Abstract
Neuronal circuits are composed of synapses that are either chemical, where signals are transmitted via neurotransmitter release and reception, or electrical, where signals pass directly through interneuronal gap junction channels. While the molecular complexity that controls chemical synapse structure and function is well appreciated, the proteins of electrical synapses beyond the gap-junction-forming Connexins are not well defined. Yet, electrical synapses are expected to be molecularly complex beyond the gap junctions. Connexins are integral membrane proteins requiring vesicular transport and membrane insertion/retrieval to achieve function, homeostasis, and plasticity. Additionally, electron microscopy of neuronal gap junctions reveals neighboring electron dense regions termed the electrical synapse density (ESD). To reveal the molecular complexity of the electrical synapse proteome, we used proximity-dependent biotinylation (TurboID) linked to neural Connexins in zebrafish. Proteomic analysis of developing and mature nervous systems identifies hundreds of Connexin-associated proteins, with overlapping and distinct representation during development and adulthood. The identified protein classes span cell adhesion molecules, cytoplasmic scaffolds, vesicular trafficking, and proteins usually associated with the post synaptic density (PSD) of chemical synapses. Using circuits with stereotyped electrical and chemical synapses, we define molecular sub-synaptic compartments of ESD localizing proteins, we find molecular heterogeneity amongst electrical synapse populations, and we examine the synaptic intermingling of electrical and chemical synapse proteins. Taken together, these results reveal a new complexity of electrical synapse molecular diversity and highlight a novel overlap between chemical and electrical synapse proteomes. Moreover, human homologs of the electrical synapse proteins are associated with autism, epilepsy, and other neurological disorders, providing a novel framework towards understanding neuro-atypical states.
Introduction
Two forms of fast synaptic transmission co-exist throughout the nervous system: chemical and electrical. While the proteomic complexity regulating neurotransmitter release and reception at chemical synapses is extensively studied1–5, the molecular composition of electrical synapse structure and function is not well understood. This is despite electrical transmission occurring throughout the animal kingdom6–9, the presence of electrical synapses during development and adulthood10–14, and their associations with human disorders including myopia, autism, seizure, and degeneration after neuronal injury15–19. Electrical synapses are clusters of tens to thousands of gap junction (GJ) channels, referred to as GJ plaques, that allow the passage of current and small molecules directly between neurons20–22. The identification of the GJ-channel forming proteins, Connexins in vertebrates and Innexins in invertebrates23–27, revealed the molecular basis for electrical transmission in synchronizing neuronal activity28, expanding receptive field sizes and dampening non-correlated noise29,30, and establishing specialized feed forward synaptic circuits31,32. In addition , electrical synapses are plastic33–36 and their ability to potentiate and depress dynamically contributes to neural computation37–40. Moreover, mutations in neural Connexins disrupt sensory systems including vision and smell, rhythmic systems including circadian and estrous cycles, and central systems including those controlling learning, memory, and fear responses41–51. Despite the breadth of knowledge indicating that electrical synapses are critical components of neural circuits throughout the brain, the molecular mechanisms by which neural Connexins localize to subcellular compartments with cell-type specificity to regulate development, plasticity, and computation remain poorly understood52. In order to localize to electrical synapses, Connexins are constantly on the move, trafficking on vesicles to sites of synapse formation53, inserting into the membrane and traversing through the GJ plaque54, until ultimately endocytosing and degrading in a process of constant turnover55. Additionally, electron microscopy of electrical synapses reveals electron dense structures within the cytoplasm, adjacent to the GJ channels56,57, termed the “electrical synapse density” (ESD)58,59. While Connexin-associated proteins within these distinct cell biological compartments are expected to regulate the structure and function of electrical transmission, molecular insight into these proteins has been hampered by a lack of methods to specifically isolate and identify the proteins of electrical synapses.
Biotinylation of proteins in proximity to the electrical synapse in vivo
To identify proteins associated with electrical synapses we employed a proximity-biotinylation based approach using the evolved protein TurboID60,61 connected to a neural Connexin (Fig. 1A). As proof of principle, we first engineered a neural Connexin linked to TurboID and tested for functionality using cell culture. Our previous work using zebrafish genetics has identified two neural Connexins (Cx34.1/gjd1a and Cx35.5/gjd2a) and a synapse-associated cytoplasmic scaffold (Zonula Occludens 1b (ZO1b/tjp1b)) that are localized to and required for electrical synapse formation and function in the zebrafish central nervous system62. Cx34.1 and Cx35.5 are homologous to the mammalian synaptic Cx3663 and ZO1b is homologous to mammalian ZO158 – these proteins in zebrafish and mammals are broadly localized to electrical synapses throughout the nervous system59,63–65. TurboID was engineered onto Cx34.1 at a position within the conserved intracellular C-terminal tail that does not disrupt known protein-protein interactions, and that when tagged with GFP allows for membrane localization in cell culture and synaptic localization in vivo66. Engineered Cx34.1-TurboID was introduced into HEK293T cells where it robustly self-biotinylated upon supplementation with exogenous biotin (Fig. S1). To determine if Cx34.1-TurboID biotinylation disrupted interaction with a known partner, mVenus was engineered onto the N-terminus of ZO1b and introduced into HEK293T cells in the absence and presence of Cx34.1-TurboID. First, we found that mVenus-ZO1b was biotinylated in the presence of Cx34.1-TurboID (Fig. S1). Second, the Cx34.1-TurboID interaction with mVenus ZO1b was preserved in the biotinylated state (Fig. S1). We conclude that the engineered chimeric neural Connexin retains aspects of its normal cell biological interactions and can biotinylate known interacting partners.
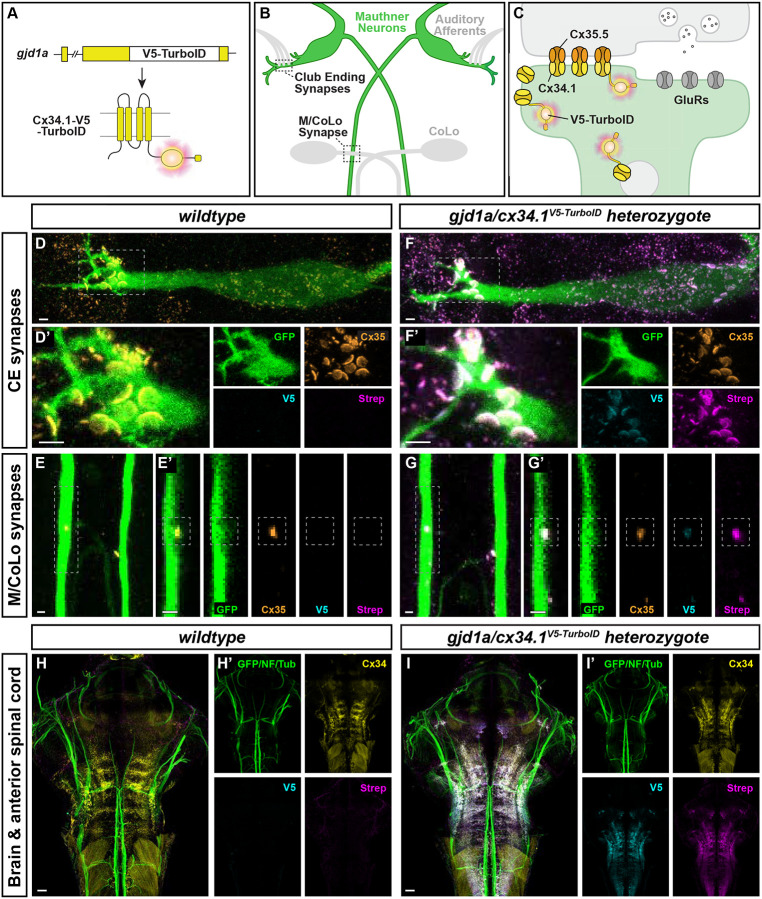
Figure 1. Connexin-TurboID localizes to electrical synapses and biotinylates proteins in vivo.
(A) Schematic diagram of the gjd1a/cx34.1 gene locus modified by in-frame insertion of V5-TurboID. The horizontal black bar represents the DNA strand, a white box represents V5-TurboID, and yellow boxes represent individual gjd1a exons. Below, a cartoon of the Cx34.1-V5-TurboID monomeric protein illustrates the insertion of the V5-TurboID cassette (magenta burst) after the four trans-membrane domains (vertical yellow cylinders) and before the C-terminal PDZ binding motif (PBM, horizontal yellow cylinder). Dark gray lines denote plasma membrane. (B) Simplified diagram illustrating the electrical synapses of interest in the Mauthner (M-) cell circuit. The image represents a dorsal view of the M-cells (green) with anterior on top. Regions in black dashed outline indicate the stereotypical synaptic contacts used for analysis in this study. Presynaptic auditory afferents (grey lines) contact the postsynaptic M-cell lateral dendrite in the hindbrain forming Club Ending (CE) synapses. In the spinal cord, the presynaptic Mauthner axons form en passant electrical synapses with the postsynaptic CoLo interneurons (grey neurons) in each spinal cord hemi-segment (1 of 30 repeating spinal segments are shown). (C) A diagram of a mixed electrical/chemical synapse as found at M-cell CEs. In the electrical component, molecularly asymmetric Connexin hemichannels (Cx35.5 [orange], Cx34.1 [yellow]) directly couple neurons by forming gap junction (GJ) channels. In the chemical component, presynaptic synaptic vesicles release neurotransmitter (grey circles) which align with postsynaptic glutamate receptors (GluRs). Cx34.1-V5-TurboID-containing hemichannels (yellow with magenta burst) are shown trafficking to electrical synapses on vesicles and localizing within the neural GJ plaque. (D-G) Enzymatically active Connexin-V5-TurboID is localized at the stereotype M-cell electrical synapses. Confocal images of M-cell circuitry and electrical synaptic contacts in 6-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae from wildtype (D, E) and gjd1a/cx34.1V5-TurboID heterozygous (F, G) siblings treated with 1mM biotin for 72 hours. Animals are stained with anti-GFP (green), anti-Cx35.5 (orange), anti-V5 (cyan), and Strep-conjugated fluorophore (magenta). Scale bar = 2 μm in all images. Anterior up. Boxed regions denote stereotyped location of electrical synapses and regions are enlarged in neighboring panels. Images of the Mauthner cell body and lateral dendrite in the hindbrain (D, F) are maximum intensity projections of ~27 μm. In D’ and F’, images are maximum-intensity projections of ~15 μm and neighboring panels show the individual channels. Images of the sites of contact of M/CoLo processes in the spinal cord (E, G) are maximum-intensity projections of ~6 μm. In E’ and G’, images are from a single 0.42 μm Z-plane and the white dashed square denotes the location of the M/CoLo site of contact. Neighboring panels show individual channels. (H, I) Enzymatically active Connexin-V5-TurboID is localized specifically in the brain similar to wildtype connexin. Confocal tile scans of zebrafish brain and anterior spinal cord from 6 dpf zf206Et zebrafish larvae from wildtype (H) or gjd1a/cx34.1V5-TurboID heterozygous (I) animals treated with 1mM biotin for 48 hours. Images are maximum intensity projections of ~82 μm. Animals are stained with anti-GFP/anti-NF/anti-tubulin (green), anti-Cx34.1 (yellow), anti-V5 (cyan), and Strep (magenta). Neighboring panels show the individual channels. Scale bars = 20 μm. Anterior up.
