Abstract
The protein picture generator (PPG) is an online service to generate pictures of a protein structure. Its design was conceived as an answer to the need expressed by a part of the community to have some means to produce simply complex pictures to insert in publications or in presentations. PPG can produce static or animated pictures. It can be accessed at http://bioserv.rpbs.jussieu.fr/cgi-bin/PPG.
INTRODUCTION
Molecular visualization has evolved much over the last three decades and has become an unavoidable means to explore at the molecular level the determinants of protein structuration, stabilization and function. Progressing with our understanding of the structures, the softwares designed for the purpose of molecular visualization have gained in complexity, covering various aspects of visualization. At the same time, the use of pictures of proteins has become a routine means for illustrating various topics in papers or in presentations. However, despite the softwares having gained in performance, ergonomics and availability on most platforms, the production of images can remain uneasy. It can require the implementation of various softwares, and the assimilation of various concepts.
So far, several attempts have been made to provide structure imaging via the Web. Different concerns underly them: some compromise has to be reached between the interactivity and the quality of the display of the structures. For example, Jmol (http://jmol.sourceforge.net), Java-based WebMol (1) or King (2), provides interactive macromolecular visualization via the net. Although impressive, they currently remain mostly limited to vector graphics. However, catalogues of static raster images are available at the PDB (3) or at other resources, such as CATH (4). These images of the protein fold do not provide details on particular sites of the structures, and it is not possible to change the orientation of the view. Tool such as the viewer of the Robetta server (5) provides an intermediate solution. It allows attainment of a good quality rendering by combining the RASMOL (6), MOLSCRIPT (7) and Raster3D (8) programs, and the view can be iteratively adjusted. However, no modification is possible of the way in which the structure is represented.
Protein picture generator (PPG) focuses on the resulting image, as the production time making interactivity is not reachable. But some means are given so that the user can view the structure in a desired orientation. Importantly, it is possible to combine different types of representation to have different levels of complexity in the picture, and to produce animations easily.
DESIGN OF THE SERVICE
The PPG interface has been designed after querying a panel of potential users about the features they think are important to produce a picture of a protein. Although rather large variations could be observed in the expected functionalities, commonly expressed needs were:
Having the possibility to render the structure using various representations, including molecular surface and hydrogen bonds;
Having various colouring schemes;
Having the possibility to focus on some part of the structure. This point covered several aspects, such as rendering only one part of the structure, viewing the structure from a particular and informative point of view, being able to combine different types of representation to illustrate different features of a structure.
Finally, some emphasis was put on the necessary ease with which such a service should, from data, be able to produce illustrations for publications or presentations. The possibility of generating animations was mentioned as a plus, as well as the possibility to insert labels.
To meet these requirements, we have chosen the following strategy:
There is a default representation for the complete structure. The default representation contains different levels: backbone, side-chains, hydrogen bonds and surface. Each can be parameterized independently (type of representation and colour pattern). It is possible to skip this default representation in order to use only more specific representations (see below).
In addition to this default representation, it is possible to specify additional representations for all or part of the structure, using different types of representation and colour patterns.
A limited number of parameters describe the ‘scene’, i.e. the way the structure is seen by the user, and the picture (file format, size and animation).
A more complete control on the rendering parameters is accessible on the form by advanced parameters, which the user does not need to manipulate at first.
Label insertion is limited to a title. Atomic labels or arrows can be inserted in the picture outside PPG.
