Abstract
The AthaMap database generates a map of cis-regulatory elements for the Arabidopsis thaliana genome. AthaMap contains more than 7.4 × 106 putative binding sites for 36 transcription factors (TFs) from 16 different TF families. A newly implemented functionality allows the display of subsets of higher conserved transcription factor binding sites (TFBSs). Furthermore, a web tool was developed that permits a user-defined search for co-localizing cis-regulatory elements. The user can specify individually the level of conservation for each TFBS and a spacer range between them. This web tool was employed for the identification of co-localizing sites of known interacting TFs and TFs containing two DNA-binding domains. More than 1.8 × 105 combinatorial elements were annotated in the AthaMap database. These elements can also be used to identify more complex co-localizing elements consisting of up to four TFBSs. The AthaMap database and the connected web tools are a valuable resource for the analysis and the prediction of gene expression regulation at http://www.athamap.de.
INTRODUCTION
The regulation of gene expression is mainly conferred by transcription factors (TFs) that bind to cis-regulatory sequences. These sequences can be used to generate hypothesis about TF that may be involved in the regulation of nearby genes (1,2). In Arabidopsis thaliana, more than 1500 TFs corresponding to ∼5% of the total genes have been identified (3). The largest families are MYB and MYB-related (190 members), AP2/EREBP (144), bHLH (139), NAC (109), C2H2(Zn) (105), HD (89), MADS (82), bZIP (81) and WRKY (72).
Since the complete sequence of the A.thaliana genome has been published (4), it was desirable to have a map of transcription factor binding sites (TFBSs) for the whole genome. The non-restrictive nature of such a map permits the identification of regulatory sequences within transcribed and coding regions as well. To accomplish such a map, the pattern search program Patser (5) and publicly available alignment matrices were used to generate the AthaMap database, the first TFBS map for the whole A.thaliana genome (6). The second release of the AthaMap database presented here has increased the data content from ∼2.4 × 106 to >7.4 × 106 putative sites. Specific care has been taken in the annotation of CAT- and TATA-boxes, which were predicted using alignment matrices from the PlantProm database (7) together with the positional information relative to transcription start sites (TSSs) or translation start sites. Because each TFBS is associated with a particular score that represents the similarity of the site to the underlying alignment matrix, a new functionality was implemented that allows the identification of highly conserved binding sites.
It is well known that the composition of binding sites in the regulatory region of a gene confers its specific expression profile (8). For example, two G-box like sequences constitute the as-1 element that is bound by bZIP TFs (9). Another example is the ocs element that occurs in certain glutathione S-transferase genes of Arabidopsis, which harbour a bZIP and DOF factor binding site in close vicinity (10–12). A wide variety of expression specificity is associated with the co-localization of MYB- and MYC-binding sites (13–16). Other examples are MADS/MADS TFBSs and those TFs that harbour two DNA-binding domains, such as AP2 (17,18).
For the identification of such co-localizing elements, a new web tool was implemented that permits a user-defined identification of pairs of TFBSs in the genome of Arabidopsis by providing distance and quality parameters. This web tool was used to identify the co-localizing sites for known interacting factors. Such combinatorial elements were annotated to the AthaMap database and can also be used for the identification of more complex elements consisting of, for example, two combinatorial elements harbouring four TFBSs.
INCREASE IN AthaMap DATA CONTENT AND FUNCTIONALITY
As summarized in Table 1, the genomic positions of more than 7.4 × 106 putative TFBSs were determined in the A.thaliana genome. These positions were identified with 42 alignment matrices for 36 TFs. For the factors bZIP910, bZIP911, PIF3, ABI4, RAV1 and MYB.PH3, two different alignment matrices were employed and they are identified by numbers in brackets behind the factor name (Table 1). The binding sites were taken directly from the published literature, which is regularly screened in the process of updating the TRANSFAC® database with plant transcription factor data (2).
Table 1.