To examine TurboID function at electrical synapses in vivo, we generated a transgenic animal expressing the Cx34.1-TurboID chimeric protein from the endogenous Cx34.1-encoding gjd1a gene locus. Cx34.1 is expressed in neurons found throughout the zebrafish central nervous system, including the retina, forebrain, hindbrain, and spinal cord, and is not expressed in non-neural cells63,67. We used CRISPR-mediated homologous recombination68 to introduce TurboID within the Cx34.1 intracellular C-terminal tail in the location that was successful in cell culture (Fig. 1A, S2) and developed a transgenic line of animals. To determine if the chimeric Cx34.1-TurboID protein localizes correctly in transgenics, we examined the electrical synapses of the Mauthner (M-) cell circuit. The M-cell is uniquely visualizable given its large size and distinct bipolar morphology with cell body and dendrites within the hindbrain and an axon extending down the length of the spinal cord (Fig. 1B)63,69. The M-cell lateral dendrites make stereotyped electrical synapses with auditory afferents of the eighth cranial nerve, termed ‘club ending’ (CE) synapses. CE synapses are ‘mixed’ electrical/chemical synapses, containing both neural GJs and glutamatergic chemical transmission (Fig. 1C)70. Additionally, the M-cell axon makes electrical synapses with Commissural Local (CoLo) interneurons found in each spinal-cord segment (M/CoLo synapses)(Fig. 1B). As with endogenous Cx34.163, Cx34.1-TurboID localized to M-cell CE and M/CoLo synapses and colocalized with Cx35.5 at the Mauthner electrical synapses (Fig. 1D–G, Fig. S3). Additionally, Cx34.1-TurboID localized to electrical synapses throughout the larval zebrafish brain (Fig. 1H,I, Fig. S3). To examine if Cx34.1-TurboID functioned in vivo, we supplemented exogenous biotin into the water of larval zebrafish during a developmental window of high synaptogenesis (4–6 days post fertilization, dpf) and found extensive biotinylation of proteins at both CE and M/CoLo synaptic locations (Fig. 1D–G) as well as throughout the developing brain (Fig. 1H,I). Animals heterozygous for the Cx34.1-TurboID insertion showed greater synaptic localization and biotinylation at M-cell synapses than their homozygous counterparts (Fig. S3, Table S1), suggesting that in heterozygotes untagged Cx34.1 facilitates synaptic localization, similar to what was shown for Cx36-GFP in mouse71. Animals that did not have exogenous biotin added showed weaker synaptic biotinylation (Fig. S3). Given these results, subsequent work used Cx34.1-TurboID heterozygote animals supplemented with biotin to increase proximity-dependent biotinylation of Connexin-associated proteins. We conclude that Cx34.1-TurboID localizes to neural gap junctions in vivo and effectively biotinylates proteins found in proximity to the electrical synapse.
Identification of neural Connexin-associated proteins
To identify neural Connexin-associated proteins in vivo we purified biotinylated proteins from Cx34.1-TurboID heterozygotes and wildtype controls, during both development (6 days post fertilization (dpf), whole embryos) and at adulthood (11.5 months, isolated brain), and analyzed them by high-resolution liquid chromatography-mass spectrometry (Fig. 2A). For both the developmental and adult datasets, three biological replicates of Cx34.1-TurboID or wildtype controls were prepared. Sex was not determined for developing animals as zebrafish do not have determinate sex until ~3 weeks of age72. Biological replicates of female and male brains were separately prepared at adulthood. Technical replicates were performed for two of the developmental datasets. In total, spanning both the developing and adult datasets, 1974 proteins were identified, with 272 proteins enriched in the Cx34.1-TurboID samples as compared to the wildtype controls (Fig. 2B, Table S2). Developmental and adult stages had 68 and 250 enriched proteins, respectively, with 53 overlapping proteins between stages (Fig. 2B,C). Of the total enriched proteins, 29 and 146 proteins in the developing and adult datasets, respectively, were significantly enriched based on a strict false discovery rate (Table S2). Additional candidate proteins considered lower-confidence, yet absent from wildtype controls, were consistently present at greater than 7-fold enrichment in all replicates of developing or adult Turbo-ID data (Fig. 2C, Table S2). No differences were observed in enriched proteins within technical replicates of the developing datasets nor between female and male adult brains (Table S2).
Figure 2. Discovery of the neural Connexin-associated proteome by biotin-proximity identification.
(A) Diagram of biotin-proximity labeling scheme. gjd1a/cx34.1V5-TurboID heterozygous larvae (developing) or adults were treated with biotin supplemented water to activate maximal biotinylation of nearby proteins. Whole larvae and brains from adults (dotted lines) were harvested and homogenized under denaturing conditions. gjd1a/cx34.1 is expressed exclusively in the nervous system providing specificity. Biotinylated proteins were captured with magnetic streptavidin beads, digested with trypsin on-bead, and analyzed by mass spectrometry to identify candidates. (B) Proteins enriched in Cx34.1-V5-TurboID samples compared to wildtype controls are depicted and grouped into grey boxes according to gene ontology (GO) analysis: Cell Adhesion, Receptors, Channels, Cell-Cell-Junction Scaffolds, Vesicular Trafficking, Signaling, Cytoskeletal Regulation, Ubiquitination, and Other. Paralogous proteins are denoted as single shapes and noted by a forward slash between name delineations. Colors denote enrichment in the developmental (yellow), adult (blue), or both datasets (green). Hexagons denote proteins associated with epithelial adherens or tight junctions (AJ/TJ), circles denote proteins associated with postsynaptic densities of chemical synapses (PSD), and squares denote the remainder of the isolated candidates. Outlines denote human disorders associated with the protein: solid outlines denote proteins associated with autism spectrum disorder (ASD), dashed outlines denote proteins associated epilepsy (Epil.), and stippled outlines denote proteins associated with Neurodevelopmental Disorders and/or Intellectual Disability (NDD/IDD). Many proteins have associations with multiple of these categories, or other neurological disorders (Table S2). (C) Enrichment of identified proteins (dots) in developing (x-axis) and adult (y-axis) datasets. Colored regions denote proteins enriched in developing (yellow), adult (blue) or both (green) time points. Proteins in white callouts have been previously localized to zebrafish electrical synapses (Cx34.1, Nbea, and ZO1) while the rest have been localized to mammalian electrical synapses or interact with mammalian Cx36. (D) Neural Connexin-associated proteome gene expression across germ layers in a whole-embryo single-cell RNA sequencing dataset (5 and 6 dpf). Clusters (Identity) are organized by germ layer along the x-axis: neurons, ectoderm (non-neural ectoderm, e.g. skin and cranial neural crest), mesoderm (e.g., skeletal muscle and blood), and endoderm (e.g. liver). A selection of Connexin-associated genes is arranged along the y-axis, grouped by GO terms. Dot size represents the percentage of cells in the identity category expressing each gene, while color indicates relative expression levels.
To assess the quality of enrichment in the dataset, we first cross referenced the identified proteins with gene expression captured in our single cell RNA-seq atlas of zebrafish development67,73. We found that enriched candidates were biased towards mRNA expression in neurons, with notable co-expression with endogenous Cx34.1/gjd1a (Fig. 2D, S4). Given the partial genome duplication of teleost lineages74, we used BLAST and the Zebrafish Information Network (ZFIN)75 to identify 216 human proteins with clear 1-to-1 orthology in the enriched protein dataset, with only 4 proteins absent from mammalian lineages (Table S2). Gene Ontology (GO) analysis76,77 of the proteins highlighted significantly enriched terms including postsynapse (p=6.19E-67), neuron-to-neuron synapse (p=1.01E-65), dendrite (p=2.27E-35), and trans-synaptic signaling (p=9.80E-33) (Table S2), suggesting we had enriched for proteins localized at neuronal synapses. In line with this enrichment, GO analysis, cross referenced with resources cataloging human-disease-associated genes78–81, revealed an abundance of proteins associated with autism spectrum disorders (ASD), epilepsy, and neurodevelopmental disorders and/or intellectual disability (Fig. 2B, Table S2). While electrical synapses are abundant throughout the brain, there are currently only ~10 proteins known to localize to vertebrate neural gap junctions52,82. Within the enriched proteins, many of these known proteins that localize to electrical synapses or interact with neural Connexins were identified (Fig. 2C). The two most enriched proteins in the datasets were Cx34.1, onto which we engineered TurboID, and ZO1b, which we have previously shown is localized to the electrical synapse and binds directly to Cx34.1 via a PDZ-PBM mediated interaction58,59,83. In addition, Neurobeachin (Nbea) was enriched in the adult dataset, and our previous work showed it is localized to electrical synapses and required for synaptic localization of both Cx34.1 and ZO1b66,84. Additionally, a number of proteins previously determined to localize to mammalian electrical synapses were enriched in the datasets, including ZO285, Afadin (Afdn)86, and Nectin187(Fig. 2C). Further, the experiment is expected to enrich for proteins within proximity of Cx34.1-TurboID from translation to degradation, therefore the candidates are likely to include proteins that are not exclusively at the synapse. Indeed, also enriched in the data are the trafficking proteins Golgi Associated PDZ And Coiled-Coil Motif Containing (Gopc) and Synergin (Synrg) that were identified to be in proximity with Cx36 when it is overexpressed in HEK293T88. We conclude that the biotin-proximity labeling approach enriched for neuronal and synaptic proteins and successfully identified Connexin-associated proteins in vivo.
Analysis of known functions of the other, enriched, candidate Connexin-associated proteins, or of human and mouse orthologs (Table S2)76, highlighted a number of putative functions that have been hypothesized to regulate electrical synapse assembly and structure52,53,89. These included neural cell adhesion molecules, neural signaling and synaptic receptors, cytoplasmic scaffolds, and cytoskeletal proteins (Fig. 2B). Additionally, Connexins are four-pass transmembrane domain proteins that oligomerize into hemichannels in the ER/Golgi, are trafficked in vesicles to sites of GJ formation, and are removed and degraded by the proteosome20,90,91. The dataset identified ER and Golgi proteins, vesicular trafficking regulators, and ubiquitination pathway proteins (Fig. 2B) that may regulate neural Connexins. Two additional major cell biological categories emerged from the analysis: (1) epithelial adherens and tight junction (AJ/TJ) proteins (Fig. 2B, hexagons) and (2) chemical synapse postsynaptic density (PSD) proteins (Fig. 2B, circles). Recent work has revealed the close association of a subset of AJ/TJ proteins with neural Connexins52,82. Regarding the ‘chemical synapse PSD’ proteins, it is noteworthy that these candidate proteins have not been studied in the context of the electrical synapse, raising the possibility that they may also be ESD proteins. Additionally, zebrafish contain an abundance of ‘mixed’ synapses wherein individual neuronal contacts exhibit simultaneous electrical and chemical transmission92,93. Mixed synapses are also found in mammalian and invertebrate neural circuits56,94, though are not well studied. Taken together, we conclude that the approach identified known neural Connexin-associated proteins and highlights further candidate molecules with a diversity of putative cell biological functions.
Localization of Electrical Synapse Density proteins in vivo
To identify proteins localized to synaptic contacts, the ESD proteins, we investigated the localization of candidate Connexin-associated proteins in vivo. We endogenously tagged candidate proteins from amongst the major identified functional classes and examined their localization in developing zebrafish. Given the general lack of antibodies available for zebrafish proteins, we used CRISPR to introduce V5-tags into proteins at their endogenous genomic loci95 and used immunofluorescence to examine the localization of the tagged proteins in relation to Cx34.1 (Fig. 3A). First, we examined the brain-wide expression patterns of candidate proteins and found a diversity of patterns, including expression throughout the brain, those with regionalized expression, including anterior (forebrain/midbrain) or posterior biases (hindbrain/spinal cord), or patterns with sparse and cell-type specific expression in subsets of neurons (Fig. 3B). In addition, protein localization was found in a diversity of other tissue types, including glia, epithelia, muscle, and vasculature (Fig. S5). Given that the approach produces mosaic embryos66,95, we examined multiple examples of each investigated protein and found consistent patterns of expression and localization (Fig. S5). Throughout the diversity of neural expression classes, regions of overlapping expression with Cx34.1 were identified for a majority of proteins examined (85%, 33/39) (Fig. 3, S5). Cases lacking overlap could be due to an inability to visualize the protein at locations of colocalization, that the proteins do not colocalize at this time of development, or that V5 tagging disrupted endogenous localization. For those with Cx34.1 overlapping localization, some proteins showed colocalization throughout the brain while others displayed only regional colocalization (Fig. 3B), suggesting that electrical synapses at distinct locations have unique proteomic makeups. From these broad categorizations of Connexin-associated proteins it appears that the candidate proteins examined show a high degree of specific neuronal expression, frequently colocalize with Cx34.1, and that distinct populations of electrical synapses may have unique molecular constituencies.
