to 500 with length 2000, but this is mainly to limit the server load and is not the limit of the PRALINE program. In addition, owing to the long running time needed for strategies, such as PRALINEPSI, an optional email notification can be requested that is delivered upon a completion of the job and contains the link to the results and some statistics on the resulting alignment.
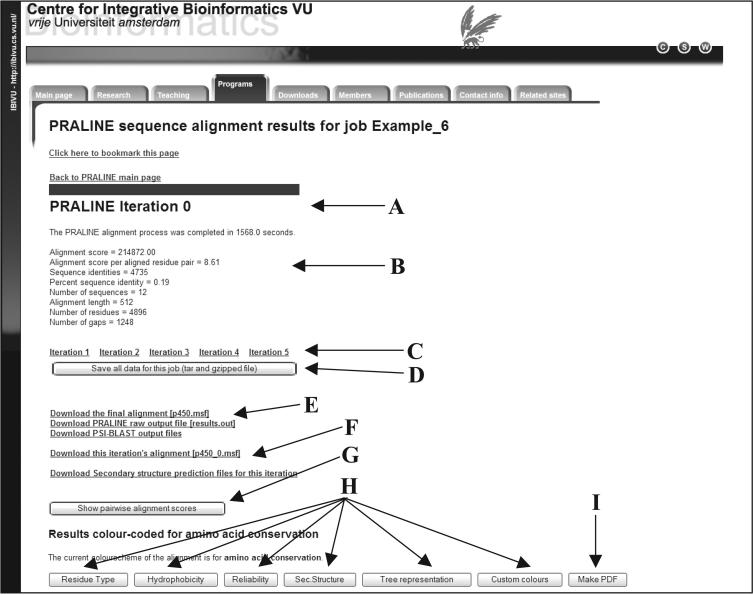
Similar to the previous version of the server (18), the gap opening and gap extension penalties and the amino acid substitution matrix can be manually set if needed [default: 12, 1 with BLOSUM62 (35)] for any of the PRALINE alignment strategies. The results page is automatically displayed once the job is complete and contains various sections depending on the options selected (Figure 1). In order to provide all generated files for the user, there is a link to download a compressed file with all the results in the job directory [Figure 1, (D)] and also individual links that allow the user to download specific files related to each sequence in the set (e.g. a PSI-BLAST profile or a secondary structure file) [Figure 1, (E)].
Figure 1.
The PRALINE results page headers. A: The subtitle indicating which iteration results are presented on this page (only available if iteration >0 is selected). B: The time taken to run the job and statistics related to the visible alignment. C: The links to all other available iteration cycle results (only available if iteration >0 is selected). D: The link to download all job files as a compressed file. E: Links to tabulated specific file types. F: Links to iteration-specific output files (only available if iteration >0 is selected). G: The button that hides/reveals the profile pre-processing scores of the sequence set (only available if profile pre-processing is selected). H: The buttons that switch between colour schemes. I: The button that generates and opens a PDF version of the alignment in the visible colour scheme.
If the iteration number selected is >0, a subtitle informs the user which iteration cycle results are presented on the page [Figure 1, (A)]. The alignment from each iteration cycle is presented on a different page and is accessible by the corresponding links [Figure 1, (C)]. In addition, it informs the user of the total time taken for the process to complete, provides some statistics related to the visible alignment [Figure 1, (B)] and if the iterations were halted due to alignment convergence or limit cycle convergence and which iteration was the last (not applicable in the Figure 1 example). In the case of iteration-specific output, such as alignment of the iteration or secondary structure prediction, additional links are displayed [Figure 1, (F)].
If profile pre-processing is selected the user has the option of viewing the profile pre-processing scores for all pairwise alignments for deriving an optimum cut-off value [Figure 1, (G)].
Finally, depending on the selected parameters of the job, a series of buttons allows switching between the available colour-coded views [Figure 1, (H)] [details about the colour schemes are described in (18)]. At any point, the visible alignment can be converted into a PDF for printing or further manipulation [Figure 1, (I)]. The remaining of the results page consists of a short description of the visible colour scheme with a key to the colours, after which the colour-coded alignment follows (an example of the conservation and the secondary structure colour-coding is shown in Figure 2).