The number of putative binding sites detected with alignment matrices for TFs from different factor families in the A.thaliana genome and annotated in the AthaMap database
| Factor | Family | Species | Number of sites | Reference for alignment matrix |
|---|---|---|---|---|
| ABF1 | bZIP | A.thaliana | 2419 | (25) |
| bZIP910[1] | bZIP | Antirrhinum majus | 345 | (26) |
| bZIP910[2] | bZIP | A.majus | 470 | (26) |
| bZIP911[1] | bZIP | A.majus | 123 | (26) |
| bZIP911[2] | bZIP | A.majus | 145 | (26) |
| TGA1 | bZIP | A.thaliana | 53 494 | (27) |
| TGA1a | bZIP | Nicotiana tabacum | 142 072 | (28) |
| O2 | bZIP | Zea mays | 173 685 | (28) |
| PIF3[1] | bHLH | A.thaliana | 1154 | (19) |
| PIF3[2] | bHLH | A.thaliana | 951 | (19) |
| DOF2 | DOF | Z.mays | 1 840 355 | (29) |
| AG | MADS | A.thaliana | 46 240 | (30) |
| AGL3 | MADS | A.thaliana | 73 298 | (31) |
| AGL15 | MADS | A.thaliana | 262 900 | (32) |
| ABI4[1] | AP2/EREBP | Z.mays | 12 830 | (33) |
| ABI4[2] | AP2/EREBP | Z.mays | 11 955 | (33) |
| ANT | AP2/EREBP | A.thaliana | 294 | (34) |
| RAV1[1] | AP2/EREBP | A.thaliana | 310 764 | (18) |
| RAV1[2] | AP2/EREBP | A.thaliana | 229 983 | (18) |
| TEIL | AP2/EREBP | N.tabacum | 602 300 | (35) |
| AtMYB15 | MYB | A.thaliana | 209 | (36) |
| AtMYB77 | MYB | A.thaliana | 17 836 | (36) |
| AtMYB84 | MYB | A.thaliana | 231 | (36) |
| CDC5 | MYB | A.thaliana | 11 574 | (37) |
| GAMYB | MYB | Hordeum vulgare | 315 722 | (38) |
| MYB.PH3[1] | MYB | Petunia hybrida | 8529 | (39) |
| MYB.PH3[1] | MYB | P.hybrida | 7638 | (39) |
| P | MYB | Z.mays | 210 035 | (40) |
| GT1 | Trihelix | Diverse species | 1 439 744 | (41) |
| PCF2 | TCP | Oryza sativa | 37 373 | (42) |
| PCF5 | TCP | O.sativa | 14 090 | (42) |
| HVH21 | HD-Kn | H.vulgare | 526 877 | (43) |
| ALFIN1 | HD-PHD | Medicago sativa | 546 159 | (44) |
| ATHB1 | HD-ZIP | A.thaliana | 66 460 | (45) |
| ATHB5 | HD-ZIP | A.thaliana | 7115 | (46) |
| ATHB9 | HD-ZIP | A.thaliana | 303 | (47) |
| HAHB4 | HD-ZIP | Helianthus annus | 90 825 | (48) |
| AGP1 | GATA | N.tabacum | 108 199 | (49) |
| ZAP1 | WRKY | A.thaliana | 4302 | (50) |
| ID1 | C2H2 (Zn) | Z.mays | 156 641 | (51) |
| TBP | Diverse species | 16 277 | (7) | |
| CBF | Diverse species | 62 033 | (7) | |
| Total number of sites | 7 413 949 | |||
The screens were performed on the most recent version of the A.thaliana genome sequence (TIGR release 5.0, January 21, 2004). The pattern search program Patser (5) was used for the identification of binding sites as described previously (6). The following command line was used to run Patser: ‘patser-v3d -A a:t 0.320 c:g 0.180 -m matrixfile -f sequencefile -c -li -d2’. For all screens, the default threshold calculated by Patser from the adjusted information content of the matrix was employed. This criterion was chosen as an objective cut-off threshold value applicable for all the matrices as it represents a measure of how far the nucleotide frequency distribution in the alignment matrix diverges from the a priori probability for the occurrence of the nucleotides in the genome (5). In the case of CAT- and TATA-boxes (CBF and TBP), only those elements that occur upstream of known TSSs or predicted translation start sites were imported into the AthaMap database. TSSs and translation start sites were annotated to the AthaMap database as provided by the TIGR.
