Abstract
Diamond STING is a new version of the STING suite of programs for a comprehensive analysis of a relationship between protein sequence, structure, function and stability. We have added a number of new functionalities by both providing more structure parameters to the STING Database and by improving/expanding the interface for enhanced data handling. The integration among the STING components has also been improved. A new key feature is the ability of the STING server to handle local files containing protein structures (either modeled or not yet deposited to the Protein Data Bank) so that they can be used by the principal STING components: JavaProtein Dossier (JPD) and STING Report. The current capabilities of the new STING version and a couple of biologically relevant applications are described here. We have provided an example where Diamond STING identifies the active site amino acids and folding essential amino acids (both previously determined by experiments) by filtering out all but those residues by selecting the numerical values/ranges for a set of corresponding parameters. This is the fundamental step toward a more interesting endeavor—the prediction of such residues. Diamond STING is freely accessible at http://sms.cbi.cnptia.embrapa.br and http://trantor.bioc.columbia.edu/SMS.
INTRODUCTION
The first appearance of the STING on the internet was in 1998. STING was hosted by the Protein Data Bank (PDB) server, located at that time at the Brookhaven National Laboratory. The initial goal of the STING was to provide a simple graphical environment for the analysis of the sequence and structure by coupling the information from those two domains. To our surprise, such a simple idea quickly attracted the attention of many users. The addition of a few extra STING components further triggered the interest of the scientific community although STING use was limited to the public PDB (1) files and only a few categories of the structural analysis involving description of the interfaces between protein chains, hydrogen bond network description and simple statistics on local residue environment. Gradually, STING was expanded by including many more components for a complex analysis (2–8) of the structure, function, stability and binding as well as for the relationship between them. During this period, STING also got several mirror sites which conveniently coveted different geographical regions of the users around the world.
We describe in this paper some of the new features available in Diamond STING Suite, the four new per-residue parameter classes and a whole new per-chain descriptor class, as well as the higher level of integration among existing and improved STING components. We also make a mention of our effort toward the integration of structural information and thermodynamic data of proteins, which is crucial for the understanding of the mechanism of protein stability.
Also, we emphasize here that the main difference that continues to work in favor of the STING and distinguishes it from other servers in this category, which are also available on the Internet (9), is the capability to present the largest number of descriptors for the sequence, structure, function, stability and binding in a concise and visually compelling manner, as well as the ability to select/focus for/on those residues that satisfy a user defined parameter/descriptor values. As we proceed to further explain in this paper, this feature presents answers to some very interesting and important questions, such as: is there a set of parameters (protein structure descriptors) that can define UNIQUELY an amino acid ensemble coinciding with the active site of a given protein or coinciding with amino acids identified experimentally as crucial for the folding/stability.
DIAMOND STING INTRINSICS
The Diamond STING components have achieved a higher level of integration among themselves when compared with the Gold STING version. As before, the main entry components in the STING Suite are the Sting Millennium (2,3), which is basically the molecular sequence and structure viewer, the JavaProtein Dossier (JPD) (4) with the Java display of the STING_DB parameters and the STING Report (8) which compiles into a single HTML document a report now containing 23 images showing parameters in context designed plots together with 17 tables. STING Report displays focused information in per-residue fashion. However, STING Report is an exhaustive, yet static presentation of the structure/function descriptors for a given residue; so a user may be able to handle all the parameters in a dynamic way by selecting the residues, e.g. those described by the parameters/descriptors within a certain user defined value range. In such a case, the STING suite entry component should be the JPD.
All the other STING components can be invoked from the ‘Modules’ and ‘Images’ menu at the sequence frame of the STING Millennium. However, a user can opt to go directly to the specific STING component (say, Graphical Contacts) and focus only on the type of information that this particular component offers.
The STING component integration is elevated by allowing a user to always receive focalized data about selected residues from any of the STING components if a residue in one of them has already been selected and/or clicked on.
STING_DB UPDATE AND RENEWAL
The STING_DB is regularly updated in synchrony with the PDB updates. This happens once a week. All the parameters for the new PDB entries are calculated by the STING server. However some missing items, namely the HSSP (10–12) related parameters are usually not available until a couple of weeks (or even a couple of months) after the new structure has been published at the PDB. They will be added as soon as the STING_DB generator identifies that the new HSSP release contains the data for this particular PDB ID.
In addition, with any new HSSP version and new SwissProt (13) version, all the data on conservation have to be re-calculated for the entire STING_DB. Since this puts an enormous load on our CPUs, the STING_DB renewal is done every two months.
NAVIGATING DIAMOND STING
Some of the Diamond STING options require a basic knowledge of the nature of the parameter chosen for inspection/calculation. In addition, some STING components require a user to be familiar with the file input options and output interpretation. In order to minimize the time required for a successful STING use, we made four different entry points a user may consult for a quicker comprehension on how to interpret and proceed with the data. These four options are: the ‘Hypervisual’ java guide with easy to use and quick to access information about any STING component and/or parameter calculated and stored in the STING_DB, the extensive HELP pages which are content sensitive, the frequently asked questions (FAQ) with which many of the users are familiar and finally the JPD and STING Report Legends containing a graphic description of the parameters, which when clicked on would display a corresponding parameter help page.