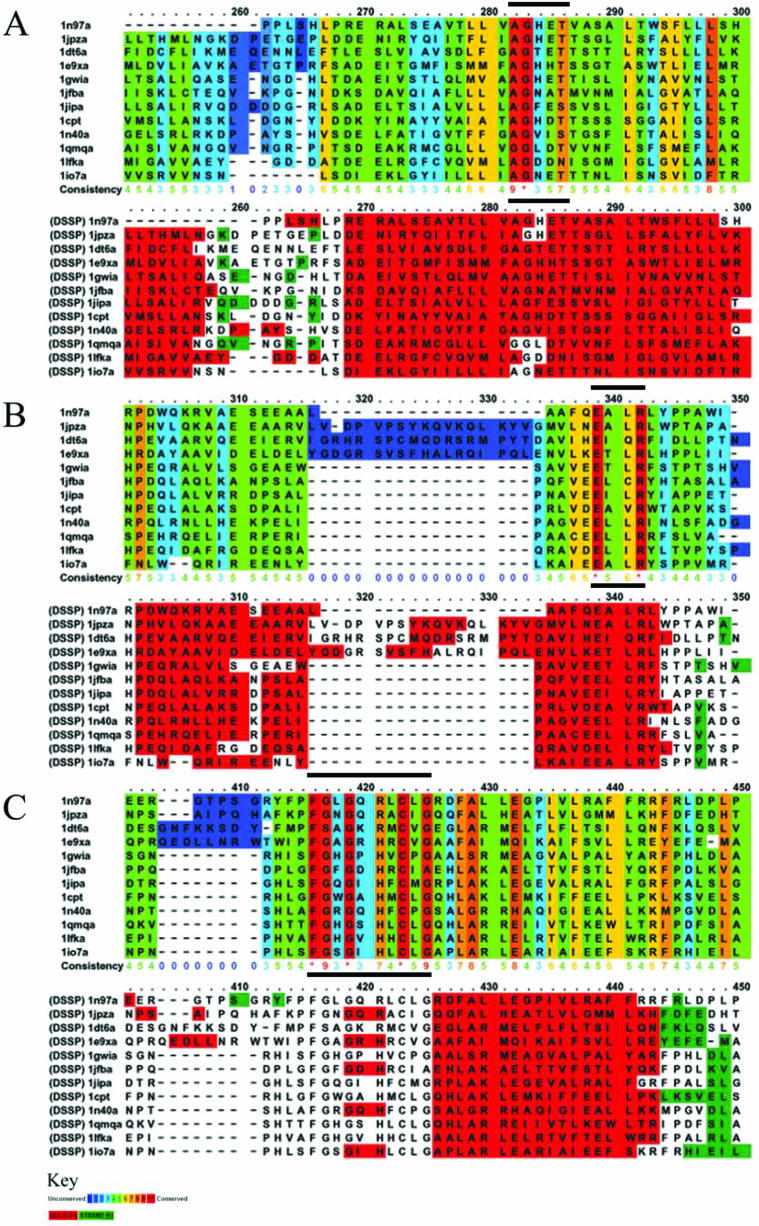
Figure 2.
The PRALINEPSI P450 alignment using both PROFsec and DSSP secondary structure integration settings. The alignment has been sectioned to focus on the regions containing the conserved motifs of the cytochrome P450 enzymes (signified by the black bars above the rulers). (A) The oxygen-binding motif, (B) the ExxR motif and (C) the haem-binding motif. For each section, the top colour scheme shows conservation levels according to the colour key and the bottom one shows the secondary structure each residue belongs to (red: helix; green: strand; and clear: coil). The ruler on top of each alignment block shows which parts of the alignment are visible.
SAMPLE OUTPUTS
Owing to the large number of possible outputs, we have provided a set of nine representative sample outputs for the P450 alignment on the server, each one representing a different combination of PRALINE strategies and settings. These examples are intended as supplementary material to this article and can be accessed through a dedicated link on the server pages or directly at http://ibivu.cs.vu.nl/programs/pralinewww/example/. They can also be used as an indication of CPU times needed by each of the PRALINE strategies.
In Figure 2, we illustrate sections of the PRALINEPSI alignment of the ‘p450’ HOMSTRAD sequence set (21% average sequence identity) using both DSSP (36) and PROFsec secondary structure integration settings. The colour schemes in the figure are for positional conservation and secondary structure. The secondary structure information for each sequence in this alignment has been derived by using DSSP, since all the sequences have a corresponding PDB structure.
The cytochrome P450 enzymes primarily act as oxidases in multi-component electron transport chains to break down naturally occurring toxins and mutagens. The structure is almost triangular, with the C-terminal part being mostly helical, while the N-terminal part is more β-sheet rich. The signature motif of P450 enzymes is the haem-binding site, which is often represented as FxxGxxxCxG (Figure 2C). Other conserved regions include the motif A(A/G)x(E/D)T (Figure 2A) where the threonine (T) residue is part of the oxygen-binding site and an invariant ExxR sequence (Figure 2B). The ExxR and the C residue at the haem-binding site are the only completely conserved amino acids in P450s. These well-documented details are straightforwardly visualized in the PRALINE output conservation colour scheme, while the secondary structure view allows us to relate them in a structural context. As stated in the literature (37), the oxygen binding and ExxR motifs are each part of two distinct C-terminal helices, while the haem-binding motif flanks the N-terminal end of the last helix. Owing to space limitations the alignment has been sectioned to concentrate on these regions, but the full alignment can be viewed online in example 9 of the supplementary material.