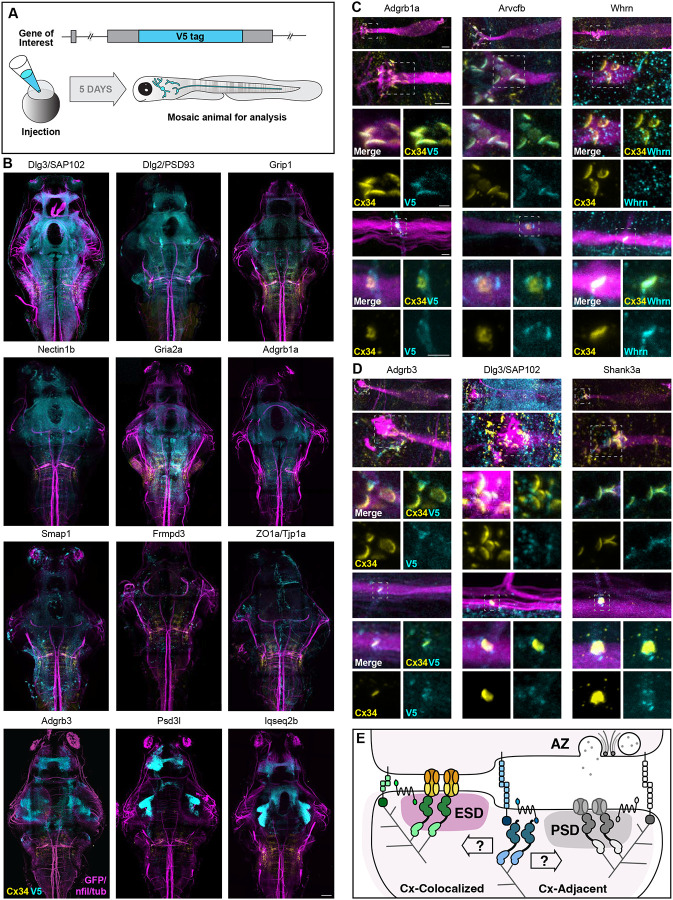
Figure 3. Identification of electrical synapse density proteins.
(A) Diagram of CRISPR-mediated epitope insertion at endogenous genes to generate animals mosaically expressing V5-tagged proteins for analysis in the zebrafish brain. The horizontal black bar represents the DNA strand of the gene of interest, a cyan box represents the insertion of the V5 epitope tag. Animals are injected at the 1-cell stage and expression is visualized five days later. (B) Confocal tile scans of zebrafish brain at 5 dpf Et(T2KHG)zf206 zebrafish larvae expressing V5-tagged proteins, as labeled. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-V5 (cyan). Scale bars = 20 μm. Anterior up. (C) Confocal images of Connexin-colocalized genes in the M-cell circuit at 5 dpf in Et(T2KHG)zf206 zebrafish larvae. Boxed regions denote stereotyped location of electrical synapses and regions are enlarged in neighboring panels. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), antiCx34.1 (yellow), and anti-V5 or anti-Whrn antibody (cyan), as labeled. Top four rows show CE synapses, anterior up. Bottom three rows show M/CoLo synapses, anterior left. Neighboring panels show individual and merged channels, as labeled. Scale bars = 2 μm. (D) Confocal images of Connexin-adjacent genes in the M-cell circuit at 5 dpf in Et(T2KHG)zf206 zebrafish larvae. Boxed regions denote stereotyped location of electrical synapses and regions are enlarged in neighboring panels. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-V5 (cyan). Top four rows show CE synapses, anterior up. Bottom three rows show M/CoLo synapses, anterior left. Neighboring panels show individual and merged channels, as labeled. Scale bars = 2 μm. (E) Model of a mixed electrical/chemical synapse illustrating Connexin-colocalized and Connexin-adjacent proteins. ESD proteins fall into classes such as cell adhesion, signaling, scaffolding, cytoskeletal regulation and colocalize with electrical synapses (green shapes). These proteins classes are analogous to proteins identified by proteomics at chemical synapse presynaptic active zones (AZ) and PSDs (grey shapes). Candidates that are adjacent to electrical synapses (blue shapes) may be exclusive to the chemical synapse, may be localized adjacent to both, and may regulate both electrical and chemical synapses assembly and function, as indicated by the opposing arrows. ESD, electrical synapse density. PSD, post-synaptic density. AZ, pre-synaptic active zone.
To investigate the subcellular localization of identified proteins we turned our attention to the M-cell circuit with its stereotypical cell biology and synapses. The M-cell CE synapses (Fig. 1B) are mixed electrical/glutamatergic chemical synaptic contacts, with EM70,96 and expansion microscopy97 showing that the chemical component of these mixed synapses residing in distinct locations surrounding the synaptic GJ plaques (Fig. S6). At M/CoLo synapses, transmission has a clear electrical component, though electrophysiology was indeterminant in relation to a chemical component at these synapses98. At both CE and M/CoLo synapses, Cx34.1 and AMPA receptors are localized to distinct subsynaptic domains where Connexin adopts a stereotypical ‘C-shaped’ pattern that stretches across the synaptic contact, whereas AMPA receptors are localized at 2–4 distinct puncta surrounding Cx34.1 localization (Fig. S6). Using the unique cell biological accessibility of the M-cell circuit, we investigated the localization of the V5-tagged Connexin-associated proteins expressed in the hindbrain and spinal cord where the circuit resides. Two major classes of specific, synaptic localization were identified: Connexin-colocalizing and Connexin-adjacent.
The Connexin-colocalizing protein class was typified by localization in a stereotypical C-shaped pattern at CE or M/CoLo synaptic contacts and extensively overlapped with Cx34.1 staining (Fig. 3C, S5). In total, 16 proteins were found to be Connexin-colocalizing: Adgrb1a, Afdna, Arvcfa, Arvcfb, Clmpb, Ctnnd2a, Ctnnd2b, Dlg1b, Jam3a, Nectin1b, Plekha5, ZO1a, ZO1b, ZO2a, ZO2b, and Whrnb. For Whrnb we obtained an antibody against the zebrafish protein99. In zebrafish, only ZO1b had previously been co-localized with neural Connexins58,59,83; in mouse, Afdn, Nectin1, ZO1, and ZO2 have been co-localized with Cx3664,85–87,96. The rest of the molecules are novel ESD proteins. It is notable that these proteins are largely, though not exclusively, associated with epithelial AJs/TJs, with little known about their function in the nervous system. The Connexin-colocalizing class represents cell adhesion molecules, cytoskeletal regulators, and cytoplasmic scaffolds (Fig. 2B). We have previously shown that ZO1b localizes to electrical synapses (Fig. S6) where it directly interacts with Cx34.1 and is required for synaptic localization and electrical transmission58,59,83, suggesting that this group of proteins may represent a signaling complex for the assembly, structure, and function of the electrical synapse.
The second synaptic class, Connexin-adjacent, contained eight proteins and included the glutamatergic neurotransmitter receptor subunit Gria2a, the receptor Adgrb3, and the cytoplasmic scaffolds Frmpd3, Grip1, Nbeaa, Sap102, Shank3a, and Syngap1b (Fig. 3D). Note that given our reliance on V5-tagging, and the lack of antibodies available that recognize these zebrafish proteins, the distribution of these proteins relative to one another at individual synapses cannot currently be determined. The Connexin-adjacent class of molecules is dominated by associations with chemical synapse PSD proteins (Fig. 2B). Yet, we have previously found that Nbeaa localizes in a Connexin-adjacent pattern at M-cell electrical synapses (Fig. S6), and despite this ‘next-to’ localization, it is required for robust synaptic Cx34.1 localization and M-cell induced escape response behavior66,84. This suggests the provocative possibility that the Connexin-adjacent class contains at least a subset of proteins with functions in electrical synapse assembly and function.
Finally, we observed that the M-cell electrical synapses displayed molecular heterogeneity. The M-cell circuit receives sensory input from auditory afferents at the CEs and transmits this information to motor circuits in the spinal cord including to the CoLo interneurons. Both Adgrb1a and Arvcfb were localized in a Connexin-colocalizing pattern at CEs in the hindbrain, but at M/CoLo synapses in the spinal cord they were localized in unique synaptic-contact-surrounding patterns (Fig. 3C, S3). This pattern appeared distinct from the Connexin-adjacent class described above. By contrast, Whrnb was found localized in a Connexin-colocalizing pattern at M/CoLo synapses but at CE contacts was localized in a Connexin-adjacent fashion. It is striking that within the M-cell escape circuit, distinct electrical synapses display molecular diversity with subcellular specificity, potentially related to the distinct functions (sensory integration at CEs, motor coordination at M/CoLos) of each synapse population.
Taken together, the synaptic proteins represent at least two distinct synaptic localization patterns, Connexin-colocalized or Connexin-adjacent. Those that are Connexin colocalized represent putative ESD proteins for which genetic, molecular, and cell biological analysis may reveal functions in electrical synapse structure and function (Fig. 3E, green). Those that are Connexin-adjacent represent putative PSD proteins, for which many are expected to have functions at the glutamatergic synapses found at Mauthner cell mixed synapses (Fig. 3E, gray). Yet the Connexin-adjacent class also highlights the emerging possibility of molecular and functional overlap between the ESD and PSD proteins (Fig. 3E, blue).
Discussion
The Connexin-associated proteome presented here reveals hundreds of new molecules we hypothesize can fulfill the varied and rich roles of electrical synapse regulation during assembly and function. These include proteins that fall into classes such as cell adhesion, signaling, scaffolding, cytoskeletal regulation, and vesicular trafficking – all categories analogous to those identified by proteomics at chemical synapse presynaptic active zones (AZ) and PSDs (Fig. 3E)100–105. Within the ~250 proteins identified here we localized sixteen proteins to Connexin-colocalized and eight to Connexin-adjacent locations at M-cell electrical synapses and observed similar distributions at other Cx34.1 puncta found throughout the brain. Localization of proteins also revealed molecular heterogeneity at M-cell synapses, highlighting that not all electrical synapses are made equal, but instead that they can have unique molecular compositions likely to affect neural gap junction structure and transmission. Additionally, the proteomic data highlights proteins that track the expected ‘life cycle’ of Connexins, from vesicular trafficking in the ER/Golgi, to synaptic associations between neurons and with cytoplasmic regulators, to protein degradation after endocytosis20. While only a subset of these molecules will be strictly ‘synaptic’, therefore being defined as ESD proteins, the entirety of the identified proteome may regulate neural Connexin biology. A key frontier that emerges is to define the electrical synapse beyond the Connexins and to understand the molecular framework that builds its structure. Critically, we must convert the gross synaptic localization patterns observed here into the sub synaptic localization at the electrical synapse. EM of electrical synapses highlights the GJ plaques containing hundreds to thousands of channels, and also reveals the directly adjacent puncta adherens, and the cytoskeletal complexity12,56,106. The molecular richness of the electrical synapse proteome is expected to define synaptic domains that regulate the structure, transmission, and plasticity of electrical transmission.
The neural Connexin-associated data here is expected to reveal only a subset of the complete electrical synapse proteome. First, proteins that are not within proximity of the Cx34.1-TurboID chimeric enzyme, or that are in compartments inaccessible to the enzyme, are not biotinylated, and therefore are not captured in the approach. In line with the expectation, N-cadherin (Cdh2) and Beta-catenin (Ctnnb2) were recently localized to M-cell electrical synapses97 but were not identified in our data. Second, more complexity is expected as we look to other electrical synapses. Here we focused on a zebrafish Cx36 homologue in fish63, yet there are five distinct Connexins that make electrical synapses in mammals21,26, and future experiments will reveal the distinct, and overlapping, proteomic compositions of the other neural Connexins. Finally, electrical synapse complexity also appears to be related to age as our data revealed differences in developing and adult animals. Such electrical synapse molecular compositions shifts could affect transmission and neural circuit function, similar to what has been observed at chemical synapses107–109. The data here reveals a molecularly rich proteome for electrical synapses, analogous to what is known for chemical synapses, with more complexity yet to be discovered.