We found that the new users appreciated the combination of three entry HELP points but always started with the ‘Hypervisual’ java guide and then continued with the extensive use of the content sensitive help pages.
DIAMOND STING HANDLING OF PUBLIC AND LOCAL PDB FORMATTED FILES
Generally, STING operates with both PDB public files and local files in PDB format. However, in order to properly handle STING_DB parameters for a local file, STING needs to pre-calculate those. Similarly, the STING JPD can handle both public and local files and this can be done for a single structure as well as for two structurally aligned proteins.
Interactive and ‘batch’ job modes
In Diamond STING, a user can initiate STING Millennium by using one of the three possible input file routes: a public PDB formatted file available at the PDB (a user can inspect any or all STING parameters as they are pre-calculated), a local PDB-formatted file (this will initiate STING relatively quickly (in ∼30 s at most) but a user will only be able to inspect a reduced number of STING parameters, and finally a TGZ file containing all STING parameters pre-calculated by the STING server upon request. The request to generate the TGZ file is a part of the separate form at STING Millennium entry page and it is one of the best in the Diamond STING offerings.
Advanced input options for local files in JPD
For the single structure, a user may submit a job to the STING Server and obtain in his/her e-mail box a message with attached TGZ file containing all parameters that JPD can display and analyze.
For two structurally aligned files the JPD can handle both two publicly available (PDB deposited) files or two local (non public) PDB formatted files or a combination of a public and a local file. If there are two local files, a user needs to submit to the STING server two separate requests and obtain two TGZ files before doing a structural alignment.
Queuing job requests
Currently the STING server has three major tasks to accomplish: the STING_DB updating (a weekly activity), the STING_DB renewal (a bi-monthly activity) and a job processing that contains a request for calculating all the STING parameters for the local user files.
In order to optimize/maximize the CPU usage at our lab, we have now implemented the STING_Que procedure that basically saves all job requests in the list, which is then executed by CRON at indicated hours [normally identified (by constant monitoring) as the lower CPU activity period]. Currently, we are dedicating four CPUs for this activity, a capacity that should provide enough processing power for generating all STING parameters for ∼300 structures per day (each structure of ∼200 AA).
ADDITIONAL STRUCTURE/FUNCTION PARAMETERS IN DIAMOND STING
We have added some extra parameters into the STING database (8).
In Table 1, we show a complete list including the newly added structure/function parameters (in their order of appearance in the actual JPD window). All these parameters are reported in per-residue fashion. In Table 2, per-chain parameters that are introduced in Diamond STING are listed.
Table 1.
Complete list of STING_DB parameters reported in ‘per-residue’ fashion) including newly added ones (11,29–32) accessible in Diamond STING by means of JPD and STING Report
| 1. ResBoxes |
| 2. Intra-chain atomic contacts [ITC] and |
| 3. The inter-chain atomic contacts [IFC] |
| 4. ITC contacts energy and |
| 5. IFC contacts energy |
| 6. Interface area [IF] |
| 7. Water contacting [WC] |
| 8. Ligand pocket forming [LP] and |
| 9. Surface forming [SF] residues |
| 10. Prosite |
| 11. ProTherm (14) |
| 12. Secondary structure indicator [PDB] |
| 13. Secondary structure indicator [DSSP](18) |
| 14. Secondary structure [STRIDE](19) |
| 15. Multiple occupancy |
| 16. Temperature factor |
| 17. Hot spots |
| 18. Sequence conservation [HSSP] |
| 19. Sequence conservation [SH2Qs] |
| 20. Solvent accessibility |
| 21. Dihedral angles |
| 22. Pockets/cavities |
| 23. Electrostatic potential |
| 24. Hydrophobicity |
| 25. Curvature |
| 26. Distance from the N-/C-terminal |
| 27. Density |
| 28. Sponge |
| 29. Order of cross presence |
| 30. Order of cross link |
| 31. Rotamers |
| 32. Space clash |
Table 2.
List of newly added parameters reported in ‘per-chain’ fashion, also accessible in Diamond STING by means of JPD and STING Report
| Total volume of the chain |
| Total surface area of the chain |
| Ratio of volume to surface |
| Ratio of total surface to surface at interface |
| Total number of |
| H-bonds |
| salt bridges |
| hydrophobic interactions |
| cystein bridges |
| aromatic stacking interactions |
| Total energy of established contacts for the chain |
| Energy of the contacts divided by the volume |
| Total surface area occupied by hot spots |
| Hot spot area divided by total surface area of molecule |
| Total pocket surface area |
| Total pocket volume area |
| Total cavity volume |
The total number of available numerical values for structure/function parameters being reported by Diamond STING is brought to 306. Although we have a total of 98 STING_DB parameters belonging to 30 distinct parameter classes, some of them are calculated with a variety of default conditions (the variable volume size for a probing sphere, the atom at which the center of the probing sphere is placed or the variable size for a sliding window).
