Abstract
Protein phosphorylation plays a fundamental role in most of the cellular regulatory pathways. Experimental identification of protein kinases' (PKs) substrates with their phosphorylation sites is labor-intensive and often limited by the availability and optimization of enzymatic reactions. Recently, large-scale analysis of the phosphoproteome by the mass spectrometry (MS) has become a popular approach. But experimentally, it is still difficult to distinguish the kinase-specific sites on the substrates. In this regard, the in silico prediction of phosphorylation sites with their specific kinases using protein's primary sequences may provide guidelines for further experimental consideration and interpretation of MS phosphoproteomic data. A variety of such tools exists over the Internet and provides the predictions for at most 30 PK subfamilies. We downloaded the verified phosphorylation sites from the public databases and curated the literature extensively for recently found phosphorylation sites. With the hypothesis that PKs in the same subfamily share similar consensus sequences/motifs/functional patterns on substrates, we clustered the 216 unique PKs in 71 PK groups, according to the BLAST results and protein annotations. Then, we applied the group-based phosphorylation scoring (GPS) method on the data set; here, we present a comprehensive PK-specific prediction server GPS, which could predict kinase-specific phosphorylation sites from protein primary sequences for 71 different PK groups. GPS has been implemented in PHP and is available on a www server at http://973-proteinweb.ustc.edu.cn/gps/gps_web/.
INTRODUCTION
Protein phosphorylation is an important and dynamic type of protein modification, orchestrating a variety of cellular signaling processes. About 2% of the human and mouse proteomes encode protein kinases (PKs) with 518 and 540 distinct PKs determined in human (1) and mouse (2), respectively. In vivo or in vitro identification of phosphorylation sites is labor-intensive, time-consuming and often limited by the availability and optimization of enzymatic reactions. Recently, several large-scale phosphoproteomic data using the mass spectrometry (MS) approach have been published for yeast (3), mouse (4) and human (5). But in these cases, it is still difficult to distinguish the kinase-specific sites on the substrates. The in silico prediction of phosphorylation sites with their specific PKs plays an important role in this field. Most of the existing systems adopt the putative rule that protein substrates are phosphorylated at the specific sites with flanking consensus sequences/motifs/functional patterns (6). Application of 3D structure conservation/similarity can significantly improve the prediction specificity (7,8), but the 3D structure information of proteins is very limited compared with the huge number of protein primary sequences available in the public databases. Thus, it would be more feasible and convenient to predict the phosphorylation sites with the specific PKs solely from the protein primary sequences.
Several such systems have been implemented over the Internet. For example, DISPHOS distinguishes the potential phosphorylation sites with position-specific amino acid frequencies and disorder information (9). Another system NetPhos outperforms the consensus sequence-based methods by training the artificial neural networks with the known data set (10). However, the above two systems provide little information about the corresponding PKs for the predicted phosphorylation sites. The enhanced version of NetPhos, NetPhosK, incorporates the functionality of providing PKs' information for ∼17 PKs (11). Scansite (12) constructs the profiles of known phosphorylation sites of ∼20 eukaryotic PKs for prediction.
In this work, we present a comprehensive PK-specific prediction server GPS (group-based phosphorylation scoring), which can predict kinase-specific phosphorylation sites in the substrate sequence for 71 PK groups, including many newly considered PKs, such as Aurora-A, Aurora-B and NIMA (NimA-like protein kinases), etc. The detailed algorithm of this system was described previously (13). We evaluate the sensitivity and specificity of different cut-off scores for each PK group by ‘leave-one-out’ validation. The default cut-off scores are chosen for the balanced pair of sensitivity and specificity. We also use the rat Spinophilin (O35274) as an example to illustrate the usage of the GPS server. Compared with the two separate in vivo or in vitro experiments (14,15) and the two in silico phosphorylation sites prediction tools ScanSite 2.0 and NetPhosK 1.0, the GPS server provides satisfying prediction performance. Thus, we propose that GPS server will be more useful and helpful in further research in the field of protein phosphorylation.
IMPLEMENTATION
First of all, we obtained the data set of phosphorylation sites with their PKs from Phospho.ELM (16), which also included the data of PhosphoBase (17). After removing the phosphorylation sites with ambiguous information of PKs, we were left with 1404 instances. We also browsed the recent publications and obtained ∼660 instances. We retrieved the sequences for the PKs of the data sets from Swiss-Prot and performed homology search in human proteome for each PK. Next, we checked the BLAST results and protein annotations manually to validate the PK subfamily information. After clustering some homology PKs with too few known phosphorylation sites into groups, we obtained 71 PK groups with 216 unique PKs.
