Abstract
While generation of high-throughput expression data is becoming routine, the fast, easy, and systematic presentation and analysis of these data in a biological context is still an obstacle. To address this need, we have developed PathwayExplorer, which maps expression profiles of genes or proteins simultaneously onto major, currently available regulatory, metabolic and cellular pathways from KEGG, BioCarta and GenMAPP. PathwayExplorer is a platform-independent web server application with an optional standalone Java application using a SOAP (simple object access protocol) interface. Mapped pathways are ranked for the easy selection of the pathway of interest, displaying all available genes of this pathway with their expression profiles in a selectable and intuitive color code. Pathway maps produced can be downloaded as PNG, JPG or as high-resolution vector graphics SVG. The web service is freely available at https://pathwayexplorer.genome.tugraz.at; the standalone client can be downloaded at http://genome.tugraz.at.
INTRODUCTION
The huge amount of large-scale gene and protein expression data requires new methods for correlation with prior biological knowledge. Intense efforts have been undertaken and a variety of different computational methods have been applied to analyze these data sets. Initial data analyses are based on tools (1) using common clustering algorithms (2–6). Subsequent analyses require tools for visualizing genes and gene products in the context of biological pathways.
In the past years, scientists have established common repositories for biological data to make it freely available to others. Consequently, relating biological data to relevant pathway overviews is now accessible through publicly available resources. These sources include large graphical collections with open source access [e.g. Kyoto Encyclopedia of Genes and Genomes KEGG (7) at http://www.genome.jp/kegg, BioCarta at http://biocarta.com/genes and GenMAPP at http://www.genmapp.org (8)]. By overlaying expression data on biological pathways, established and novel relationships among genes can be explored. These pathways give key information about the functional and metabolic organization of cellular and biological systems within organisms.
Scientists working with large-scale expression data have already tried to verify and visualize their data in the context of biological pathways, but the analysis is hampered by a lack of systematic approaches. Growing trends of using freely available software packages in bioinformatics along with the increasing diversity of different operating systems are more and more claiming for platform-independent web solutions which are able to meet these requirements. Although some tools are available (9–13), limitations in performance, usability and functionality still remain. A user-friendly graphical user interface is critical for optimal usability by a broad range of users. Furthermore, there should be several methods available for downloading user-relevant information in a textual and graphical way.
For this purpose, we have developed a web-based service PathwayExplorer, which provides comprehensive and easily accessible representations of expression profiles onto major regulatory, metabolic and cellular pathways. The integrated pathway resources include KEGG, BioCarta and GenMAPP.
METHODS
We used state-of-the-art Java technologies to develop PathwayExplorer. It is an entirely Java-based application client using a three-tiered-architecture that ensures a clean separation between the presentation front-end, business and database back-end layer. The business layer, a Java application client conforming to the J2EE specification, performs the calculations and search functions and can be accessed by the presentation layer in two ways: (i) an application client using SOAP (simple object access protocol); and (ii) a web browser-based application client running on a web server using JSP technology. The database layer called PathwayDB is based on an Oracle DBMS (data base management system), which is also portable to freely available MySQL or PostgreSQL DBMS. Frequently changing information is kept in flat files, which are obtained and constantly updated from NCBI (ftp://ftp.ncbi.nih.gov/refseq/LocusLink/) (14) and KEGG (ftp://ftp.genome.ad.jp/pub/kegg/ligand/) (7).
PathwayDB minimizes the ambiguity among its gene identifiers. The fact that almost all identifiers are relationally and hierarchically linked allows it to specify the gene element nodes with only one kind of identifier, which constitutes the top of the hierarchical identifier tree. All the gene identifiers that lie below the root identifier can be linked to it later by using external data sources. We have successfully integrated KEGG, BioCarta and GenMAPP pathways into PathwayDB by using only the minimum information necessary. This comprises information from parsed SMBL (Systems Biology Markup Language) (15) files obtained from KEGG, which were converted into the PathwayExplorer application client (16) format. This was performed because of the lack of the SMBL format for encoding feasible graphical visualization, which is essential for the graphical evaluation of mapped pathways. The EC (17) (Enzyme Commission) defines the root identifier, which can hold several gene identifiers from all available organisms, i.e. the LocusLinks (they can contain again several gene identifiers, such as RefSeq or UniGene IDs) or the official gene identifiers for other organisms. To integrate BioCarta and GenMAPP into PathwayDB, the PathwayExplorer application client was once again used for automatically parsing the HTML pages holding the necessary pathway information.