DESIGN OF THE SOFTWARE
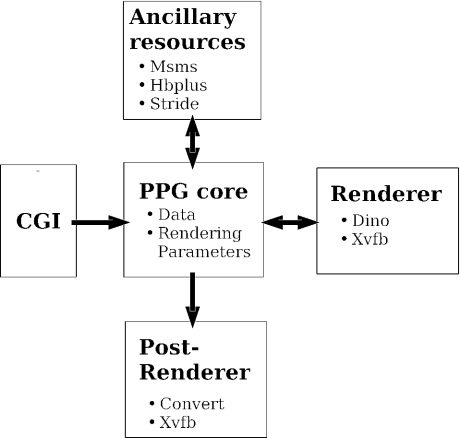
The software underlying the service consists of four components as illustrated Figure 1. The core component consists of a python PPG class that embeds all the information necessary to produce the image: data and rendering parameters. At the creation of an instance of the class, a dictionary of all the parameters assigned to default values is created. A second component is a common gateway interface (CGI) whose role is to get the values from the web form and transmit it to the PPG core instance. The third component is a renderer. Currently, PPG relies on Dino (9). The communication between the PPG core instance and the renderer is based on the generation by the PPG instance of a script. It requires the invocation of ancillary programs, such as stride (10), to determine the secondary structures, msms (11) to compute the solvent accessible surface or hbplus (12) to identify hydrogen bonds. The last component manages the post-rendering of the pictures. It organizes the insertion of the title, as well as the format conversion from the default png format generated. It is based on the convert program of the ImageMagick suite (http://www.imagemagick.org). To satisfy the condition that images are rendered in a batch mode (non interactive), we use the virtual display facility provided by Xvfb (http://www.xfree86.org). Coming with such a design, it is possible to trigger the rendering by a mechanism different from CGI, for instance interactively using a command line. Besides, it is conceivable to implement an image production mechanism based on a renderer different from Dino.
Figure 1.
Flowchart of the PPG.
IMPLEMENTATION
Input form
The input is split into four sections consistent with the design of the service (see above).
Data input
PPG supports only the PDB format. It can be specified on the form as a PDB identifier or as one file to upload.
Default representation
This section manages a series of parameters that provide the necessary information to generate a drawing of the complete structure. In order to provide a means of generating simply various representations, the information is split as follows: display of the backbone, the side-chains, the hetero groups (including solvent), the hydrogen bonds and the molecular surface. Each of these subpart is managed independently from the others, and for each, a display mask, a type of representation and a colour pattern can be specified. It is possible to combine them in various ways, and it is possible to invalidate each by setting its display mask to the ‘None’ value.
In addition, this section also groups scene parameters, structure orientation adjustment parameters, picture production parameters and the title. The only scene parameters are the definition of a centre of the view, a focus, a view angle and a stereo mode. The ‘centre’ and the ‘focus’ provide one means of controlling simply the orientation of the structure relative to the user. They define one axis that goes from the ‘centre’ to the user eye via the ‘focus’. The centre is displayed at the centre of the picture, which positions the ‘focus’ at the centre of image, front. Each of the centre or the focus can be specified on the form by coordinates or by naming one residue or atom of the structure. By default, the centre is set to the coordinates of the centre of mass of the structure, and the focus is set along the structure z-axis. The ‘view angle’ corresponds to the classical angle of the field of view of a perspective projection. By default, it is assigned to a value computed from the atomic coordinates of the structure to adjust the ratio between the size of the structure and the size of the picture. Stereo pairs can be produced by splitting the image into two.
Since the adjustment of the structure orientation can be complex, it is possible to specify rotations that will be applied once the transformations based on the ‘centre’ and the ‘focus’ have been performed. To make this process more interactive, a previewer based on Jmol (http://jmol.sourceforge.net) has been installed. Its aim is to determine rotation values that lead to the desired orientation. The user can then simply report them in the main PPG form.
Concerning the picture, it is possible to specify a colour for the background, a size (among three only, since most image manipulation softwares, including presentation or publishing softwares, are able to refine this parameter), and a format for the image (among png, postscript, jpeg, gif and mpeg). The gif and mpeg formats correspond to animated images. Four animation modes are proposed: rock, rotation around the x- or y-axis, Z translation. Finally, it is possible to produce one, two or three images corresponding to perpendicular views of the structure.
Supplementary representations
This section allows the definition of supplementary representations for all or part of the structure. It is possible to define up to four selections to which a specific drawing mode and colouring pattern can be applied. Selections are currently defined using a language similar to that of Dino. More details are given in the PPG help page. If their display value is set to ‘None’, the colouring pattern specified for the selection will apply to the default representation. This provides a means of generating complex colouring schemes at the level of the complete structure.
Advanced parameters
This section groups all the parameters that are not of primary importance to the production of the picture. It is intended to provide some means of customizing the default values, such as the colours. Focus and animation options offer a fine tuning of the structure orientation and image animation.
Some examples on how to use these parameters are given in the gallery at the end of the help page of the service.