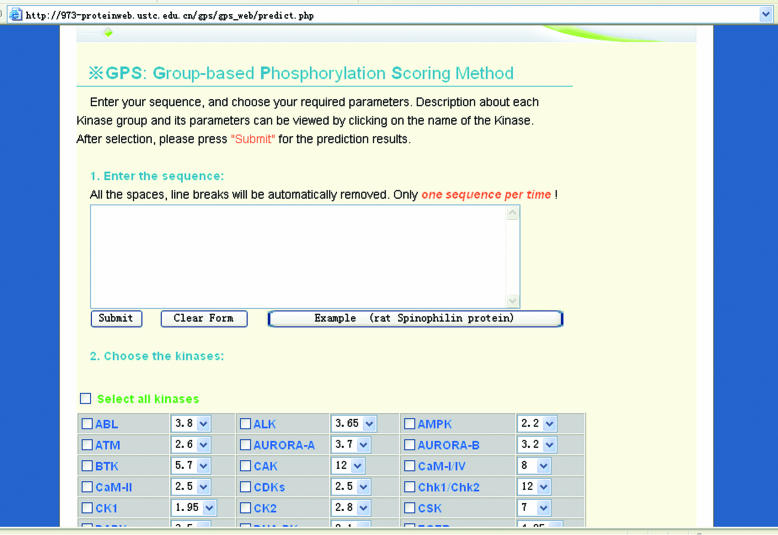
The GPS www server is implemented in PHP + MySQL and the prediction page is shown in Figure 1. The detailed information for each PK group can be visited by clicking on the PK group's name. Several pairs of sensitivity and specificity at different cut-off values for each PK group by the ‘leave-one-out’ validation are also listed for each PK group. Users can choose their required cut-off or select zero for the full list of the scores of the S/T or Y sites.
Figure 1.
The prediction page of GPS www server. The detailed information for each PK can be viewed by clicking on the PK names.
USAGE
Here, we present the rat Spinophilin protein (Swiss-Prot accession no: O35274) as an example to demonstrate the simplicity and precision in the usage of GPS web server. Rat Spinophilin (also called neural tissue-specific F-actin binding protein II or Neurabin-II) is an 817 amino acid, actin- and protein phosphotase-1 (PP1)-binding protein that is ubiquitously expressed but enriched in dendritic spines of rat brain and adherents junction of rat liver (14,15,18–22). Spinophilin binds to and bundles F-actin in vivo, involving in the regulation of dendritic spine morphogenesis (14,19). Spinophilin also modulates excitatory synaptic transmission by binding PP1, redirecting it to postsynaptic densities, and regulating its dephosphorylation activity toward glutamate receptors (18,20,21). Moreover, Spinophilin knock-out mice show impaired synaptic transmission and long-term depression. In addition, young Spinophilin-deficient mice exhibit a significant increase in spine density and enhanced filopodial formation (21).
The phosphorylated Spinophilin plays important roles in spine morphology. The N-terminal 221 residues of Spinophilin (actin-binding domain) can be phosphorylated by at least four PKs, including PKA (14), Ca2+/calmodulin-dependent PK II (CaMKII/CaM-II) (22), cyclin-dependent PK5 (Cdk5) and ERK2 (MAPK1) (15). Phosphorylation of Spinophilin by PKA and CaM-II disrupts its association with the F-actin (14,22). It is proposed that ERK2 phosphorylates the actin-binding domain of Spinophilin to reduce its interaction with actin filaments (15). The phosphorylation sites on Spinophilin were experimentally identified by tryptic phosphopeptide mapping, site-directed mutagenesis, microsequencing analysis and phosphospecific antibodies (14,15,22). We scanned our training data set and found that the CaM-II sites have been used in the current GPS server. The sites for the other three PKs are not included in our training data set. Therefore, we choose to predict the phosphorylation sites on Spinophilin for kinases PKA, ERK2 and Cdk5. The GPS server clusters all CDK PKs, except p34Cdc2 (Cdk1) as the PK group ‘CDKs’, and all MAP kinases as the PK group ‘MAPK’.