The AthaMap database is based on the in silico determination of binding sites and does not distinguish between experimentally verified and predicted sites. Therefore, it is desirable to discriminate between higher and lower conserved binding sites. A criterion for the conservation of a site is the individual score of a TFBS determined by using Patser (5). In general, only TFBSs with a specific score above a threshold score determined for each matrix were imported into the AthaMap database and are displayed as putative binding sites. A high score close to the possible maximum score represents a highly conserved binding site whereas a low score close to the threshold stands for a less conserved site. Maximum score, threshold score and specific score of a site are identified in a tool tip box in the AthaMap database to evaluate individual TFBSs (6).
To permit the exclusive display of higher conserved TFBSs, a new function was implemented in the AthaMap database that allows the user to restrict the number of sites shown by the quality of their scores. With the new ‘Restriction’ function on the ‘Search’ page of AthaMap, the user is able to restrict the sites displayed to those that are closer to the maximum score. This requires an input value as a percentage, which is then applied to the difference between maximum score and threshold score. For example, if the restrictive value is set to 20% then only sites with a score of at least 6 will be displayed for a matrix with a maximum score of 10 and a threshold score of 5, while normally all sites with a score of at least 5 would be shown. A user-defined increase in the threshold score of TFBSs displayed in the AthaMap database may eliminate putative false positive TFBSs.
A WEB TOOL FOR THE IDENTIFICATION OF CO-LOCALIZING TFBS
Gene expression specificity is often mediated by the interaction between TFs that recognize closely spaced binding sites (8). The importance of combinatorial control for gene expression makes it desirable to identify co-localizing TFBSs in the genome based on user provided parameters.
For this, a new ‘co-localization’ web tool was implemented on the AthaMap website that permits the selection of two TFs and the designation of a specific minimum and maximum spacer of up to 50 bp between two TFBSs. The user may select two different TFs or two identical TFs. Furthermore, one can increase the threshold score of the TFBSs individually to obtain combinatorial elements that show a higher conservation of underlying binding sites. The result of the co-localization analysis is shown on the same page and gives the total number of co-localizing TFBSs detected, the chosen parameters for the co-localization analysis and the number of sites used in the analysis. The spacer between two binding sites is defined by the distance between the most 5′ positions of both TFBSs. This permits the identification of overlapping sites that may be relevant for longer matrices with non-overlapping core sequences. To avoid identical hits at the same chromosomal position when using TFs of the same family, it is suggested to select a minimum spacer length that is as long as the matrix of one of the two factors. In addition, even known TSSs can be selected to identify TFBSs in close vicinity to the TSSs.
Owing to the large number of putative binding sites for some factors, the co-localization analysis had to be limited to ∼200 000 TFBSs for each factor to permit a co-localization analysis in a reasonable time. The number of TFBSs of 10 matrices was limited to higher conserved sites by increasing their threshold scores in the co-localization analysis. This applies to the matrices of factors AGL15, ALFIN1, DOF2, GAMYB, HVH21, P, RAV1, TEIL and GT1. The applied parameters can be found on the AthaMap website. With these restrictions, co-localization analyses are generally executed in <1 min.