A notable revelation from the Connexin-associated proteomic data was the identification of chemical synapse PSD molecules, including glutamate receptors, scaffolds, cell adhesion molecules, and regulators. These observations force us to consider where the boundaries lie between different synapse types (Fig. 3E). Electrical and excitatory chemical co-transmission is a common synaptic motif that can be found at mixed synapses between two neurons, as found in the M-cell circuit and other locations, or in cases where separate electrical and glutamatergic synapses are made between two presynaptic neurons and a single postsynaptic partner11,92,94. In both cases, postsynaptic electrical and chemical components could co-mingle in close proximity, or in trafficking pathways, and this could explain the observed PSD proteins in the Connexin-TurboID data. Alternatively, so-called PSD proteins may not be as exclusive as the name implies, and perhaps they also function in the regulation of electrical synapses. Notably, this proteomic work identified Neurobeachin (Nbea), which is linked to autism and epilepsy110–112 and is required for the localization of both electrical and chemical synapse proteins66,84,113–116. The notion of synapse coordination marks a frontier where we must define the proteins that belong exclusively to the electrical or chemical synapse and those that ‘moonlight’ and perform essential functions at both. Future imaging with higher spatial resolution (e.g. immuno-EM and expansion microscopy) combined with genetic, biochemical, and functional work, will reveal the mechanistic functions of synapse specific and synapse coordinating proteins.
Broadening our molecular understanding of the electrical synapse creates fundamental new insights into the complexity of neural circuits and new possibilities related to human neurological disease states. In line with this notion, neural Connexin mutants in mouse and zebrafish have revealed complex behavioral defects related to vision, smell, circadian rhythms, estrous cycles, learning and memory, fear responses, and motor coordination41–51,63. Additionally, human mutations affecting Cx36/GJD2 are a leading cause of myopia15 and have been proposed to underlie autism16, while hyperfunction of electrical synapses may lead to seizure17,18. Here, the Connexin-associated proteome identified a myriad of proteins in which the human orthologues are linked to a variety of neurological disorders, with particular enrichment for autism, epilepsy, and neurodevelopmental and intellectual disabilities. Future experiments to reveal the mechanisms by which the Connexin-associated proteins regulate electrical synapse assembly, structure, plasticity, and coordination with chemical synapses, highlights an enticing new direction to understand human neural circuit disorders.
Methods
Cell culture, transfection, and immunoprecipitation
HEK293T/17 verified cells purchased from ATCC (CRL-11268; STR profile, amelogenin: X) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC) with 10 % fetal bovine serum (FBS, Gibco) at 37 °C in a humidified incubator in the presence of 5 % CO2. Low passage cells (less than 20 passages) were used for transfection experiments and were monitored for mycoplasma contamination using the Universal Mycoplasma Detection Kit (ATCC, 30–1012K).
Sequence encoding gjd1a/cx34.1 fused in-frame to the V5-TurboID encoding sequence60,61 was constructed into a pCMV expression vector using Gibson cloning. The V5-TurboID was placed internally (following Asp278 in Cx34.1) to preserve availability of the PDZ binding domain66. The plasmid directing expression of mVenus-Tjp1b/ZO1b-Beta-8XHIS was generated in a previous study83. Low passage HEK293T/17 cells were seeded at a density of 1 × 106 cells/well of a six-well dish 24 hours prior to transfection. Plasmids were co-transfected using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. If needed, empty pCMV plasmid was co-transfected to preserve the total concentration of transfected plasmid DNA, as denoted in the figure legend. Prior to collection 36–48 hours post-transfection, cells were incubated with culture medium supplemented with 0.5mM biotin at 37 °C in a humidified incubator in the presence of 5 % CO2. Cells were rinsed with 1x PBS, collected into a 1.5ml Eppendorf, and pelleted by centrifugation at 5,000 × g for 5 min at 4°C. Pellets were lysed in 0.20 ml solubilization buffer (50 mM Tris [pH7.4], 100 mM NaCl, 5 mM EDTA, 1.5 mM MgCl2, 1% Triton X-100) plus a protease inhibitor cocktail (Pierce) for 1hr with rocking at 4°C and centrifuged at 20,000 × g for 30 min at 4°C. Equal amounts of extract were immunoprecipitated with 2.0 μl rabbit anti-GFP (Abcam, ab290) overnight with rocking at 4°C. Immunocomplexes were captured with 25 μl prewashed Protein A/G agarose beads for 1 hour with rocking at 4°C, washed three times with lysis buffer, and boiled for 5 min in the presence of LDS-PAGE loading dye containing 200 mM DTT. Samples were resolved by SDS-PAGE using an 8–16% gradient gel and analyzed by Western blot using rabbit anti-GFP (Invitrogen, 11122) followed by IRDye 680LT goat anti-rabbit secondary antibody (LI-COR), IRDye 680LT (LI-COR) conjugated-rabbit anti-Cx34.1–3A4 (Fred Hutch Antibody Technology Facility, clone 3A4), and Strep-IRDye 800CW (LI-COR) and visualized with the near-infrared Odyssey system (LI-COR).
Zebrafish care
With approval from the Institutional Animal Care and Use Committee (IACUC AUP 21–42), zebrafish (Danio rerio) were bred and maintained in the University of Oregon fish facility at 28 °C on a 14 hours on and 10 hours off light cycle. Animals were housed in groups according to genotype, with no more than 25 animals per tank. Development time points were assigned according to standard developmental staging117. All fish used for this project were maintained in the ABC background developed at the University of Oregon, and most contained the enhancer trap transgene Et(T2KHG)zf206 unless otherwise noted98. All immunohistochemistry was performed at 5 dpf, at which stage zebrafish sex is indeterminate72.
Genome engineering of gjd1a-V5-TurboID fish by homologous recombination
A single guide RNA (sgRNA) targeting exon 2 of gjd1a/cx34.1 was designed using the CRISPRscan algorithm118 and synthesized as previously described62 using the T7 megascript kit (ThermoFisher). The CRISPR target sequence used to create gjd1a-V5-TurboID (PAM underlined) is: 5’- GCGGCCCAAGTTGGGCACGGAGG −3’.
A plasmid was built to contain coding sequence for the V5 affinity tag and TurboID60,61, flanked by homologous arms cloned from genomic DNA of ABC fish carrying the enhancer trap transgene Et(T2KHG)zf206. The 5’ homologous arm was 418bp (Chr5: 36,996,725–36,997,142) and the 3’ homologous arm was 297bp (Chr5: 36,997,143–36,997,439). Two silent mutations were placed into the CRISPR target site of the repair template (5’-GCGGCCCAAGTTGGGCACtGAtG) to prevent re-cleavage of the repaired site. A double-stranded DNA (dsDNA) repair template was generated by PCR from the plasmid template using primers modified with 5’-biotin and phosphorothioate bonds (denoted by *) for the first five nucleotides of the primer: FP: 5’ Biotin-C*A*A*A*G*ACGACCGCGAATGTCTG-3’; RP: 5’ Biotin-G*T*A*A*C*ATGAGCCCCTGCTACGTTC-3’. The 1723 bp amplicon was treated with DpnI at 37 °C for 30 min, then 80 °C for 20min, column purified and resuspended in RNase-free water.
Injection mixes were prepared in a pH 7.5 buffer solution of 300 mM KCl and 4 mM HEPES and contained a final concentration of 12 μM sgRNA, 10uM Cas9 protein (IDT), and 25nM dsDNA template. Injection mixes were incubated at 37 °C for 5 min immediately prior to injection to promote formation of the Cas9 and sgRNA complex. Parent fish containing the enhancer trap transgene Et(T2KHG)zf206 were previously sequenced for those containing exact homologous arms. Embryos generated from this select parent pool were injected with 1 nL at the one-cell stage119. Samples of injected sibling embryos were either processed for immunofluorescence to confirm mosaic expression of V5 at M/CoLo electrical synapses or harvested for PCR analysis of genomic DNA to confirm template integration. The remaining injected embryos were raised to adulthood and outcrossed to wild-type animals. An animal carrying the homologously recombined V5-TurboID in-frame insertion was identified by PCR and immunofluorescent staining of the V5-tagged allele, and the allele was verified by Sanger sequencing Gt(gjd1a-V5-TurboIDb1445).
Preparation of Larval Fish
Heterozygous Gt(gjd1a-V5-TurboIDb1445) containing the enhancer trap transgene Et(T2KHG)zf206 were in-crossed, grown to adulthood, and genotyped to identify homozygous and wildtype siblings. Homozygous Gt(gjd1a-V5-TurboIDb1445) were crossed with wild type ABC fish to yield heterozygous offspring. The wild type siblings were crossed with wild type ABC fish to yield offspring for control comparisons. Crosses were set up between only one male and one female to produce a clutch, such that several siblings from each clutch could be sacrificed to verify the genotype of the entire clutch. At day 4 dpf larvae were switched to embryo medium containing 1mM biotin. At day 6 dpf larvae were euthanized with tricaine methanesulfonate (ms-222), collected, extensively rinsed in embryo medium (supplemented with ms-222) to remove exogenous biotin, and pooled together by genotype. Larvae (at least 1200 per genotype) were flash frozen in liquid nitrogen and stored at −80 °C until use. Larval preparations were collected on three separate dates.
Preparation of Adult Fish Brains
Heterozygous Gt(gjd1a-V5-TurboIDb1445) containing the enhancer trap transgene Et(T2KHG)zf206 were in-crossed, grown to adulthood, and genotyped to identify heterozygous and wildtype siblings. Six males and six females of each genotype (11.5 months old siblings) were sorted into individual static tanks containing fish water supplemented with 1mM biotin. At 74 hours post-biotin, fish were euthanized and fin clipped for subsequent genotype verification. Brains were harvested, snap frozen in liquid nitrogen, and stored at −80 °C until use120.
Preparation of Homogenates
Frozen larvae for each genotype were resuspended in 4mL 1X RIPA (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 0.1 % SDS, 0.5 % sodium deoxycholate, 1.0 % NP-40 substitute, 1mM PMSF, and protease inhibitors [ThermoFisher A32955]) in a 5ml LoBind eppie. Samples were sonicated for 15 sec at 35% amplitude (1 sec on, 1 sec off) on ice, resting 15 sec on ice and repeated twice (total of 45 sec). Samples rotated overnight at 4 °C.
For adult brains, two brains were removed from the −80 °C freezer and pooled together as one biological sample, with a total of three biological replicates for each genotype/sex combination. The two brains were homogenized using a glass Dounce homogenizer in 750ul Homogenization buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1mM PMSF, and protease inhibitors [ThermoFisher A32955]). The homogenate was transferred to a 2mL LoBind eppie and an equal volume of 2X RIPA buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 0.2% SDS, 1.0% sodium deoxycholate, 2.0% NP-40 substitute, 1mM PMSF, and protease inhibitors [ThermoFisher A32955]) was added (final concentration of detergents: 0.1% SDS, 0.5% sodium deoxycholate, 1.0% NP-40). Samples were sonicated for 15 sec at 35% amplitude (1 sec on, 1 sec off) on ice. Samples rotated overnight at 4 °C.