Acknowledgments
The authors would like to thank the Vrije Universiteit Amsterdam for funding this project. Special thanks are also due to Drs Franca Fraternali, Jens Kleinjung and John Romein for help with debugging and server testing. Funding to pay the Open Access publication charges of this article was provided by the Vrije Universiteit Amsterdam.
Conflict of interest statement. None declared.
REFERENCES
- 1.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 2.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G., Dunbrack R.L., Jr Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar R.C., Sjolander K. A comparison of scoring functions for protein sequence profile alignment. Bioinformatics. 2004;20:1301–1308. doi: 10.1093/bioinformatics/bth090. [DOI] [PubMed] [Google Scholar]
- 6.Heringa J. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput. Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
- 7.Heringa J. Local weighting schemes for protein multiple sequence alignment. Comput. Chem. 2002;26:459–477. doi: 10.1016/s0097-8485(02)00008-6. [DOI] [PubMed] [Google Scholar]
- 8.Simossis V.A., Kleinjung J., Heringa J. Homology-extended sequence alignment. Nucleic Acids Res. 2005;33:816–824. doi: 10.1093/nar/gki233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S.F., Koonin E.V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung R., Yona G. Protein family comparison using statistical models and predicted structural information. BMC Bioinformatics. 2004;5:183. doi: 10.1186/1471-2105-5-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginalski K., Pas J., Wyrwicz L.S., von Grotthuss M., Bujnicki J.M., Rychlewski L. ORFeus: detection of distant homology using sequence profiles and predicted secondary structure. Nucleic Acids Res. 2003;31:3804–3807. doi: 10.1093/nar/gkg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginalski K., von Grotthuss M., Grishin N.V., Rychlewski L. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–W581. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2004;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 15.von Ohsen N., Sommer I., Zimmer R., Lengauer T. Arby: automatic protein structure prediction using profile–profile alignment and confidence measures. Bioinformatics. 2004;20:2228–2235. doi: 10.1093/bioinformatics/bth232. [DOI] [PubMed] [Google Scholar]
- 16.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuguchi K., Deane C.M., Blundell T.L., Overington J.P. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simossis V.A., Heringa J. The PRALINE online server: optimising progressive multiple alignment on the web. Comput. Biol. Chem. 2003;27:511–519. doi: 10.1016/j.compbiolchem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 20.Lin K., Simossis V.A., Taylor W.R., Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics. 2005;21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- 21.Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 23.Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 24.Simossis V.A., Heringa J. The influence of gapped positions in multiple sequence alignments on secondary structure prediction methods. Comput. Biol. Chem. 2004;28:351–366. doi: 10.1016/j.compbiolchem.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Heringa J. Computational methods for protein secondary structure prediction using multiple sequence alignments. Curr. Protein Pept. Sci. 2000;1:273–301. doi: 10.2174/1389203003381324. [DOI] [PubMed] [Google Scholar]
- 26.Lüthy R., McLachlan A.D., Eisenberg D. Secondary structure-based profiles: use of structure-conserving scoring tables in searching protein sequence databases for structural similarities. Proteins. 1991;10:229–239. doi: 10.1002/prot.340100307. [DOI] [PubMed] [Google Scholar]
- 27.Przybylski D., Rost B. Alignments grow, secondary structure prediction improves. Proteins. 2002;46:197–205. doi: 10.1002/prot.10029. [DOI] [PubMed] [Google Scholar]
- 28.Pollastri G., Przybylski D., Rost B., Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 29.Cuff J.A., Barton G.J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 30.Frishman D., Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 31.Frishman D., Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 32.Simossis V.A., Heringa J. Integrating protein secondary structure prediction and multiple sequence alignment. Curr. Protein Pept. Sci. 2004;5:249–266. doi: 10.2174/1389203043379675. [DOI] [PubMed] [Google Scholar]
- 33.Simossis V.A., Kleinjung J., Heringa J. An overview of multiple sequence alignment. In: Baxevanis A.D., editor. Current Protocols in Bioinformatics. NY: John Wiley; 2003. pp. 3.7.1–3.7.25. [DOI] [PubMed] [Google Scholar]
- 34.Pearson W.R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 35.Dayhoff M.O., Barker W.C., Hunt L.T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz de Montellano P.R., editor. Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd edn. NY: Plenum Press; 1995. [Google Scholar]