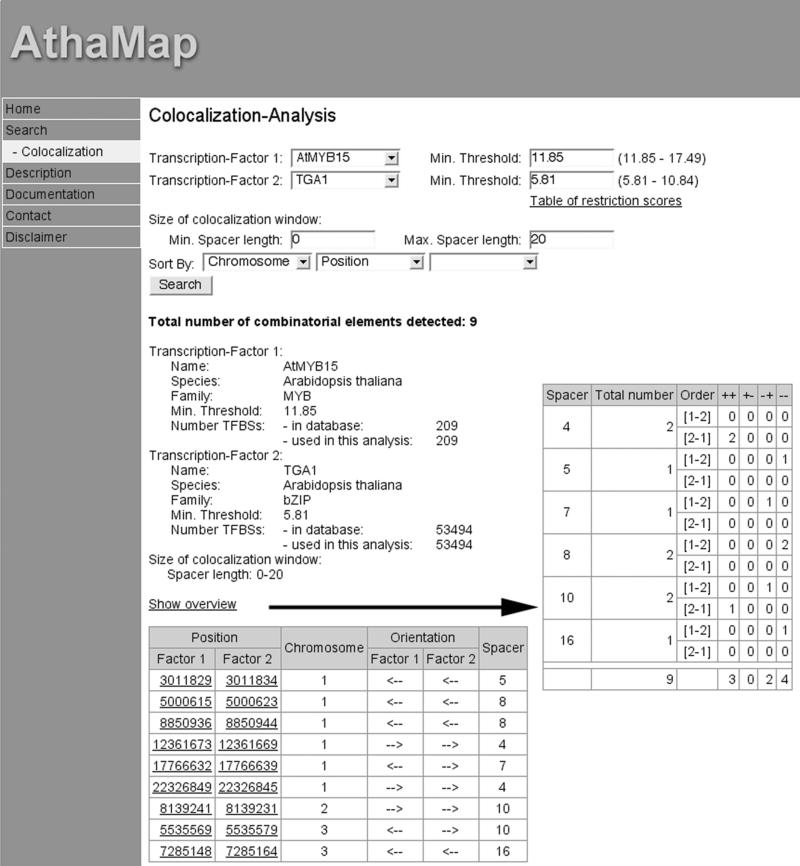
Figure 1 shows a modified screenshot of a result page for a co-localization analysis with AtMYB15 and TGA1, which are both factors from A.thaliana (Table 1). As user-defined parameters, a minimum spacer of 0 nt and a maximum spacer of 20 nt between the binding sites and the default threshold of the alignment matrices (11.85 and 5.81, respectively) were selected. The total number of co-localizing sites detected is nine (Figure 1, combinatorial elements). A result table shows the positions of the co-localizing binding sites, the chromosome and the orientation of the respective site with an arrow. Furthermore, the spacer length of the individual co-localizing element is shown. Each position is linked to an AthaMap sequence window that opens and shows the co-localizing sites highlighted within their genomic context (data not shown).
Figure 1.
Modified screenshots of the web tool for the identification of co-localizing TFBSs in the A.thaliana genome. The results for a co-localization analysis between TFBSs for TGA1 and AtMYB15 using the default parameters are shown. The arrow points to a result window when ‘Show overview’ is selected. See the text for details.
On the result page, when selected, a feature ‘Show overview’ displays a table with a summary of the co-localization analysis (Figure 1, arrow). The inserted table displays the total number of sites that were obtained with all spacer lengths between the selected minimum and the maximum spacer. Here, the user can readily see if a preferred spacer length is detected for binding sites of two TFs. This new tool will be very helpful to identify co-localizing binding sites for TFs that were shown experimentally to interact with each other. Furthermore, genes harbouring a similar architecture of cis-regulatory elements may be identified.
ANNOTATION OF COMBINATORIAL ELEMENTS IN THE AthaMap DATABASE
The well-known examples for combinatorial elements in plants are the as-1 element that is bound by two dimers of bZIP transcription factors, the endosperm or ocs element that is recognized by a member of the bZIP and DOF TF family, and promoters that harbour MYC/MYB or MADS/MADS TF binding sites (9,12,16,17). Based on the approximate spacing between these elements, co-localizing sites were determined with the above described web tool and annotated as bZIP/bZIP, bZIP/DOF, MYC/MYB and MADS/MADS combinatorial elements. A second class of co-localizing TFBSs consists of sites for factors that harbour two DNA-binding domains, such as RAV1 (18). RAV1 belongs to the AP2/EREBP superfamily of TFs that comprises the subfamilies AP2, EREBP and RAV-like (3). RAV1 has two different DNA-binding domains and for each of them the binding specificity was identified (18) and annotated as RAV1[1] and RAV1[2] in the AthaMap database. All the putative RAV combinatorial elements were derived from a co-localization of RAV1[1] and RAV1[2]. Table 2 lists the total number of combinatorial elements identified in the A.thaliana genome and annotated in the AthaMap database. The factors used for the determination of combinatorial sites and the distances between putative binding sites are shown. A total of 183 159 combinatorial elements were annotated in the AthaMap database. These elements are identified in the AthaMap database by the factor family names and are displayed with a double line in the sequence window. For the AP2/EREBP member RAV1 the two different alignment matrices were employed for co-localization analysis. Each combinatorial RAV element consists of two TFBSs that correspond to both matrices.
Table 2.