The following day samples were centrifuged 14,000 × g, 20min, 4°C. The supernatant was desalted over a PD-10 column (Cytiva) pre-equilibrated with 1X RIPA buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 0.1 % SDS, 0.5 % sodium deoxycholate, 1.0 % NP-40 substitute) following the manufacturer’s protocol. The protein concentration of the eluent was determined using the Pierce BCA Protein Assay Kit. Equal amounts of total protein were aliquoted into a 5mL LoBind eppie and prewashed Streptavidin magnetic beads (Pierce Streptavidin Magnetic Beads, Cat# 88817) were added. For larvae, three separate dates of collection represent the three biological replicates for each genotype. Collection date #1 contained one replicate for each genotype (15 mg total protein/300ul prewashed Streptavidin magnetic beads). Collection dates #2 and #3 contained two technical replicates for each genotype (7mg total protein/150ul prewashed Streptavidin magnetic beads). For adults, one date of collection represents the three biological replicates for each genotype for each sex (1mg total protein/150ul prewashed Streptavidin magnetic beads). Samples were rocked overnight at 4 °C.
The following day, magnetic streptavidin beads were washed using a magnetic stand once with wash buffer 1 (50mM Tris pH 7.4, 2% SDS), twice with wash buffer 2 (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1.0% NP-40 substitute), once with wash buffer 3 (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1.-% NP-40), and three times with wash buffer 4 (50mM ammonium bicarbonate in mass spectrometry grade water). Samples were immediately shipped on wet ice overnight to the OHSU Proteomics Shared Resource mass spectrometry facility for analysis.
Mass Spectrometry
Magnetic streptavidin bead samples were washed twice with 100ul of 100mM ammonium bicarbonate and resuspended in a final volume of 100ul. Trypsin (12.5ul of 80ng/μl trypsin in 50mM TEAB (1ug) was added to each sample, mixed gently and incubated at 37C overnight with shaking on a Thermomixer at 800rpm. The following day samples were placed on a magnetic stand for 2min, the supernatant removed, filtered through a 0.22um Millipore filter and dried. The dried filtered sample was dissolved in 20ul of 5% Formic acid and injected into QExactive HF and run with a 90min LC/MS method. Steps were performed at the Oregon Health Sciences University (OHSU) Proteomic Shared Resource.
Mass Spectrometry Data Analysis
The binary instrument files were processed with the PAW pipeline (https://github.com/pwilmart/PAW_pipeline)121. Binary files were converted to text files using MSConvert122. Python scripts extracted fragment ion spectra in MS2 format123. The Comet search engine (version 2016.03)124 was used: 1.25 Da monoisotopic peptide mass tolerance, 0.02 Da monoisotopic fragment ion tolerance, fully tryptic cleavage with up to three missed cleavages, variable oxidation of methionine residues, and static alkylation of cysteines. Searches used UniProt proteome UP000000437 (zebrafish, taxon ID 7955) canonical FASTA sequences (20,603 proteins). Six additional protein sequences and common contaminants (174 sequences excluding any albumins) were added, and sequence-reversed entries were concatenated for a final protein FASTA file of 52,824 sequences (https://github.com/pwilmart/fasta_utilities).
Top-scoring peptide spectrum matches (PSMs) were filtered to a 3% false discovery rate (FDR) using interactive delta-mass and conditional Peptide-prophet-like linear discriminant function scores125. Incorrect delta-mass and score histogram distributions were estimated using the target/decoy method126. The filtered PSMs were assembled into protein lists using basic and extended parsimony principles and required two distinct peptides per protein. The final list of identified proteins, protein groups, and protein families were used to define unique and shared peptides for quantitative use. Total (summed) corrected spectral counts were computed for each protein where shared peptide counts were fractionally split between the proteins they mapped to based on relative unique count totals. The overall protein FDR was 1.1%.
The protein corrected spectral count values for each biological sample in each biological condition were compared for differential protein expression using the Bioconductor package edgeR127 exact test within Jupyter notebooks. For developmental samples or adult brain samples, a minimum average spectral count of 1.5 was required for quantification. A count value of one was added to all spectral count totals for each protein to remove any zero count values. Result tables contained typical proteomics summaries, spectral count totals, and statistical testing results.
To aid interpretation of results, the zebrafish identified protein sequences were matched against the human canonical reference proteome use BLAST and Python scripts (https://github.com/pwilmart/PAW_BLAST and https://github.com/pwilmart/annotations). Additional UniProt annotation information was added for the human ortholog proteins (GO terms and Reactome pathways). STRING and g:Profiler were used for Gene Ontology (GO) analysis76,77. Resources including SFARI78, OMIM79, and Genecards80 that catalog human-disease-associated genes were used to for gene annotation.
scRNA-seq analysis
The single-cell RNA sequencing (scRNAseq) data used in this study originates from the Farnsworth et al. 2020 dataset (5 dpf)73 and the Posner et al. 2024 dataset (6 dpf)128. Both datasets were processed using the pipeline described in Lukowicz-Bedford et al. 202267. Briefly, pooled larval samples were collected in two replicates at each time point. Standard protocols for cell dissociation from whole larvae were followed73. The dissociated cells were processed on the 10X Chromium platform using v2 chemistry for the 5 dpf dataset and v3 chemistry for the 6 dpf dataset, targeting approximately 10,000 cells per run. Aligned reads were mapped to the zebrafish genome (GRCz11) using the 10X Cellranger pipeline (version 3.1), with an updated GTF file that captures Connexin-encoding gene expression67. Cells were processed in Seurat (v5) for R (v4.3.2) with standard quality control, normalization, integration, and analysis procedures129. Analysis code and data sets can be found at www.adammillerlab.com/resources.
Germ layer clusters were identified using established tissue markers from Farnsworth et al. 2022. Mesoderm clusters included putative skeletal muscle, blood, and kidney cells (markers: myod1, smyhc1, myhz2, tnnc1b, gata1a, hbae1.1, cahz, slc4a1a, slc5a9, prr15lb, aqp8b, pdzk1ip1). Endoderm clusters included putative liver cells (foxa3, fabp10a, prox1a, cp). Non neural ectoderm clusters included putative skin and cranial neural crest cells (trpv6, grhl1, gjb8, foxi3a, sox10, dlx2a, foxd3, crestin). Neural ectoderm clusters were identified based on neural markers (elavl3, elavl4, snap25a).
Cas9-mediated genome engineering of V5-tagged mosaics
The CRISPR target sequences used to insert V5 coding sequence for each gene (PAM underlined) are noted in Table S3. The V5-tagged single-stranded donor oligos (ssODN) were designed to repair into endogenous loci, as listed to Table S3. The ssODN contained ~24–36 bp homology arms, tandem V5 sequences, and a 5x-glycine linker between V5 tags and between the V5 tags and endogenous gene sequence. If the inserted sequence did not disrupt the endogenous sgRNA recognition site, silent mutations were designed in the CRISPR/PAM sites of the ssODN to prevent further double stranded breaks after repair.
Injection mixes were prepared in a pH 7.5 buffer solution of 300 mM KCl and 4 mM HEPES and contained a final concentration of 2uM ssODN, 100–1000pg sgRNA, and 8 μM Cas9 protein (IDT) and were incubated at 37°C for 5 min prior to injection to promote formation of the Cas9 and sgRNA complex. Embryos containing the enhancer trap transgene Et(T2KHG)zf206 were injected with 1 nL at the one-cell stage95. At 3–4 dpf, injected embryos were screened by high resolution melting (HRM) to confirm active CRISPRing using gene-specific HRM primers as denoted in Table S3. At 5 dpf, injected larvae were screened by immunohistochemistry for mosaic expression of V5-tagged proteins.
Immunohistochemistry and confocal imaging
Anesthetized, 5 dpf larvae were fixed for 3 hours in 2% trichloroacetic acid in PBS130. Fixed tissue was washed in PBS containing 0.5% Triton X-100, followed by standard blocking and antibody incubations. Primary antibody mixes included combinations of the following: rabbit anti-Cx34.1 3A4 (Fred Hutch Antibody Technology Facility, clone 3A4, 1:200), mouse IgG1 anti-NF(RM044, Invitrogen, 13–0500, 1:200), mouse IgG2a anti-V5 (Invitrogen, R960–25, 1:500), mouse IgG2b anti-acetylated tubulin (Millipore Sigma, T7451–25UL, 1:1000) and chicken anti-GFP (Abcam, ab13970, 1:500). All secondary antibodies were raised in goat (Invitrogen, conjugated with Alexa-405-Plus, −488, −555, −633, or −647 fluorophores, 1:500). Tissue was cleared stepwise in a 25%, 50%, 75% glycerol series, dissected, and mounted in ProLong Gold antifade reagent (ThermoFisher). Using a Leica SP8 Confocal microscope, images were acquired using a 405-diode laser and a white light laser set to 499, 553, 598, and 631 nm, depending on the fluorescent dye imaged. Laser line data was collected sequentially using custom detection filters based on the dye. Images of the Club Endings (CEs) were collected using a 63x, 1.40 numerical aperture (NA), oil immersion lens, images of M/CoLo synapses were collected using a 40x, 1.20 NA, water immersion lens, and tile scans were collected with either objective protocol, as noted in figure legend. The optimal optical section thickness was calculated by the Leica software based on the pinhole, emission wavelengths, and NA of the lens. For whole brain images, tile scans were stitched together using the LAS X software (Leica). Within each experiment where fluorescence intensity was to be quantified, all animals were stained together using the same antibody mix, processed at the same time, and all confocal settings (laser power, scan speed, gain, offset, objective, and zoom) were identical. The M/CoLo synapses analyzed began at approximately somite 10 within the spinal cord. Similar numbers of neighboring, caudal somites were imaged and analyzed in all animals. Multiple animals per genotype were analyzed to account for biological variation, and fluorescence intensity values for each region of each animal were an average across multiple synapses to account for technical variation.
Analysis of confocal imaging
For fluorescence intensity quantitation, FiJi software was used to process and analyze confocal images131. To quantify staining at CE synapses, confocal z-stacks of the Mauthner soma and lateral dendrite (centered around the lateral dendritic bifurcation) were cropped to 200 × 200 pixels. Based on GFP staining, a FIJIscript cleared the region outside of the Mauthner cell, and for each channel a standard threshold was set to remove background staining. The image was then transformed into a max intensity projection, synapse thresholds were set to WT levels, and the integrated density of each channel within the club ending synapses was measured. To quantify staining at M/CoLo synapses, a standard region of interest (ROI) surrounding each M/CoLo site of contact was drawn around the synapse, and the mean fluorescence intensity was measured. Values were normalized to the animals with the highest value in the group (denoted as a grey bar in the figure), and n represents the number of fish used. Figure images were created using FiJi and Illustrator (Adobe).
Statistical analysis of confocal imaging
Standard deviation, standard error of the mean, and the final statistical analysis was performed using Prism (GraphPad) software. Dunnett’s post-hoc multiple comparisons test followed all ANOVA. Statistical tests performed and p values are noted in the figure legends.
Supplementary Material
Figure S1. Connexin-V5-TurboID biotinylates expected proteins in cell culture. HEK293T/17 cells were transfected with plasmids to express Cx34.1-V5-TurboID or empty vector (−) together with mVenus-ZO1b (+) or empty vector (−). Prior to harvest, transfected cells were treated for 10 min with fresh cell culture medium supplemented with 1mM biotin. Lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblot for the presence of mVenus-ZO1b using anti-GFP antibody (green), Cx34.1 protein using Cx34.1-specific antibody (yellow), or biotin using Strep-conjugated fluorophore (magenta) as shown in the channel separated images (middle and right panels). Biotinylated transfected proteins appear white in the merged images (left panels). Total extracts (bottom panels, 5% input) were blotted for transfected proteins to demonstrate uniform antibody recognition and equivalent expression of proteins. Results are representative of three independent experiments.
