Abstract
OrfPredictor is a web server designed for identifying protein-coding regions in expressed sequence tag (EST)-derived sequences. For query sequences with a hit in BLASTX, the program predicts the coding regions based on the translation reading frames identified in BLASTX alignments, otherwise, it predicts the most probable coding region based on the intrinsic signals of the query sequences. The output is the predicted peptide sequences in the FASTA format, and a definition line that includes the query ID, the translation reading frame and the nucleotide positions where the coding region begins and ends. OrfPredictor facilitates the annotation of EST-derived sequences, particularly, for large-scale EST projects. OrfPredictor is available at https://fungalgenome.concordia.ca/tools/OrfPredictor.html.
INTRODUCTION
The generation of expressed sequence tags (ESTs) was originally proposed as a strategy for cDNA characterization over a decade ago (1). Subsequent improvements in sequencing methods and dramatically reduced unit costs have increased the attractiveness of the EST-based research, such that it is now one of the most widely employed methods used for gene discovery and genome characterization. Consequently, the number of organisms with EST sequences deposited in the GenBank dbEST database is increasing rapidly (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). To maximize the value of these ESTs, NCBI has built UniGenes that incorporated EST data for a number of species (http://www.ncbi.nlm.nih.gov/RefSeq/), and The Institute for Genomics Research (TIGR) has been working on the Gene Indices for more than 70 species (http://www.tigr.org/tdb/tgi/).
Annotating EST and cDNA sequences often involves the identification of potential protein-coding regions. Two tools that have been designed for locating protein-coding regions in cDNA and EST sequences are ORFfinder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi) and ESTScan (http://www.ch.embnet.org/software/ESTScan.html), respectively. ORFfinder processes individual cDNA sequences to identify the coding regions for GenBank submission. It provides all the six frame translations and identifies all the possible coding regions. The ESTScan server is designed for processing a batch of EST sequences for identifying the protein-coding regions with a function for correcting insertions or deletions, but it is only trained for mammals and yeast. We tested ESTScan with our Aspergillus niger EST sequences and found that the results were not satisfactory. For example, using a full-length cDNA sequence encoding glucoamylase, a well-characterized enzyme in A.niger, we found that ESTScan could not identify its correct coding region and had the undesired side effect of inserting nucleotides even when the test sequence was correct.
We have implemented a web server called OrfPredictor for the prediction of protein-coding regions within EST-derived sequences. The algorithm uses the translation reading frames predicted by using BLASTX (2) as a guide for the identification of the coding region in sequences that have a hit (3) and predicts a coding region ab initio for sequences without a hit.
OVERVIEW OF THE ALGORITHM AND IMPLEMENTATION
All eukaryotic mRNAs contain a contiguous sequence of nucleotides coding for protein synthesis. A mature eukaryotic mRNA molecule, starting from the 5′ end, typically consists of a 5′ cap, a 5′-untranslated region (5′-UTR), a protein-coding region [open reading frame (ORF)] and a 3′-UTR followed by a poly(A) tail. The protein-coding region extends from the start codon AUG (ATG in a cDNA) and continues until the reading frame defined by the start codon is terminated by one of three translation stop codons, UGA, UAA or UAG.
Most cDNA libraries are constructed using oligo(dT) primers to direct first-strand synthesis by reverse transcriptase. Essentially, all clones in oligo(dT) primed cDNA libraries will therefore include information for the 3′ end of the processed transcript and a poly(A) region. ESTs are single-pass sequencing reads obtained from either the 5′ or 3′ end of the cDNA insert. The high-quality sequence obtained using systems, such as ABI 3730XL, is typically 700–800 nt per read. Given that the 3′- and 5′-UTRs are typically much shorter than 500 nt, most ESTs and the consensus sequences (contigs) generated by an EST assembler are expected to include some coding sequence useful for predicting gene function.