Combinatorial TFBS in the A.thaliana genome annotated in the AthaMap database
| Combinatorial element | Distance between sites (bp) | Factor binding sites employeda | Number of elements annotated |
|---|---|---|---|
| RAV | 3–20 | RAV1[1]/RAV1[2] | 28 535 |
| bZIP/bZIP | 10–15 | TGA1a/TGA1a | 1037 |
| bZIP/DOF | 3–40 | TGA1a/DOF2 | 84 389 |
| MYC/MYB | 3–40 | TGA1a/all MYB | 38 065 |
| MADS/MADS | 10–100 | All MADS/all MADS | 31 133 |
| Total number of combinatorial elements | 183 159 | ||
aOwing to the palindromic nature of the TGA1a and MADS box matrices, TFBSs frequently occur in sense and antisense at the same position. This leads to redundant combinatorial elements for which only one was annotated in the database and is displayed.
MYC (bHLH) TFs apparently recognize binding sites that are identical or are very closely related to bZIP-binding sites (19–21). Hence, annotated bZIP sites were employed for the identification of MYC-binding sites in combinatorial elements. The identification of functional MYC/MYB-binding sites by employing bZIP sites can be shown for the gene encoding BANYULS that is induced by the interacting TFs TT8 (MYC) and TT2 (MYB) (16,22,23). When the Arabidopsis genome identification number of the Banyuls gene (AT1G61720.1) is used for a search in the AthaMap database, a putative MYC/MYB combinatorial element is detected upstream of the TATA-box (data not shown). This combinatorial element corresponds to the previously determined MYC and MYB regulatory sites in the Banyuls promoter (24). Table 3 summarizes several known or experimentally predicted combinatorial elements detected in the AthaMap database.
Table 3.
Examples of known and experimentally predicted combinatorial elements identified by co-localization analysis and annotated in the AthaMap database
| Combinatorial element | Gene | AGI | Reference |
|---|---|---|---|
| MYC/MYB | Banyuls | AT1G61720.1 | (24) |
| MYC/MYB | TT3 | AT5G42800.1 | (52,53) |
| MADS/MADS | Apetala3 | AT3G54340.1 | (54) |
| MADS/MADS | Agamous | AT4G18960.1 | (55) |
| bZIP/DOF | GST8 | AT1G78380.1 | (56) |
The element can be displayed when entering the Arabidopsis Genome Identifier (AGI) in the search window of the AthaMap database.
As a further asset of the AthaMap database, these annotated combinatorial elements can be included in the user-defined identification of co-localizing TFBSs as well. Therefore, more complex arrangements of regulatory elements consisting of up to four individual binding sites can be detected.
AVAILABILITY
The AthaMap resources are freely available for non-commercial users at http://www.athamap.de.
Acknowledgments
This work was carried out in the Intergenomics Braunschweig Bioinformatics Competence Center and was supported by the German Ministry of Education and Research (BMBF grant no. 031U110C/031U210C). Funding to pay the Open Access publication charges for this article was provided by BMBF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hehl R., Wingender E. Database-assisted promoter analysis. Trends Plant Sci. 2001;6:251–255. doi: 10.1016/s1360-1385(01)01954-9. [DOI] [PubMed] [Google Scholar]
- 2.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 4.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 5.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 6.Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004;32:D368–D372. doi: 10.1093/nar/gkh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahmuradov I.A., Gammerman A.J., Hancock J.M., Bramley P.M., Solovyev V.V. PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003;31:114–117. doi: 10.1093/nar/gkg041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh K.B. Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol. 1998;118:1111–1120. doi: 10.1104/pp.118.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krawczyk S., Thurow C., Niggeweg R., Gatz C. Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 2002;30:775–781. doi: 10.1093/nar/30.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Chen W., Foley R.C., Buttner M., Singh K.B. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., Chao G., Singh K.B. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen W., Singh K.B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 13.Goff S.A., Cone K.C., Chandler V.L. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- 14.Chung S.K., Parish R.W. Studies on the promoter of the Arabidopsis thaliana cdc2a gene. FEBS Lett. 1995;362:215–219. doi: 10.1016/0014-5793(95)00211-q. [DOI] [PubMed] [Google Scholar]
- 15.Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 17.Davies B., Egea-Cortines M., de Andrade Silva E., Saedler H., Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- 18.Kagaya Y., Ohmiya K., Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Garcia J.F., Huq E., Quail P.H. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 20.Toledo-Ortiz G., Huq E., Quail P.H. The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boter M., Ruiz-Rivero O., Abdeen A., Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004;18:1577–1591. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix–loop–helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi H., Hong J., Ha J., Kang J., Kim S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Garcia J.F., Moyano E., Alcocer M.J., Martin C. Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J. 1998;13:489–505. doi: 10.1046/j.1365-313x.1998.00050.x. [DOI] [PubMed] [Google Scholar]