Figure S2. Generation scheme and verification of gjd1a/cx34.1V5-TurboID fish. (A) Schematic diagram of the gjd1a/cx34.1 gene locus on chromosome 5:36,974,931–36,997238 (GRCz11 Ensembl) showing the exon structure located on the forward strand. Horizontal black bar represents the DNA strand, and boxes represent individual exons as labeled. The CRISPR location is indicated by cartoon scissors in Exon 2. The native CRISPR sequence is listed (PAM is underlined) and the silent mutations incorporated into the repair template are indicated in red font. Areas used as homologous arms (HA) in the dsDNA repair template are represented by cyan boxes (5’ HA, 418bp) and orange boxes (3’ HA, 297bp). The inserted V5-TurboID cassette (1008bp) is represented by a grey box. The red stars indicate the dsDNA template is modified by 5’ biotin plus phosphorothioate bonds between the first five nucleotides. Lengths of DNA regions are indicated. Arrows indicate the location of the flanking primers used to genotype the fish (as shown in panel C). (B) Amino acid sequence of the Cx34.1 wildtype C-terminal tail compared to the predicted amino acid sequence of the Cx34.1-V5-TurboID protein produced in gjd1a/cx34.1V5-TurboID fish. PDZ binding motif (PBM) is underlined. To the right, a cartoon of the Cx34.1-V5-TurboID monomer illustrates the in-frame insertion of the V5-TurboID cassette after the four trans-membrane domains (vertical orange bars) and before the C-terminal PBM (horizontal orange bar). (C) Genotyping of wildtype (Wt) versus gjd1a/cx34.1V5-TurboID homozygous (Hom) fish by PCR using flanking primers indicated in (A). PCR products were resolved by agarose gel and detected by SYBR Safe stain. Bands were gel purified and isolated DNA was sequenced to verify precise genome modification.
Figure S3. Characterization of electrical synapse biotinylation in gjd1a/cx34.1V5-TurboID fish. (A-L) Confocal images of M-circuit neurons and stereotypical electrical synaptic contacts in 6-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae from wildtype (A-D), gjd1a/cx34.1V5-TurboID heterozygous (E-H) and gjd1a/cx34.1V5-TurboID homozygous (I-L) siblings either untreated (A-J) or treated for 72 hours with 1mM biotin (C-L). Animals are stained with anti-GFP (green), anti Cx35.5 (orange), anti-V5 (cyan), and Strep-conjugated fluorophore (magenta). Scale bar = 2 μm in all images. Anterior up. Boxed regions denote stereotyped location of electrical synapses and regions are enlarged in neighboring panels. Images of the M-cell body and lateral dendrite in the hindbrain (A, C, E, G, I, K) are maximum intensity projections of ~20–27 μm. In A’, C’, E’, G’, I’, and K’, images are maximum-intensity projections of ~12–15 μm and neighboring panels show the individual channels. Images of the sites of contact of M/CoLo processes in the spinal cord (B, D, F, H, J, L) are maximum-intensity projections of ~6–7 μm. In B’, D’, F’, H’, J’ and K’, images are from a single 0.42 μm Z-plane and the white dashed square denotes the location of the M/CoLo site of contact. Neighboring panels show individual channels. (M) Quantification of Cx35.5 (orange), V5 (cyan), and Strep (magenta) fluorescence intensities at CE synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the genotype indicated by the grey bar. Circles represent the normalized value of each individual animal. Mean is shown ± SEM. For untreated CE synapses, wt n=3; gjd1a/cx34.1V5−TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. For biotin treated CE synapses, wt n=4; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. **** indicates p<0.0001, *** indicates p<0.0005, ** indicates p<0.0065, and ns is not significant by ANOVA with Dunnett’s test. (N) Quantification of Cx35.5 (orange), V5 (cyan), and Strep (magenta) fluorescence intensities at M/CoLo synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the genotype indicated by the grey bar. Circles represent the normalized value of each individual animal. Mean is shown ± SEM. For untreated M/CoLo synapses, wt n=3; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=3. For biotin treated M/CoLo synapses, wt n=4; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. **** indicates p<0.0001, ** indicates p<0.0025, * indicates p<0.0399, and ns is not significant by ANOVA with Dunnett’s test.
Figure S4. scRNA sequencing analysis of all proteome candidates demonstrates high neural expression. Neural Connexin-associated proteome gene expression across germ layers in a whole-embryo single-cell RNA sequencing dataset (5 and 6 dpf). All genes corresponding to Connexin-associated proteins are listed alphabetically along the y-axis. Clusters are organized by germ layer along the x-axis: neurons, ectoderm (non-neural ectoderm, e.g. skin and cranial neural crest), mesoderm (e.g., skeletal muscle and blood), and endoderm (e.g. liver). Marker genes for specific germ layers are highlighted: elavl3 (neurons), crestin (cranial neural crest, ectoderm), grhl1 (skin, ectoderm), gata1a (blood, mesoderm), myod1 (skeletal muscle, mesoderm), and fabp10a (liver, endoderm). Dot size represents the percentage of cells in the category expressing each gene, while color indicates relative expression levels.
Figure S5. Mosaic V5-gene verification of candidate genes in developing zebrafish. Additional confocal images of V5-tagged candidate genes in 5-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae. Genes are alphabetically ordered. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-V5 (cyan). Neighboring panels show individual and merged channels, as labeled. Images are mainly tile scans of the whole brain or focused on hindbrain and spinal cord circuitry related to the M-cell, with examples from additional brain regions.
Figure S6. Connexin-colocalized and connexin-adjacent models of electrical synapses. (A,B) Model of the M-cell CE mixed electrical/chemical synapses illustrating Connexin-colocalized (A) or Connexin-adjacent (B) patterns. Gap junction plaques made of Connexin protein are yellow, while synapse-localizing proteins are shown in cyan. (C-H) Confocal images of Connexin-associated genes in the Mauthner circuit in 5-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-GluR2/3 (cyan, C,D), anti-ZO1 (cyan, E,F), or anti-Nbea (cyan, G,H). Scale bar = 2 μm.
Acknowledgments
Research was supported by NIH grants R21NS117967 and R01NS105758 to ACM, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institute of Health under Award Number F32HD102182 to EAM, Dow Graduate Research and Lester Wolfe Fellowships to T.C.B., and the NIH National Institute of General Medical Sciences (NIGMS) Graduate Training in Genetics Grant T32GM007413 to MH, RML, and WEC. Mass spectrometric analysis was performed by the OHSU Proteomics Shared Resource by Kilsun Kim (sample preparation), Ashok Reddy (data collection), and Phillip Wilmarth (data analysis), with partial support from NIH core grants P30EY010572, P30CA069533 and OHSU Emerging Technology Fund. We thank the University of Oregon zebrafish facility staff for superb fish care, especially through the challenges of the global pandemic. We thank the administrative staff at the University of Oregon Institute of Neuroscience. We thank Ava Komons and the entire Miller lab for critical contributions and feedback on the manuscript.
References:
- 1.Südhof T. C. Towards an Understanding of Synapse FormaDon. Neuron 100, 276–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia P. H., Li P. & Shen K. Cellular and molecular mechanisms underlying presynapse formaDon. J. Cell Biol. 203, 11–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y. & Garner C. C. Molecular Mechanisms of PresynapDc DifferenDaDon. Annu. Rev. Cell Dev. Biol. 24, 237–262 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Sanes J. R. & Lichtman J. W. InducDon, assembly, maturaDon and maintenance of a postsynapDc apparatus. Nat. Rev. Neurosci. 2, 791–805 (2001). [DOI] [PubMed] [Google Scholar]
- 5.McAllister A. K. Dynamic Aspects of CNS Synapse FormaDon. Annu. Rev. Neurosci. 30, 425–450 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connors B. W. & Long M. A. ELECTRICAL SYNAPSES IN THE MAMMALIAN BRAIN. Annu. Rev. Neurosci. 27, 393–418 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Marder E., GuDerrez G. J. & Nusbaum M. P. ComplicaDng connectomes: Electrical coupling creates parallel pathways and degenerate circuit mechanisms. Dev. Neurobiol. 77, 597–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller A. C. & Pereda A. E. The electrical synapse: Molecular complexiDes at the gap and beyond. Dev. Neurobiol. 77, 562–574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ammer G., Vieira R. M., Fendl S. & Borst A. Anatomical distribuDon and funcDonal roles of electrical synapses in Drosophila. Curr. Biol. CB 32, 2022–2036.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Connors B. W. Synchrony and so much more: Diverse roles for electrical synapses in neural circuits. Dev. Neurobiol. 77, 610–624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereda A. E. Electrical synapses and their funcDonal interacDons with chemical synapses. Nat. Rev. Neurosci. 15, 250–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigulinsky C. L. et al. Network Architecture of Gap JuncDonal Coupling among Parallel Processing Channels in the Mammalian ReDna. J. Neurosci. Off. J. Soc. Neurosci. 40, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan K., Lu Z. & Meinertzhagen I. A. The CNS connectome of a tadpole larva of Ciona intesDnalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 5, e16962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook S. J. et al. Whole-animal connectomes of both CaenorhabdiDs elegans sexes. Nature 571, 63–71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sande E. van der et al. The Role of GJD2(Cx36) in RefracDve Error Development. Invest. Ophthalmol. Vis. Sci. 63, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welsh J. P., Ahn E. S. & Placantonakis D. G. Is auDsm due to brain desynchronizaDon? Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 23, 253–263 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Cunningham M. O. et al. Glissandi: Transient fast electrocorDcographic oscillaDons of steadily increasing frequency, explained by temporally increasing gap juncDon conductance. Epilepsia (2012) doi: 10.1111/j.1528-1167.2012.03530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traub R. D., Bibbig A., LeBeau F. E. N., Buhl E. H. & Whikngton M. A. CELLULAR MECHANISMS OF NEURONAL POPULATION OSCILLATIONS IN THE HIPPOCAMPUS IN VITRO. Annu. Rev. Neurosci. (2004) doi: 10.1146/annurev.neuro.27.070203.144303. [DOI] [PubMed] [Google Scholar]
- 19.Talhouk R. S., Zeinieh M. P., MikaD M. A. & El-Sabban M. E. Gap juncDonal intercellular communicaDon in hypoxia-ischemia-induced neuronal injury. Progress in Neurobiology Preprint at 10.1016/j.pneurobio.2007.10.001 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Goodenough D. A., Goliger J. A. & Paul D. L. CONNEXINS, CONNEXONS, AND INTERCELLULAR COMMUNICATION. Annu. Rev. Biochem. 65, 475–502 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Connors B. W. & Long M. A. ELECTRICAL SYNAPSES IN THE MAMMALIAN BRAIN. Annu. Rev. Neurosci. 27, 393–418 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Meier C. & Dermietzel R. Electrical synapses–gap juncDons in the brain. Results Probl. Cell Differ. 43, 99–128 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Bhalacharya A., Aghayeva U., Berghoff E. G. & Hobert O. PlasDcity of the Electrical Connectome of C. elegans. Cell (2019) doi: 10.1016/j.cell.2018.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phelan P. Innexins: Members of an evoluDonarily conserved family of gap-juncDon proteins. Biochimica et Biophysica Acta - Biomembranes vol. 1711 225–245 (2005). [DOI] [PubMed] [Google Scholar]
- 25.ShruD S., Schulz D. J., Lel K. M. & Marder E. Electrical coupling and innexin expression in the stomatogastric ganglion of the crab Cancer borealis. J. Neurophysiol. 112, 2946–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söhl G., Maxeiner S. & Willecke K. Expression and funcDons of neuronal gap juncDons. Nat. Rev. Neurosci. 6, 191–200 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Hall D. H. Gap juncDons in C. elegans: Their roles in behavior and development. Dev. Neurobiol. 77, 587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennel M. V. L. & Zukin R. S. Electrical Coupling and Neuronal SynchronizaDon in the Mammalian Brain. Neuron Preprint at 10.1016/S0896-6273(04)00043-1 (2004). [DOI] [PubMed] [Google Scholar]