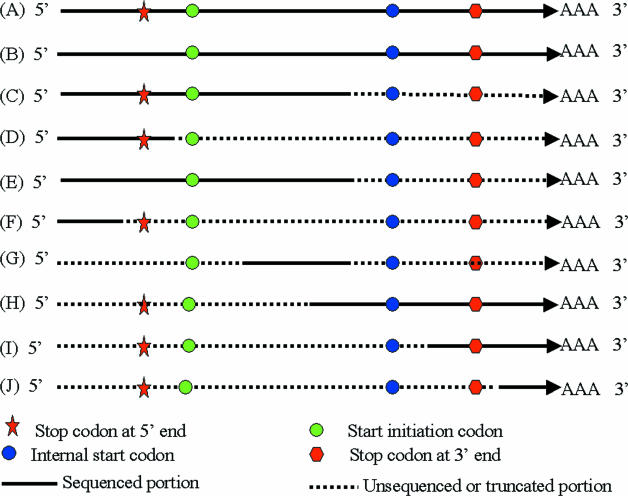
In the annotation and analysis of ESTs, overlapping EST sequences are often assembled into contigs to remove redundancy, reduce the frequency of sequencing errors and extend the length of sequence derived from each mRNA species. Assuming the ESTs are sequenced from the 5′ end, sequence information from contigs and individual ESTs fall into 10 categories (Figure 1) as follows:
A full-length sequence that includes the 5′-UTR with one or more stop codons (5′ stop codon), translation start codon, complete protein-coding region, translation stop codon and the 3′-UTR. The protein-coding ORF may have internal ATG codons and the 3′-UTR may possess multiple stop codons. The 5′-UTR may be truncated.
A full-length sequence as defined for category (A), except that it does not contain any 5′ stop codons.
A partial sequence that has a portion of the 5′-UTR, one or more 5′ stop codons, the start codon and a portion of the coding region.
A sequence that contains only 5′-UTR sequence and there is a 5′ stop codon.
A sequence that contains a 5′-UTR sequence, the start codon and a portion of the protein-coding ORF.
A sequence that contains only 5′-UTR sequence without a 5′ stop codon.
A sequence that contains a portion of the protein-coding region, but does not contain the start codon or the stop codon.
A sequence with the potential protein-coding region truncated at its 5′ end, one or more ATG codons in the truncated ORF, the stop codon and a 3′-UTR.
A sequence that contains a portion of the potential protein-coding region and the 3′-UTR sequence.
A sequence that contains a portion of the 3′-UTR and a poly(A) sequence at its 3′ end.
Figure 1.
Categories of information derived from the EST sequences. (A) A typical full-length cDNA sequence including one or more stop codons in the 5′-UTR, a start codon and a stop codon. The coding region may contain multiple ATG codons encoding methionine and the 3′-UTR may harbor additional stop codons. (B) A full-length cDNA without a stop codon in the 5′-UTR. (C) A sequence containing a 5′-UTR with a stop codon and a portion of the coding region. (D) A sequence containing a 5′-UTR with a stop codon. (E) A sequence containing a 5′-UTR without a 5′ stop codon, and a portion of the coding region. (F) A sequence containing a portion of 5′-UTR without a 5′ stop codon. (G) A sequence containing the internal portion of a coding region with or without internal ATG codons. (H) A sequence containing a portion of the coding region with an internal ATG codon, a 3′ stop codon and 3′-UTR. (I) A sequence containing a portion of the coding region with no internal ATG codons, a 3′ stop codon and a 3′-UTR. (J) A sequence containing a 3′-UTR without a 3′ stop codon. Red star: stop codon at 5′ end; green circle: start codon; blue circle: internal ATG codon; red hexagon: stop codon; solid line: sequenced portion of the full-length cDNA; and dashed line: unsequenced or truncated portion of the full-length cDNA.
For sequences generated by sequencing cDNA inserts from their 3′ ends, the categories of their reverse complementary sequences only include (A), (B), (H), (I) and (J).
Algorithm and implementation
Most ESTs encompass only a portion of the mRNA sequence. Therefore, it is more challenging to predict the coding region within an EST than it is to predict the coding region of a fully sequenced cDNA. Distinguishing the translation start codon from other ATG codons remains a difficult task. Identifying start codons is further complicated because there is not a universal consensus sequence surrounding eukaryotic start codons, although the conserved consensus sequence, GCCRCCaugG (R: purine; aug: start codon) is present in mammals (4). However, BLASTX using a nucleotide query against a protein database is able to reliably identify protein-coding regions within a DNA sequence if sufficient similarity exists between the translated query and an entry in the database (3). Sequencing errors may disrupt the conceptual translation of ORFs, BLASTX could also detect frame shifts if there are insertions/deletions in the coding regions of the query sequences. When significant BLASTX alignments can be generated our algorithm uses them as a guide to identify the translation reading frames and coding regions. For EST-derived sequences without a database match, their frames and coding sequences are predicted based on the presence and the location of intrinsic signals in a sequence that include start codons, 5′ or/and 3′ stop codons and stretches of poly(A) (Figure 1).