- 27.Schindler U., Beckmann H., Cashmore A.R. TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell. 1992;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izawa T., Foster R., Chua N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- 29.Yanagisawa S., Schmidt R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313x.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- 30.Huang H., Mizukami Y., Hu Y., Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H., Tudor M., Weiss C.A., Hu Y., Ma H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 1995;28:549–567. doi: 10.1007/BF00020401. [DOI] [PubMed] [Google Scholar]
- 32.Tang W., Perry S.E. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J. Biol. Chem. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- 33.Niu X., Helentjaris T., Bate N.J. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell. 2002;14:2565–2575. doi: 10.1105/tpc.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nole-Wilson S., Krizek B.A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 2000;28:4076–4082. doi: 10.1093/nar/28.21.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosugi S., Ohashi Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28:960–967. doi: 10.1093/nar/28.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero I., Fuertes A., Benito M.J., Malpica J.M., Leyva A., Paz-Ares J. More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- 37.Hirayama T., Shinozaki K. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1996;93:13371–13376. doi: 10.1073/pnas.93.23.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gubler F., Raventos D., Keys M., Watts R., Mundy J., Jacobsen J.V. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- 39.Solano R., Nieto C., Avila J., Canas L., Diaz I., Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grotewold E., Drummond B.J., Bowen B., Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 41.Ouwerkerk P.B., Trimborn T.O., Hilliou F., Memelink J. Nuclear factors GT-1 and 3AF1 interact with multiple sequences within the promoter of the Tdc gene from Madagascar periwinkle: GT-1 is involved in UV light-induced expression. Mol. Gen. Genet. 1999;261:610–622. doi: 10.1007/s004380050003. [DOI] [PubMed] [Google Scholar]
- 42.Kosugi S., Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313x.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 43.Krusell L., Rasmussen I., Gausing K. DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare) FEBS Lett. 1997;408:25–29. doi: 10.1016/s0014-5793(97)00382-7. [DOI] [PubMed] [Google Scholar]
- 44.Bastola D.R., Pethe V.V., Winicov I. Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 1998;38:1123–1135. doi: 10.1023/a:1006081926699. [DOI] [PubMed] [Google Scholar]
- 45.Sessa G., Morelli G., Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johannesson H., Wang Y., Engström P. DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 2001;45:63–73. doi: 10.1023/a:1006423324025. [DOI] [PubMed] [Google Scholar]
- 47.Sessa G., Steindler C., Morelli G., Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 1998;38:609–622. doi: 10.1023/a:1006016319613. [DOI] [PubMed] [Google Scholar]
- 48.Palena C.M., Gonzalez D.H., Chan R.L. A monomer–dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem. J. 1999;341:81–87. [PMC free article] [PubMed] [Google Scholar]
- 49.Sugimoto K., Takeda S., Hirochika H. Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J. 2003;36:550–564. doi: 10.1046/j.1365-313x.2003.01899.x. [DOI] [PubMed] [Google Scholar]
- 50.de Pater S., Greco V., Pham K., Memelink J., Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–4631. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozaki A., Hake S., Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004;32:1710–1720. doi: 10.1093/nar/gkh337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sainz M.B., Grotewold E., Chandler V.L. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell. 1997;9:611–625. doi: 10.1105/tpc.9.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong X., Braun E.L., Grotewold E. Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol. 2001;127:46–57. doi: 10.1104/pp.127.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tilly J.J., Allen D.W., Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- 55.Bomblies K., Dagenais N., Weigel D. Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 1999;216:260–264. doi: 10.1006/dbio.1999.9504. [DOI] [PubMed] [Google Scholar]
- 56.Perl-Treves R., Foley R.C., Chen W., Singh K.B. Early induction of the Arabidopsis GSTF8 promoter by specific strains of the fungal pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2004;17:70–80. doi: 10.1094/MPMI.2004.17.1.70. [DOI] [PubMed] [Google Scholar]