- 29.CurD S. & O’Brien J. CharacterisDcs and plasDcity of electrical synapDc transmission. BMC Cell Biol. 17 Suppl 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien J. Design principles of electrical synapDc plasDcity. Neuroscience LeGers Preprint at 10.1016/j.neulet.2017.09.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereda A. E. et al. Gap juncDon-mediated electrical transmission: Regulatory mechanisms and plasDcity. Biochimica et Biophysica Acta - Biomembranes vol. 1828 Preprint at 10.1016/j.bbamem.2012.05.026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloomfield S. A. & Völgyi B. The diverse funcDonal roles and regulaDon of neuronal gap juncDons in the reDna. Nat. Rev. Neurosci. 10, 495–506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas J. S., Zavala B. & Landisman C. E. AcDvity-dependent long-term depression of electrical synapses. Science 334, 389–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X. D., Korn H. & Faber D. S. Long-term potenDaDon of electrotonic coupling at mixed synapses. Nature 348, 542–5 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Pereda A., Triller A., Korn H. & Faber D. S. Dopamine enhances both electrotonic coupling and chemical excitatory postsynapDc potenDals at mixed synapses. Proc. Natl. Acad. Sci. 89, 12088–12092 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereda A. E. & Faber D. S. AcDvity-dependent short-term enhancement of intercellular coupling. J. Neurosci. Off. J. Soc. Neurosci. 16, 983–92 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcamí P. & Pereda A. E. Beyond plasDcity: the dynamic impact of electrical synapses on neural circuits. Nature Reviews Neuroscience Preprint at 10.1038/s41583019-0133-5 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Jabeen S. & Thirumalai V. The interplay between electrical and chemical synaptogenesis. Journal of Neurophysiology Preprint at 10.1152/jn.00398.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas J. S., Greenwald C. M. & Pereda A. E. AcDvity-dependent plasDcity of electrical synapses: increasing evidence for its presence and funcDonal roles in the mammalian brain. BMC Cell Biol. 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Brien J. The ever-changing electrical synapse. Current Opinion in Neurobiology vol. 29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frisch C. et al. SDmulus complexity dependent memory impairment and changes in motor performance arer deleDon of the neuronal gap juncDon protein connexin36 in mice. Behav. Brain Res. 157, 177–185 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Brunal A. A., Clark K. C., Ma M., Woods I. G. & Pan Y. A. Effects of ConsDtuDve and Acute Connexin 36 Deficiency on Brain-Wide SuscepDbility to PTZ-Induced Neuronal HyperacDvity. Front. Mol. Neurosci. 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quint W. H. et al. Loss of Gap JuncDon Delta-2 (GJD2) gene orthologs leads to refracDve error in zebrafish. Commun. Biol. 4, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobson G. M. et al. Connexin36 knockout mice display increased sensiDvity to pentylenetetrazol-induced seizure-like behaviors. Brain Res. 1360, 198–204 (2010). [DOI] [PubMed] [Google Scholar]
- 45.ChrisDe J. M. et al. Connexin36 mediates spike synchrony in olfactory bulb glomeruli. Neuron 46, 761–772 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Buhl D. L., Harris K. D., Hormuzdi S. G., Monyer H. & Buzsáki G. SelecDve impairment of hippocampal gamma oscillaDons in connexin-36 knock-out mouse in vivo. J. Neurosci. Off. J. Soc. Neurosci. 23, 1013–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long M. A., Jutras M. J., Connors B. W. & Burwell R. D. Electrical synapses coordinate acDvity in the suprachiasmaDc nucleus. Nat. Neurosci. 8, 61–66 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Allen K., Fuchs E. C., Jaschonek H., Bannerman D. M. & Monyer H. Gap juncDons between interneurons are required for normal spaDal coding in the hippocampus and short-term spaDal memory. J. Neurosci. 31, 6542–6552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Güldenagel M. et al. Visual transmission deficits in mice with targeted disrupDon of the gap juncDon gene connexin36. J. Neurosci. Off. J. Soc. Neurosci. 21, 6036–44 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell R. E. et al. Gap juncDons between neuronal inputs but not gonadotropinreleasing hormone neurons control estrous cycles in the mouse. Endocrinology 152, 2290–2301 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Bissiere S. et al. Electrical Synapses Control Hippocampal ContribuDons to Fear Learning and Memory. Science 331, 87–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MarDn E. A., Lasseigne A. M. & Miller A. C. Understanding the Molecular and Cell Biological Mechanisms of Electrical Synapse FormaDon. Front. Neuroanat. 14, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw R. M. et al. Microtubule plus-end-tracking proteins target gap juncDons directly from the cell interior to adherens juncDons. Cell 128, 547–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laird D. W. Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berthoud V. M., Minogue P. J., Laing J. G. & Beyer E. C. Pathways for degradaDon of connexins and gap juncDons. Cardiovasc. Res. 62, 256–267 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Sotelo C. & Korn H. Morphological correlates of electrical and other interacDons through low-resistance pathways between neurons of the vertebrate central nervous system. Int. Rev. Cytol. VOL 55, 67–107 (1978). [DOI] [PubMed] [Google Scholar]
- 57.Llinas R., Baker R. & Sotelo C. Electrotonic coupling between neurons in cat inferior olive. J. Neurophysiol. 37, 560–71 (1974). [DOI] [PubMed] [Google Scholar]
- 58.Marsh A. J., Michel J. C., Adke A. P., Heckman E. L. & Miller A. C. Asymmetry of an Intracellular Scaffold at Vertebrate Electrical Synapses. Curr. Biol. 27, 3561–3567.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lasseigne A. M. et al. Electrical synapDc transmission requires a postsynapDc scaffolding protein. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Branon T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho K. F. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Shah A. N., Davey C. F., Whitebirch A. C., Miller A. C. & Moens C. B. Rapid reverse geneDc screening using CRISPR in zebrafish. Nat. Methods 12, 535–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller A. C. et al. A geneDc basis for molecular asymmetry at vertebrate electrical synapses. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X. et al. Neuronal connexin36 associaDon with zonula occludens-1 protein (ZO-1) in mouse brain and interacDon with the first PDZ domain of ZO-1. Eur. J. Neurosci. 19, 2132–2146 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciolofan C. et al. AssociaDon of connexin36 and zonula occludens-1 with zonula occludens 2 and the transcripDon factor zonula occludens-1-associated nucleic acid-binding protein at neuronal gap juncDons in rodent reDna. Neuroscience 140, 433–51 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MarDn E. A. et al. Neurobeachin controls the asymmetric subcellular distribuDon of electrical synapse proteins. Curr. Biol. 33, 2063–2074.e4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukowicz-Bedford R. M., Farnsworth D. R. & Miller A. C. Connexinplexity: the spaDal and temporal expression of connexin genes during vertebrate organogenesis. G3 Genes Genomes Genet. 12, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoshijima K., Jurynec M. J. M. J. & Grunwald D. J. D. J. Precise EdiDng of the Zebrafish Genome Made Simple and Efficient. 36, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimmel C. B., Hala K. & Metcalfe W. K. Early axonal contacts during development of an idenDfied dendrite in the brain of the zebrafish. Neuron 4, 535–545 (1990). [DOI] [PubMed] [Google Scholar]
- 70.Yao X.-H. et al. Electrical coupling regulates layer 1 interneuron microcircuit formaDon in the neocortex. Nat. Commun. 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Helbig I. et al. In vivo evidence for the involvement of the carboxy terminal domain in assembling connexin 36 at the electrical synapse. Mol. Cell. Neurosci. 45, 47–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson C. A. et al. Wild Sex in Zebrafish: Loss of the Natural Sex Determinant in DomesDcated Strains. GeneMcs 198, 1291–1308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farnsworth D. R., Saunders L. M. & Miller A. C. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howe K. et al. The zebrafish reference genome sequence and its relaDonship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bradford Y. M. et al. Zebrafish informaDon network, the knowledgebase for Danio rerio research. GeneMcs 220, iyac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szklarczyk D. et al. The STRING database in 2023: protein–protein associaDon networks and funcDonal enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kolberg L. et al. g:Profiler—interoperable web service for funcDonal enrichment analysis and gene idenDfier mapping (2023 update). Nucleic Acids Res. 51, W207–W212 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abrahams B. S. et al. SFARI Gene 2.0: a community-driven knowledgebase for the auDsm spectrum disorders (ASDs). Mol. AuMsm 4, 1–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amberger J. S., Bocchini C. A., Scol A. F. & Hamosh A. OMIM.org: leveraging knowledge across phenotype–gene relaDonships. Nucleic Acids Res. 47, D1038–D1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stelzer G. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinforma. 54, 1.30.1–1.30.33 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Chi W. & Kiskinis E. IntegraDve analysis of epilepsy-associated genes reveals expression phenotype correlaDons. Sci. Rep. 14, 3587 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagy J. I., Pereda A. E. & Rash J. E. Electrical synapses in mammalian CNS: Past eras, present focus and future direcDons. Biochimica et Biophysica Acta - Biomembranes Preprint at 10.1016/j.bbamem.2017.05.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michel J. C. et al. Electrical synapse structure requires disDnct isoforms of a postsynapDc scaffold. PLoS Genet. 19, e1011045 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miller A. C., Voelker L. H., Shah A. N. & Moens C. B. Neurobeachin is required postsynapDcally for electrical and chemical synapse formaDon. Curr. Biol. 25, 16–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X., Lu S. & Nagy J. I. Direct associaDon of connexin36 with zonula occludens-2 and zonula occludens-3. Neurochem. Int. 54, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X., Lynn B. D. & Nagy J. I. The effector and scaffolding proteins AF6 and MUPP1 interact with connexin36 and localize at gap juncDons that form electrical synapses in rodent brain. Eur. J. Neurosci. 35, 166–181 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Nagy J. I. & Lynn B. D. Structural and Intermolecular AssociaDons Between Connexin36 and Protein Components of the Adherens JuncDon–Neuronal Gap JuncDon Complex. Neuroscience 384, 241–261 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Tetenborg S. et al. RegulaDon of Cx36 trafficking through the early secretory pathway by COPII cargo receptors and Grasp55. Cell. Mol. Life Sci. 81, 431 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meng L. & Yan D. NLR-1/CASPR Anchors F-AcDn to Promote Gap JuncDon FormaDon. Dev. Cell 55, 574–587.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flores C. E. et al. Trafficking of gap juncDon channels at a vertebrate electrical synapse in vivo. Proc. Natl. Acad. Sci. 109, E573–E582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laird D. W. The life cycle of a connexin: gap juncDon formaDon, removal, and degradaDon. J. Bioenerg. Biomembr. 28, 311–318 (1996). [DOI] [PubMed] [Google Scholar]
- 92.Rash J. E. et al. Heterotypic gap juncDons at glutamatergic mixed synapses are abundant in goldfish brain. Neuroscience 285, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Menelaou E., Kishore S. & McLean D. L. Mixed synapses reconcile violaDons of the size principle in zebrafish spinal cord. eLife 11, e64063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nagy J. I., Pereda A. E. & Rash J. E. On the occurrence and enigmaDc funcDons of mixed (chemical plus electrical) synapses in the mammalian CNS. Neurosci. LeG. 695, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wierson W. A. et al. Efficient targeted integraDon directed by short homology in zebrafish and mammalian cells. eLife 9, 1–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pereda A. et al. Connexin35 mediates electrical transmission at mixed synapses on Mauthner cells. J. Neurosci. Off. J. Soc. Neurosci. 23, 7489–7503 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cárdenas-García S. P., Ijaz S. & Pereda A. E. The components of an electrical synapse as revealed by expansion microscopy of a single synapDc contact. eLife 13, e91931 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Satou C. et al. FuncDonal role of a specialized class of spinal commissural inhibitory neurons during fast escapes in zebrafish. J. Neurosci. Off. J. Soc. Neurosci. 29, 6780–6793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Toro S., Phillips J. & Westerfield M. Whirlin proteins localize at the outer limiDng membrane and subapical region of zebrafish reDna. Invest. Ophthalmol. Vis. Sci. 54, 287 (2013). [Google Scholar]