Our algorithm uses the following rules to locate protein-coding regions and predict the translation reading frame. For cases where BLASTX identified a significant database match (E-value lower than a user chosen threshold), the frame assignment in the BLASTX output will be used and Rules 1–9 are applied. If there is a conflict, Rules 1 and 2 will override the other rules. For sequences that do not produce a significant BLASTX alignment Rules 3–10 are used to identify the potential coding regions.
Rule 1: The predicted coding region must contain at least a portion of the translated query aligned by using BLASTX.
Rule 2: If there is a frame shift, the first frame assignment in the BLASTX alignment is used.
Rule 3: When there are no internal stop codons within a potential protein-coding region that is flanked by translation start and stop codons, the predicted coding region extends from the start codon to the stop codon (Figure 1A).
Rule 4: A sequence that contains a poly(A) signature but does not contain a stop codon does not include any portion of the coding region (Figure 1J).
Rule 5: If there are one or more ATG codons in a sequence and they are all downstream from one or more stop codons, the first ATG following the last 5′ stop codon is selected as the start codon (Figure 1A and C).
Rule 6: To be considered a potential coding region, an ORF that is flanked by a 5′ stop codon and a 3′ stop codon must be at least 90 nt (code for a protein that has at least 30 amino acids).
Rule 7: If a sequence includes a poly(A) signature preceded by one or more 3′ stop codons, but does not include a 5′ stop codon, the sequence upstream of the stop codons is considered the coding region (Figure 1B, H and I).
Rule 8: If a sequence lacks a poly(A) signature and encodes an ORF without any stop codons, it is assumed that the entire sequence is the coding region (Figure 1E, F and G). Although in rare cases (such as in Figure 1F), the 5′-UTR will be considered as a coding sequence.
Rule 9: For cases like that presented in Figure 1D, it is impossible to know if the stop codon is a 5′ stop codon or a 3′ stop codon, if it lacks a poly(A) signature. However, because cDNA clones are more likely to be truncated at their 5′ end, the program assumes in this case that the sequence upstream of the stop codon is the coding sequence.
Rule 10: The longest stretch of ORF present in the six possible reading frames is selected as the coding region.
Input
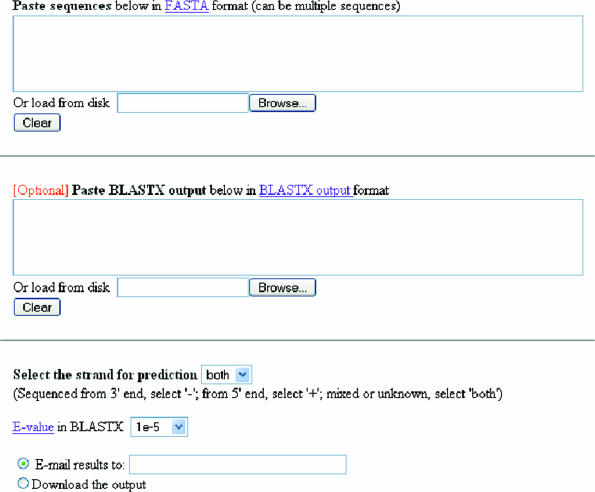
The server provides a user interface for copy and paste, or for loading the users' sequences and BLASTX outputs. It also allows for inputting other parameters (Figure 2). These various inputs are summarized as follows:
A sequence file in the FASTA format. The poly(A) or poly(T) signatures that identify mRNA poly(A) tails should be retained, as they are used to determine the strand to be used for coding region and reading frame prediction.