Since both of these pathway resources use many different gene identifiers, LocusLink was again used as root identifier. The LocusLinks are linked with the user-defined gene identifier groups (UniGene, GeneOntology, GenBank and/or RefSeq), which are used then to align the mapped gene IDs.
PROGRAM DESCRIPTION
Accessibility
PathwayExplorer is a web-based service constantly available at https://pathwayexplorer.genome.tugraz.at, with a public, login-free data repository for uploading data sets.
The PathwayExplorer standalone client application can perform the same mapping operations on an independent, local-platform computer system. In this case, instead of uploading the expression data to the web server, the pathway information from PathwayDB is downloaded to the user's local computer system. The standalone client connects to PathwayDB through a SOAP interface. The standalone client is available at the PathwayExplorer homepage or at http://genome.tugraz.at/Software/PathwayExplorer/Setup.html.
Input
As input, PathwayExplorer receives a common tab-delimited text file containing expression profiles with the gene identifier as first column, the gene name as an optional second column and any experiment or time point data as further columns. Possible gene identifiers for organisms using LocusLinks are GenBank accession numbers, RefSeq IDs, UniGene IDs and Gene Ontology IDs (e.g. see Table 1). For all other organisms, systematic gene identifiers are possible. The RefSeq IDs are used as the initial default gene identifier group, and this can be changed later. The uploaded gene-expression data sets can be stored in either a public or a login-requiring repository where they can be modified or deleted again.
Table 1.
The number of unique gene identifiers (e.g. Homo sapiens) available for mapping expression profiles in PathwayExplorer
| Pathway resource | No. of pathways | Unique RefSeq accession no. | Unique GenBank accession no. | Unique UniGene accession no. | Unique GO accession no. |
|---|---|---|---|---|---|
| KEGG | 120 | 4099 | 26589 | 2827 | 2561 |
| BioCarta | 311 | 2209 | 15889 | 1438 | 1671 |
| GenMAPP | 82 | 6374 | 38171 | 4527 | 2857 |
| Sum | 513 | 8947 | 55111 | 6276 | 3623 |
The table shows the sum of non-redundant accession numbers available.
Calculations and visualization
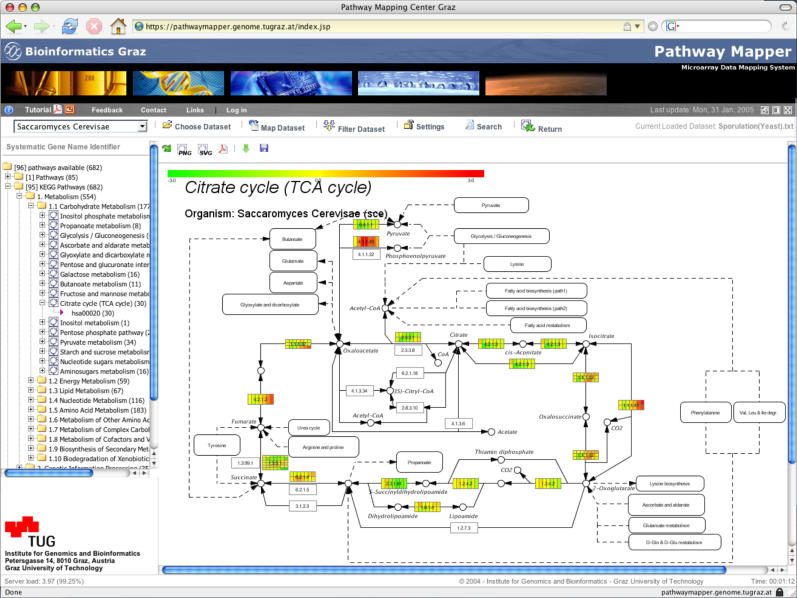
An example for mapping a public data set from a yeast sporulation study (18) is given in Figure 1. In order to map data sets onto pathways, the user is requested to select the organism and the data set to be mapped. The loaded data set remains in the background as long as it has not been closed, and subsequently every pathway which becomes opened is then automatically mapped with this data set. To restrict the uploaded data set to certain criteria before mapping (e.g. to use only expression profiles of differentially expressed gene or proteins), filter options can be applied: (i) to filter out expression profiles with too many missing data points; (ii) to filter out weakly expressed profiles (based on a certain standard deviation threshold); and (iii) to filter out genes whose expression values do not meet a certain threshold.