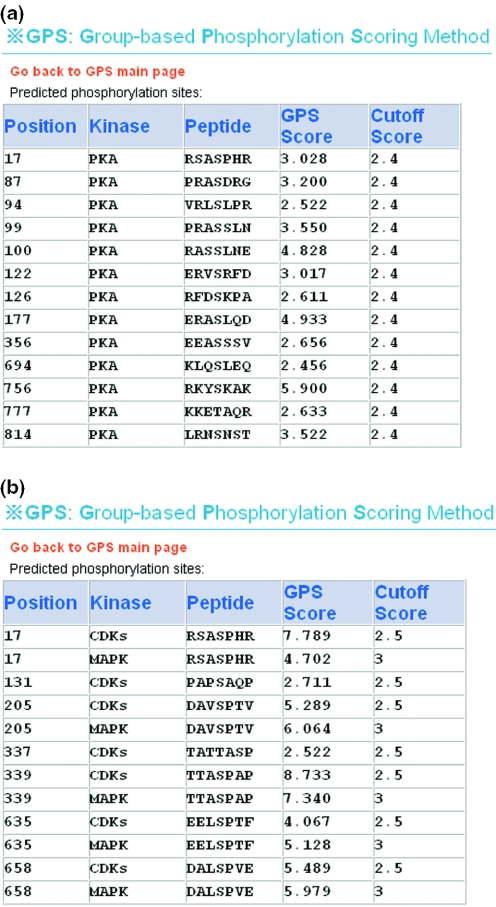
We obtained the primary sequence of rat Spinophilin (O35274) from the Swiss-Prot database and pasted it into the ‘Prediction’ section of the GPS server (Figure 1). The default parameters were chosen. The prediction result of PKA (default cut-off value: 2.4; sensitivity: 88.9%; and specificity: 90.6%) is shown in Figure 2a, and the prediction results of CDKs (default cut-off value: 2.5; sensitivity: 94.4%; and specificity: 91.7%) and MAPK (default cut-off value: 2.5; sensitivity: 83.0%; and specificity: 91.9%) are shown in Figure 2b. For comparison, the prediction results from ScanSite 2.0 and NetPhosK 1.0 are also listed in Table 1. Both ScanSite 2.0 and NetPhosK 1.0 do not provide prediction for MAPK1, so we choose p38MAPK (MAPK5) since the substrate specificity is very similar among the MAPK family. We followed the low stringency for ScanSite 2.0 and default threshold 0.5 for NetPhosK 1.0. The total information on phosphorylation sites of Spinophilin is listed in Table 1.
Figure 2.
Prediction results of GPS server for the rat Spinophilin (O35274). (a) The prediction results of kinase PKA for Spinophilin. There are 13 predicted hits (S17, S87, S94, S99, S100, S122, S126, S177, S356, S694, S756, T777 and S814). (b) The prediction results of kinases CDKs and MAPK for Spinophilin. There are seven predicted hits (S17, S131, S205, T337, S339, S635 and S658) for CDKs and five predicted hits (S17, S205, S339, S635 and S658) for MAPK.
Table 1.
The experimental verified and predicted phosphorylation sites of the rat Spinophilin (O35274)
| Spinophilin (O35274) | PMID | Phosphorylation sites | ||
|---|---|---|---|---|
| PKA | Cdk5/CDKs | p38MAPK/MAPK | ||
| See Hsieh-Wilson et al. (14) | 12417592 | S94, S100 and S177 | — | — |
| See Futter et al. (15) | 15728359 | — | S17 | S15 and S205 |
| ScanSite 2.0 | 12824383 | S17, S59, S94, S100, S177, S694 and S756 | S17 and S339 | — |
| NetPhosK 1.0 | 15174133 | S59, S87, S99, S126, S694, S756 and S814 | S17 and S635 | S17, S635 |
| GPS server 1.10 | — | S17, S87, S94, S99, S100, S122, S126, S177, S356, S694, S756, T777 and S814 | S17, S131, S205, T337, S339, S635 and S658 | S17, S205, S339, S635 and S658 |
PKA phosphorylates Spinophilin at three major sites, S94, S100 and S177 (14). Both ScanSite and GPS could predict the three sites properly while NetPhosK could not. Since only the N-terminal sequence (1–221 amino acids) was used in the experimental identification, it is possible that PKA may have additional phosphorylation sites in rest of the Spinophilin sequence with unknown functions. For the N-terminal sequence of Spinophilin, ScanSite predicts five sites as positive hits (S17, S59, S94, S100 and S177) while GPS claims eight potential sites (S17, S87, S94, S99, S100, S122, S126 and S177). In the other region, ScanSite and NetPhosK predict two (S694 and S756) and three sites (S694, S756 and S814), respectively, while GPS claims three potential sites (S356, T777 and S814).