Output
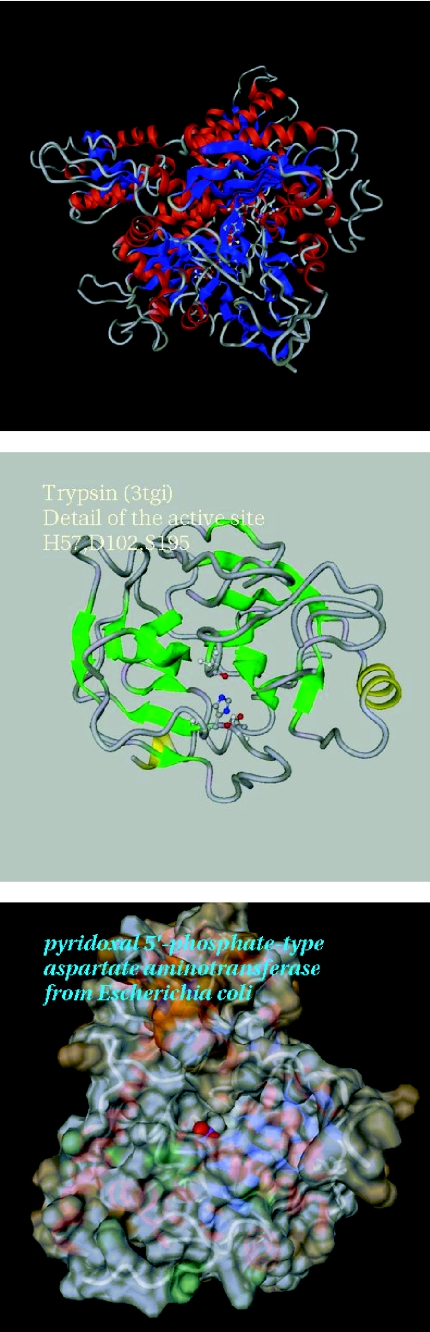
Figure 2 shows some examples of pictures that can be produced by PPG. PPG returns one, two or three images in one of the available formats: postscript, png, jpeg, gif or mpeg. From these formats, users can easily insert the pictures in a document or a presentation. In addition, a copy of the script used to generate the image can also be downloaded. It can provide a starting point for further improvements using Dino.
Figure 2.
Examples of pictures generated by PPG. Top: PDB entry 1ggm, middle: PDB entry 3tgi, bottom: PDB entry 1art. Details on how the pictures were generated can be found in the help page of PPG.
DISCUSSION AND FUTURE WORK
PPG was designed to produce simply complex images, and its default parameter values have been chosen for the production of schematic view of the complete structure. Nevertheless, PPG offers a large range of possibilities. Some of them are illustrated in the help page of the service. Some limitations come from PPG design. First, the PPG offers no interactivity in image production, even if it is possible to compare the images produced under different orientations to refine progressively the view. Presently, the main view control parameter is the specification of the focus, supplemented by explicit rotation values. Even if efficient, small adjustments are often necessary to reach the desired orientation. Work is in under progress to identify some further means of specifying at a high level of molecular description the desired orientation of the structure. In particular, the problem arises for structures having multiple units, when one wishes to illustrate properties at an interface. Also, PPG currently allows the view of one structure at a time. Even if this limitation can be circumvented by merging several files into one, one additional perspective is to make the PPG usable for more than one structure at a time. In particular, it seems desirable to have some means of imaging the comparison of structures. It remains that PPG design makes simple to consider further extensions. Interestingly, Dino can output scripts for the povray ray tracer (http://www.povray.org), which opens the door to very high quality imaging.
Acknowledgments
The authors thank all the persons that have, by their advices and comments, helped designing the protein picture generator. Particular thanks to C. Etchebest. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Walther D. WebMol: a Java-based PDB viewer. Trends Biochem. Sci. 1997:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 2.Richardson D.C., Richardson J.S. MAGE, PROBE, and Kinemages. In: Rossmann M.G., Arnold E., editors. International Tables for Crystallography. Vol. F. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2001. pp. 727–730. [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH: a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 5.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Suppl. 2):W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 7.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 8.Merritt E.A. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 9.Philippsen A. DINO: Visualizing Structural Biology. 2003. Available online at: http://www.dino3d.org. [Google Scholar]
- 10.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 11.Sanner M.F., Olson A.J., Spehner J.-C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 12.McDonald I.K., Thornton J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]