Figure 1.
PathwayExplorer example: a screenshot of a pathway mapped with expression data. (i) The toolbar frame (the row including the organism field) offers various setup and visualization options. (ii) Hierarchical tree frame (on the left) enables browsing through all available pathway sections. (iii) The main frame (in the center) displays the citrate cycle pathway extracted from KEGG, which mapped with a yeast sporulation data set with seven different time points (0, 0.5, 2, 5, 7, 8 and 11.5 h). The color-coded boxes represent mapped genes. If more than just one gene ID (e.g. RefSeq) matches to a box, each box is split up into several horizontal elements. According to the number of experiments/time points (in this case seven time points), the boxes are split again into vertical columns which display the expression level of each time point. To visualize a mapped expression profile in a different way, one has to choose the corresponding horizontal row of a box (see Figure 2).
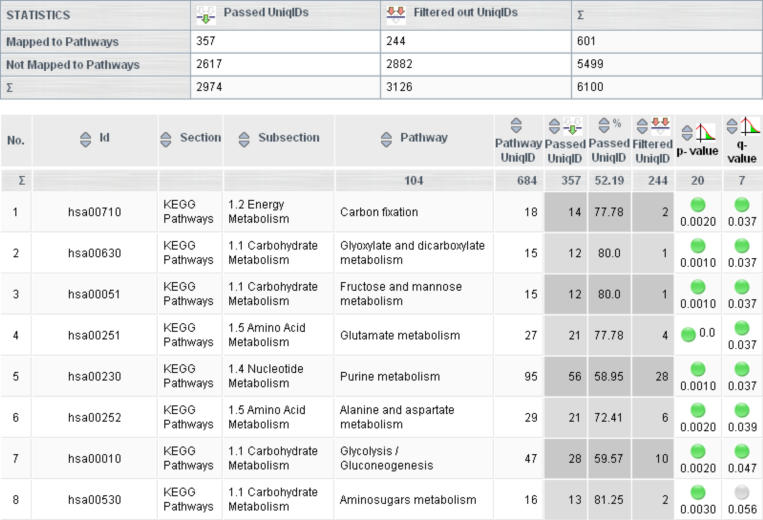
After filtering the data set, PathwayExplorer provides two mapping options: (i) mapping the data set to a single pathway by choosing one in the hierarchical tree or (ii) Mapping the data set onto all available pathways at once. Option (ii) generates a list (Figure 3), which ranks all mapped pathways by their number of mapped genes and allows for sorting the list based on different criteria, such as (a) pathway name; (b) unique gene identifiers available in each pathway; (c) the number of gene identifiers which has passed the filter criteria and was mapped to the pathway; (d) the number of genes which would have been mapped to the pathway if they had passed the filter criteria; and (e) the right-tailed P-value of a Fisher's exact test (f) the false discovery rate (FDR) (19) corrected Q-value.
Figure 3.
(a) Shows the overall statistic of the filtered unique identifiers of the expression data set mapped on all pathways. (b) The ranking list of all mapped pathways is also displayed and can be sorted by different criteria, allowing easy navigation through all pathways. By selecting the pathway of interest, the data set will be automatically mapped on the pathway and the expression values will be displayed in a selectable and intuitive color code (see Figure 1). The last two columns display the P-value and the FDR (19) corrected Q-value.