- 100.Morciano M., Beckhaus T., Karas M., Zimmermann H. & Volknandt W. The proteome of the presynapDc acDve zone: from docked synapDc vesicles to adhesion molecules and maxi-channels. J. Neurochem. 108, 662–675 (2009). [DOI] [PubMed] [Google Scholar]
- 101.Morciano M. et al. ImmunoisolaDon of two synapDc vesicle pools from synaptosomes: a proteomics analysis. J. Neurochem. 95, 1732–1745 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Collins M. O. et al. Molecular characterizaDon and comparison of the components and mulDprotein complexes in the postsynapDc proteome. J. Neurochem. 97, 16–23 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Emes R. D. et al. EvoluDonary expansion and anatomical specializaDon of synapse proteome complexity. Nat. Neurosci. 11, 799–806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bayés À. et al. EvoluDon of complexity in the zebrafish synapse proteome. Nat. Commun. 8, 14613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heo S. et al. IdenDficaDon of long-lived synapDc proteins by proteomic analysis of synaptosome protein turnover. Proc. Natl. Acad. Sci. 115, E3827–E3836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kimmel C. B., Sessions S. K. & Kimmel R. J. Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron. J. Comp. Neurol. 198, 101–120 (1981). [DOI] [PubMed] [Google Scholar]
- 107.Dahlhaus M. et al. The SynapDc Proteome during Development and PlasDcity of the Mouse Visual Cortex *. Mol. Cell. Proteomics 10, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.VanGuilder H. D., Yan H., Farley J. A., Sonntag W. E. & Freeman W. M. Aging alters the expression of neurotransmission-regulaDng proteins in the hippocampal synaptoproteome. J. Neurochem. 113, 1577–1588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.VanGuilder H. D. et al. Hippocampal dysregulaDon of synapDc plasDcity-associated proteins with age-related cogniDve decline. Neurobiol. Dis. 43, 201–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Castermans D. et al. The neurobeachin gene is disrupted by a translocaDon in a paDent with idiopathic auDsm. J. Med. Genet. 40, 352–356 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Creemers J. W. M., Nuytens K. & Tuand K. Neurobeachin Gene in AuDsm. in Comprehensive Guide to AuMsm 825–844 (2014). doi: 10.1007/978-1-4614-4788-7_42. [DOI] [Google Scholar]
- 112.Nuytens K. et al. Haploinsufficiency of the auDsm candidate gene Neurobeachin induces auDsm-like behaviors and affects cellular and molecular processes of synapDc plasDcity in mice. Neurobiol. Dis. 51, 144–151 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Nair R. et al. Neurobeachin regulates neurotransmiler receptor trafficking to synapses. J. Cell Biol. 200, 61–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang X. et al. Neurobeachin: A protein kinase A-anchoring, beige/Chediak-higashi protein homolog implicated in neuronal membrane traffic. J. Neurosci. Off. J. Soc. Neurosci. 20, 8551–8565 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su Y. et al. Neurobeachin is essenDal for neuromuscular synapDc transmission. J. Neurosci. Off. J. Soc. Neurosci. 24, 3627–3636 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Niesmann K. et al. DendriDc spine formaDon and synapDc funcDon require neurobeachin. Nat. Commun. 2, 557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 118.Moreno-Mateos M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPRCas9 targeDng in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Burger A. et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, (2016). [DOI] [PubMed] [Google Scholar]
- 120.Michel J. C. & Miller A. C. IsolaDon of Immunocomplexes from Zebrafish Brain. BioProtoc. 13, e4646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilmarth P. A., Riviere M. A. & David L. L. Techniques for accurate protein idenDficaDon in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2, 223–234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chambers M. C. et al. A cross-pla{orm toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McDonald W. H. et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and idenDficaDons. Rapid Commun. Mass Spectrom. RCM 18, 2162–2168 (2004). [DOI] [PubMed] [Google Scholar]
- 124.Eng J. K., Jahan T. A. & Hoopmann M. R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Keller A., Nesvizhskii A. I., Kolker E. & Aebersold R. Empirical StaDsDcal Model To EsDmate the Accuracy of PepDde IdenDficaDons Made by MS/MS and Database Search. Anal. Chem. 74, 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
- 126.Elias J. E. & Gygi S. P. Target-decoy search strategy for increased confidence in largescale protein idenDficaDons by mass spectrometry. Nat. Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 127.Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differenDal expression analysis of digital gene expression data. BioinformaMcs 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Posner M. et al. Loss of αBa-crystallin, but not αA-crystallin, increases age-related cataract in the zebrafish lens. Exp. Eye Res. 244, 109918 (2024). [DOI] [PubMed] [Google Scholar]
- 129.Hao Y. et al. DicDonary learning for integraDve, mulDmodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.MarDn E. A., Ijaz S., Pereda A. E. & Miller A. C. TrichloroaceDc Acid FixaDon and AnDbody Staining of Zebrafish Larvae. Bio-Protoc. 12, e4289 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schindelin J. et al. Fiji: an open-source pla{orm for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Connexin-V5-TurboID biotinylates expected proteins in cell culture. HEK293T/17 cells were transfected with plasmids to express Cx34.1-V5-TurboID or empty vector (−) together with mVenus-ZO1b (+) or empty vector (−). Prior to harvest, transfected cells were treated for 10 min with fresh cell culture medium supplemented with 1mM biotin. Lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblot for the presence of mVenus-ZO1b using anti-GFP antibody (green), Cx34.1 protein using Cx34.1-specific antibody (yellow), or biotin using Strep-conjugated fluorophore (magenta) as shown in the channel separated images (middle and right panels). Biotinylated transfected proteins appear white in the merged images (left panels). Total extracts (bottom panels, 5% input) were blotted for transfected proteins to demonstrate uniform antibody recognition and equivalent expression of proteins. Results are representative of three independent experiments.
Figure S2. Generation scheme and verification of gjd1a/cx34.1V5-TurboID fish. (A) Schematic diagram of the gjd1a/cx34.1 gene locus on chromosome 5:36,974,931–36,997238 (GRCz11 Ensembl) showing the exon structure located on the forward strand. Horizontal black bar represents the DNA strand, and boxes represent individual exons as labeled. The CRISPR location is indicated by cartoon scissors in Exon 2. The native CRISPR sequence is listed (PAM is underlined) and the silent mutations incorporated into the repair template are indicated in red font. Areas used as homologous arms (HA) in the dsDNA repair template are represented by cyan boxes (5’ HA, 418bp) and orange boxes (3’ HA, 297bp). The inserted V5-TurboID cassette (1008bp) is represented by a grey box. The red stars indicate the dsDNA template is modified by 5’ biotin plus phosphorothioate bonds between the first five nucleotides. Lengths of DNA regions are indicated. Arrows indicate the location of the flanking primers used to genotype the fish (as shown in panel C). (B) Amino acid sequence of the Cx34.1 wildtype C-terminal tail compared to the predicted amino acid sequence of the Cx34.1-V5-TurboID protein produced in gjd1a/cx34.1V5-TurboID fish. PDZ binding motif (PBM) is underlined. To the right, a cartoon of the Cx34.1-V5-TurboID monomer illustrates the in-frame insertion of the V5-TurboID cassette after the four trans-membrane domains (vertical orange bars) and before the C-terminal PBM (horizontal orange bar). (C) Genotyping of wildtype (Wt) versus gjd1a/cx34.1V5-TurboID homozygous (Hom) fish by PCR using flanking primers indicated in (A). PCR products were resolved by agarose gel and detected by SYBR Safe stain. Bands were gel purified and isolated DNA was sequenced to verify precise genome modification.
Figure S3. Characterization of electrical synapse biotinylation in gjd1a/cx34.1V5-TurboID fish. (A-L) Confocal images of M-circuit neurons and stereotypical electrical synaptic contacts in 6-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae from wildtype (A-D), gjd1a/cx34.1V5-TurboID heterozygous (E-H) and gjd1a/cx34.1V5-TurboID homozygous (I-L) siblings either untreated (A-J) or treated for 72 hours with 1mM biotin (C-L). Animals are stained with anti-GFP (green), anti Cx35.5 (orange), anti-V5 (cyan), and Strep-conjugated fluorophore (magenta). Scale bar = 2 μm in all images. Anterior up. Boxed regions denote stereotyped location of electrical synapses and regions are enlarged in neighboring panels. Images of the M-cell body and lateral dendrite in the hindbrain (A, C, E, G, I, K) are maximum intensity projections of ~20–27 μm. In A’, C’, E’, G’, I’, and K’, images are maximum-intensity projections of ~12–15 μm and neighboring panels show the individual channels. Images of the sites of contact of M/CoLo processes in the spinal cord (B, D, F, H, J, L) are maximum-intensity projections of ~6–7 μm. In B’, D’, F’, H’, J’ and K’, images are from a single 0.42 μm Z-plane and the white dashed square denotes the location of the M/CoLo site of contact. Neighboring panels show individual channels. (M) Quantification of Cx35.5 (orange), V5 (cyan), and Strep (magenta) fluorescence intensities at CE synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the genotype indicated by the grey bar. Circles represent the normalized value of each individual animal. Mean is shown ± SEM. For untreated CE synapses, wt n=3; gjd1a/cx34.1V5−TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. For biotin treated CE synapses, wt n=4; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. **** indicates p<0.0001, *** indicates p<0.0005, ** indicates p<0.0065, and ns is not significant by ANOVA with Dunnett’s test. (N) Quantification of Cx35.5 (orange), V5 (cyan), and Strep (magenta) fluorescence intensities at M/CoLo synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the genotype indicated by the grey bar. Circles represent the normalized value of each individual animal. Mean is shown ± SEM. For untreated M/CoLo synapses, wt n=3; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=3. For biotin treated M/CoLo synapses, wt n=4; gjd1a/cx34.1V5-TurboID heterozygotes n=4, gjd1a/cx34.1V5-TurboID homozygotes n=4. **** indicates p<0.0001, ** indicates p<0.0025, * indicates p<0.0399, and ns is not significant by ANOVA with Dunnett’s test.
Figure S4. scRNA sequencing analysis of all proteome candidates demonstrates high neural expression. Neural Connexin-associated proteome gene expression across germ layers in a whole-embryo single-cell RNA sequencing dataset (5 and 6 dpf). All genes corresponding to Connexin-associated proteins are listed alphabetically along the y-axis. Clusters are organized by germ layer along the x-axis: neurons, ectoderm (non-neural ectoderm, e.g. skin and cranial neural crest), mesoderm (e.g., skeletal muscle and blood), and endoderm (e.g. liver). Marker genes for specific germ layers are highlighted: elavl3 (neurons), crestin (cranial neural crest, ectoderm), grhl1 (skin, ectoderm), gata1a (blood, mesoderm), myod1 (skeletal muscle, mesoderm), and fabp10a (liver, endoderm). Dot size represents the percentage of cells in the category expressing each gene, while color indicates relative expression levels.
Figure S5. Mosaic V5-gene verification of candidate genes in developing zebrafish. Additional confocal images of V5-tagged candidate genes in 5-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae. Genes are alphabetically ordered. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-V5 (cyan). Neighboring panels show individual and merged channels, as labeled. Images are mainly tile scans of the whole brain or focused on hindbrain and spinal cord circuitry related to the M-cell, with examples from additional brain regions.
Figure S6. Connexin-colocalized and connexin-adjacent models of electrical synapses. (A,B) Model of the M-cell CE mixed electrical/chemical synapses illustrating Connexin-colocalized (A) or Connexin-adjacent (B) patterns. Gap junction plaques made of Connexin protein are yellow, while synapse-localizing proteins are shown in cyan. (C-H) Confocal images of Connexin-associated genes in the Mauthner circuit in 5-day-post-fertilization, Et(T2KHG)zf206 zebrafish larvae. Animals are stained with anti-GFP/anti-NF/anti-tubulin (magenta), anti-Cx34.1 (yellow), and anti-GluR2/3 (cyan, C,D), anti-ZO1 (cyan, E,F), or anti-Nbea (cyan, G,H). Scale bar = 2 μm.