BLASTX output for all the EST-derived query sequences. Although it is optional, the user is encouraged to provide a pre-run BLASTX output. The user can choose a cut-off E-value when setting up the BLASTX run. If a BLAST output file is provided by a user, the frame used in BLASTX for alignments will be used for the prediction of the protein-coding region. For query sequences without a BLASTX hit or for which the BLASTX output is not provided, predictions will be performed based on the intrinsic signals of the query sequences using the rules described above. Users can also use our TargetIdentifier server (https://fungalgenome.concordia.ca/tools/TargetIdentifier.html) to obtain BLASTX outputs for their query sequences. TargetIdentifier server uses the UniProt/Swiss-Prot protein database.
E-value: The user can also set a threshold E-value for their BLASTX file. If the E-value in the BLASTX file is larger than the user selected threshold, the query sequence will be taken as ‘no hit’. The default threshold is 1 × 10−5.
Strand: The user can choose which strand will be used for prediction. If the sequences were obtained by sequencing cDNAs from the 5′ ends, the ‘+’ strand should be chosen. If the sequences were obtained by sequencing cDNAs from their 3′ ends, the ‘−’ strand should be chosen. If the file contains sequences obtained by sequencing from both ends, both strands should be used for prediction. In this case, the default setting, ‘both’, should be used.
Options for user to select how to obtain the output. Users can select download or use email for receiving their results.
Figure 2.
The OrfPredictor server interface for loading data and choosing other parameters.
Output
Two files are generated. One file is in the FASTA format. It contains the following information for each input sequence. An identifier for the sequence, the reading frame for the predicted coding region, the location of the beginning and end of the predicted coding region, a flag shown as ‘FS’ that locates any translation frame shifts detected in the BLASTX alignment and the predicted protein sequence. The other file contains the query identifiers for those sequences that do not have predicted protein-coding regions (Figure 1J).
EVALUATIONS OF ACCURACY
We evaluated the accuracy of OrfPredictor using 2127 Arabidopsis cDNA sequences that have annotated protein sequences in GenBank, and 4289 A.niger and 3065 Phanerochaete chrysosporium sequences assembled from ESTs with Phrap (http://www.phrap.org/phredphrap/phrap.html). We first compared the predicted Arabidopsis protein sequences obtained when BLASTX alignments were used as a guide with the annotated protein sequences in GenBank, and confirmed that our program was able to predict the protein-coding regions with 100% accuracy. Then, we compared the ab initio predicted protein sequences with the results obtained by using BLASTX. For the 2127 Arabidopsis cDNA sequences, only one sequence was predicted incorrectly. We then examined the prediction accuracy using the A.niger and P.chrysosporium sequences, which had a BLASTX hit in the NCBI nr database with an E-value ≤1 × 105. The ab initio predicted frames were then compared with the frames identified by using BLASTX. We found that the reading frame predicted ab initio was identical with the frames predicted by using BLASTX for 3943 (91.9%) of the A.niger sequences and 2867 (93.5%) of the P.chrysosporium sequences. The sequences used for the accuracy evaluation can be downloaded from the following website https://fungalgenome.concordia.ca/tools/supplement/.
SUMMARY
We implemented a web server, OrfPredictor, for predicting protein-coding regions in EST-derived sequences. OrfPredictor uses the reading frame predicted by using BLASTX when a significant alignment is produced, whereas for sequences that do not return a significant BLASTX alignment protein-coding regions are predicted ab initio. The predicted protein sequences can then be used as the input for additional annotation tools, such as InterProScan (5), for identifying protein families, domains and functional sites, the Conserved Domain Search service (6) for the detection of structural and functional domains, and SignalP (7) for locating potential signal peptides.
Acknowledgments
We thank Jian Sun for assisting with the EST assembly and Wei Ding for assisting with the development of the server interface. This project was supported by Genome Quebec and Genome Canada. Funding to pay the Open Access publication charges for this article was provided by Genome Quebec and Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Adams M.D., Kelley J.M., Gocayne J.D., Dubnick M., Polymeropoulos M.H., Xiao H., Merril C.R., Wu A., Olde B., Moreno R.F., et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Gish W., State D.J. Identification of protein coding regions by database similarity search. Nature Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 4.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. reviews 0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zdobnov E.M., Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A., Bryant S.H. CD-search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]