With a right-tailed Fisher's exact test, we test whether the proportion of mapped genes within the set of differentially expressed genes is significantly larger than the proportion of mapped genes that are not differentially expressed (see Table 2). We use a Fisher's exact test because the number of counts might be smaller than five for any of the fields in the contingency table. Multiple hypotheses correction (19) is needed to control the number of false positives, since many hypotheses are tested simultaneously.
Table 2.
2 × 2 contingency table for the Fisher's exact test
| Genes that are differentially expressed (passed the filter) | Gene that are not differentially expressed (filtered out) | ||
|---|---|---|---|
| Mapped genes | A | B | A+B |
| Unmapped genes | C | D | C+D |
| A+C | B+D | Total number of genes |
The null hypothesis of the right-tailed Fisher's exact test states that the proportion of A/C is smaller or equal to the proportion of B/D. If the right-tailed P-value is <5%, we reject the null hypothesis, which means that the proportion of differentially expressed genes is significantly greater than those that are not differentially expressed.
The graphical visualization of the displayed pathway image can be changed to the SVG view using the freely available SVG Viewer plug-in from Adobe (http://www.adobe.com). This enables on-line zooming of the pathway graphic.
Output
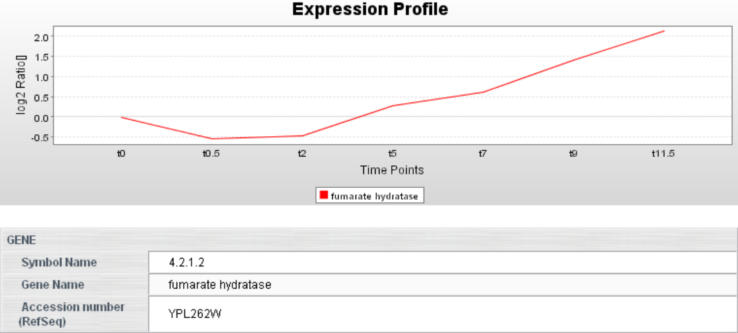
PathwayExplorer provides graphical and textual output. It generates a gene cluster for each mapped pathway, which can be downloaded in the same tab-delimited text format as the uploaded data set. Each mapped expression profile can be displayed (Figure 2) by selecting the corresponding box (Figure 1) on the pathway image. The generated ranking list for all mapped pathways (Figure 3) can also be downloaded as tab-delimited text file and can be used for statistical analyses.
Figure 2.
By selecting one row (if there are more than one) of a mapped gene box (Figure 1), the corresponding gene and expression profile information will be displayed. To obtain additional information links to GenBank, Entrez and OMIM are provided. Only one mapped expression profile from the uploaded data set can be displayed at once.
A special feature is the PDF generator, which can be applied for every single pathway, irrespective of whether the genes were mapped to the pathway or not. This generator creates a PDF document of the currently loaded pathway, which can be downloaded or directly displayed in the user's web browser. If genes were mapped to the pathway, the PDF generator additionally adds the expression profiles to the document. In the case of human expression data sets, additional information about each gene is directly extracted from the OMIM (Online Mendelian Inheritance in Man) database. This feature offers the user a special opportunity to get a quick and comprehensive overview of the current pathway plus detailed information about each mapped gene. The graphical output of PathwayExplorer can be directly downloaded in PNG or SVG graphic format.
CONCLUSION
We have developed PathwayExplorer, a web server providing comprehensive and facile mapping of gene or protein expression profiles. The profiles are simultaneously mapped onto the major regulatory, metabolic and cellular pathways available from the KEGG, BioCarta and GenMAPP pathway resources. The server accepts expression data files in a tab-delimited text format and generates high-resolution vector graphic images of mapped pathways. It enables further very compact representations of expression profiles within all available pathways. PathwayExplorer not only unifies the access to different pathway resources, but also combines gene identifiers arbitrarily selectable by the user.