Since GPS has no entries for Cdk5 and ERK2 (MAPK1), we choose their PKs group CDKs and MAPK instead. Both ScanSite and NetPhosK have Cdk5 without ERK2. Hence, we choose p38MAPK (MAPK5) as a homolog PK to predict its sites on Spinophilin, based on the hypothesis that PKs in one subfamily exhibit very similar substrate specificity. Cdk5 and ERK2 can phosphorylate Spinophilin at S17, and S15 and S205, respectively. ScanSite and NetPhosK could accurately predict S17 properly, while GPS predicted three sites (S17, S131 and S205). For p38MAPK/MAPK, only GPS could correctly predict one site S205. Since we used PK groups of CDKs and MAPK to predict the Spinophilin, the prediction result suggests that Spinophilin may also be phosphorylated by other CDKs and MAPK kinases. The example used here shows that GPS could be a good complementary tool for the experimental work and for other in silico prediction tools, e.g. ScanSite and NetPhosK.
Acknowledgments
The authors thank Drs T.J. Gibson and F. Diella for providing the data set of Phospho.ELM for this work. The authors would also like to thank the editor and the two anonymous referees for their constructive comments on the manuscript. The work was supported by grants from Chinese Natural Science Foundation (39925018 and 30121001), Chinese Academy of Science (KSCX2–2-01), Chinese 973 project (2002CB713700) and American Cancer Society (RPG-99–173-01) to X.Y. X.Y. is a GCC Distinguished Cancer Research Scholar. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health (DK56292).
Conflict of interest statement. None declared.
REFERENCES
- 1.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Caenepeel S., Charydczak G., Sudarsanam S., Hunter T., Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl Acad. Sci. USA. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ficarro S.B., McCleland M.L., Stukenberg P.T., Burke D.J., Ross M.M., Shabanowitz J., Hunt D.F., White F.M. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 4.Ballif B.A., Villen J., Beausoleil S.A., Schwartz D., Gygi S.P. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villen J., Li J., Cohn M.A., Cantley L.C., Gygi S.P. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreegipuu A., Blom N., Brunak S., Jarv J. Statistical analysis of protein kinase specificity determinants. FEBS Lett. 1998;430:45–50. doi: 10.1016/s0014-5793(98)00503-1. [DOI] [PubMed] [Google Scholar]
- 7.Brinkworth R.I., Breinl R.A., Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc. Natl Acad. Sci. USA. 2003;100:74–79. doi: 10.1073/pnas.0134224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., Shakhnovich E.I., Mirny L.A. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc. Natl Acad. Sci. USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iakoucheva L.M., Radivojac P., Brown C.J., O'Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 11.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 12.Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F.F., Xue Y., Chen G.L., Yao X. GPS: a novel group-based phosphorylation predicting and scoring method. Biochem. Biophys. Res. Commun. 2004;325:1443–1448. doi: 10.1016/j.bbrc.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh-Wilson L.C., Benfenati F., Snyder G.L., Allen P.B., Nairn A.C., Greengard P. Phosphorylation of spinophilin modulates its interaction with actin filaments. J. Biol. Chem. 2003;278:1186–1194. doi: 10.1074/jbc.M205754200. [DOI] [PubMed] [Google Scholar]
- 15.Futter M., Uematsu K., Bullock S.A., Kim Y., Hemmings H.C., Jr, Nishi A., Greengard P., Nairn A.C. Phosphorylation of spinophilin by ERK and cyclin-dependent PK 5 (Cdk5) Proc. Natl Acad. Sci. USA. 2005;102:3489–3494. doi: 10.1073/pnas.0409802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diella F., Cameron S., Gemund C., Linding R., Via A., Kuster B., Sicheritz-Ponten T., Blom N., Gibson T.J. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreegipuu A., Blom N., Brunak S. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen P.B., Ouimet C.C., Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl Acad. Sci. USA. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satoh A., Nakanishi H., Obaishi H., Wada M., Takahashi K., Satoh K., Hirao K., Nishioka H., Hata Y., Mizoguchi A., et al. Neurabin-II/spinophilin. An actin filament-binding protein with one pdz domain localized at cadherin-based cell–cell adhesion sites. J. Biol. Chem. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh-Wilson L.C., Allen P.B., Watanabe T., Nairn A.C., Greengard P. Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry. 1999;38:4365–4373. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- 21.Feng J., Yan Z., Ferreira A., Tomizawa K., Liauw J.A., Zhuo M., Allen P.B., Ouimet C.C., Greengard P. Spinophilin regulates the formation and function of dendritic spines. Proc. Natl Acad. Sci. USA. 2000;97:9287–9292. doi: 10.1073/pnas.97.16.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman S.D., Futter M., Snyder G.L., Allen P.B., Nairn A.C., Greengard P., Hsieh-Wilson L.C. Spinophilin is phosphorylated by Ca2+/calmodulin-dependent protein kinase II resulting in regulation of its binding to F-actin. J. Neurochem. 2004;90:317–324. doi: 10.1111/j.1471-4159.2004.02491.x. [DOI] [PubMed] [Google Scholar]