PathwayExplorer is at present limited by common gene identifiers, such as RefSeq, GenBank, Gene Ontology or UniGene, but due to PathwayExplorer's flexible design, future requirements can be easily integrated. PathwayExplorer is thus endowed with a wide range of functionality giving the user multiple options to extract biological information in a comprehensive systematic and intuitive way. The online accessibility and the intuitive interface of PathwayExplorer should make it a valuable tool for a broad range of users.
Acknowledgments
The authors thank all members of the Institute for Genomics and Bioinformatics and external scientists who contributed to this work and James McNally for critically reviewing the manuscript. This work was supported by the Austrian Science Fund (Grant SFB Biomembranes F718) and the bm:bwk, GEN-AU:BIN, Bioinformatics Integration Network. B.M. was supported by a grant from the Austrian Academy of Sciences (OEAW). Funding to pay the Open Access publication charges for this article was provided by GEN-AU:BIN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sturn A., Mlecnik B., Pieler R., Rainer J., Truskaller T., Trajanoski Z. Client-server environment for high-performance gene expression data analysis. Bioinformatics. 2003;19:772–773. doi: 10.1093/bioinformatics/btg074. [DOI] [PubMed] [Google Scholar]
- 2.Tamayo P., Slonim D., Mesirov J., Zhu Q., Kitareewan S., Dmitrovsky E., Lander E.S., Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen X., Fuhrman S., Michaels G.S., Carr D.B., Smith S., Barker J.L., Somogyi R. Large-scale temporal gene expression mapping of central nervous system development. Proc. Natl Acad. Sci. USA. 1998;95:334–339. doi: 10.1073/pnas.95.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M.P., Grundy W.N., Lin D., Cristianini N., Sugnet C.W., Furey T.S., Ares M., Jr, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellenberg K., Hauser N.C., Brors B., Neutzner A., Hoheisel J.D., Vingron M. Correspondence analysis applied to microarray data. Proc. Natl Acad. Sci. USA. 2001;98:10781–10786. doi: 10.1073/pnas.181597298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M., Goto S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 9.Pandu R., Guru R.K., Mount D.W. Pathway Miner: extracting gene association networks from molecular pathways for predicting the biological significance of gene expression microarray data. Bioinformatics. 2004;20:2156–2158. doi: 10.1093/bioinformatics/bth215. [DOI] [PubMed] [Google Scholar]
- 10.Chung H.J., Kim M., Park C.H., Kim J., Kim J.H. ArrayXPath: mapping and visualizing microarray gene-expression data with integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2004;32:W460–W464. doi: 10.1093/nar/gkh476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirava M., Schäfer T., Eiglsberger M., Kaufmann M., Kohlbacher O., Bornber-Bauer E., Lenhof HP. BioMiner—modeling, analyzing, and visualizing biochemical pathways and networks. Bioinformatics. 2002;18:219–230. doi: 10.1093/bioinformatics/18.suppl_2.s219. [DOI] [PubMed] [Google Scholar]
- 12.Goesmann A., Haubrock M., Meyer F., Kalinowski J., Giegerich R. PathFinder: reconstruction and dynamic visualization of metabolic pathways. Bioinformatics. 2002;18:124–129. doi: 10.1093/bioinformatics/18.1.124. [DOI] [PubMed] [Google Scholar]
- 13.Pan D., Sun N., Cheung K.H., Guan Z., Ma L., Holford M., Deng X., Zhao H. PathMAPA: a tool for displaying gene expression and performing statistical tests on metabolic pathways at multiple levels for Arabidopsis. BMC Bioinformatics. 2003;32:W460–W464. doi: 10.1186/1471-2105-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hucka M., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 16.Trost E., Hackl H., Maurer M., Trajanoski Z. Java editor for biological pathways. Bioinformatics. 2003;19:786–787. doi: 10.1093/bioinformatics/btg052. [DOI] [PubMed] [Google Scholar]
- 17.Kotera M., Okuno Y., Hattori M., Goto S., Kanehisha M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 18.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]