One of the new features in Diamond STING is the integration between structural parameters and thermodynamic parameters in ProTherm (14), which compiles a large amount of experimental thermodynamic data on protein stability and the effects of amino acid mutations. Thermodynamic data are crucial for the understanding of the mechanism of protein stability and the function of proteins such as enzymatic reaction and ligand binding. We have created links from per-residue structural parameters to the corresponding thermodynamic data for the mutations of that residue, as well as the opposite link from particular thermodynamic data in ProTherm to STING Report. This will enable users to interpret the structural characteristics around that residue in terms of thermodynamic effects of the mutation, as well as to interpret the thermodynamic data in terms of structural characteristics (see for an example the JPD description of the 1stn.pdb and corresponding ProTherm information).
Selecting parameter range in Diamond STING
The select feature continues to be one of the major pillars of the STING Suite. In JPD, residue selection can be performed according to multiple criteria. The selection of amino acids can be made with any combination of conditions, permitting powerful identification of functionally/structurally important regions or sites. Consequently in JPD a user can make decisions, backed by evidence from the set of protein descriptors, about the possible role of specific amino acids in defining the function of the protein. At the same time, a user may also see the candidate residues which contribute critically to the protein stability or binding or even judiciously evaluate the type of effect one mutation might have on function/stability/binding.
EXAMPLES OF THE DIAMOND STING APPLICATION
In order to emphasize the Diamond STING capabilities and its potential impact on the biological side of a problem under study in the following exercises, we present two interesting examples.
In the first example, we asked a very simple question: is there a set of parameters (protein structure descriptors) that can define UNIQUELY an amino acid ensemble coinciding with the active site of a given protein? An in silico experiment done by JPD and STING_DB allowed us to test this hypothesis and to confirm the existence of such a set of parameters on HIV-integrase.
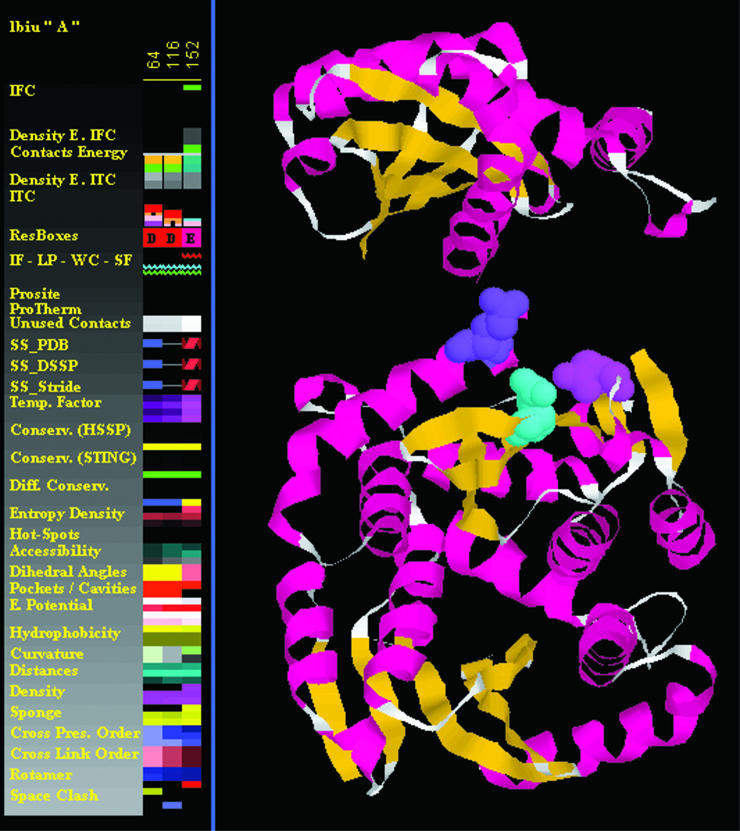
The HIV-1 integrase is an essential enzyme in the life cycle of the virus responsible for catalyzing the insertion of the viral genome into the host cell chromosome; it provides an attractive target for an antiviral drug design. The evidence was obtained from site-directed mutagenesis experiments that even the most conservative substitutions of any of the three absolutely conserved carboxylate residues, D64, D116 and E152 (the so-called DDE motif), abolished catalytic activity (15).
By combining selected ranges for a number of structure parameters, we have obtained at the end of a Select procedure (see Figure 1) an ensemble of amino acids which coincides with the three critical residues identified experimentally as the active site of the HIV integrase. The Select procedure used the following parameters and their numerical values/regions:
Conservation: SH2Qs, relative entropy <30
Physical–chemical: electrostatic potential, average <−20 kT/J/mol
Geometric: pocket/cavity in complex, volume >0.
Figure 1.
The HIV-integrase (1biu.pdb) active site residues: Asp_64 (Cyan), Asp_116 (Magenta) and Glu_152 (purple). Those three residues were selected by means of JPD and its select feature, using a set of parameters and the range for corresponding values: (i) conservation: SH2Qs: relative entropy <30; (ii) physical–chemical: electrostatic potential: average <−20 kT/J/mol; (iii) geometric: pocket/cavity in complex: volume >0.
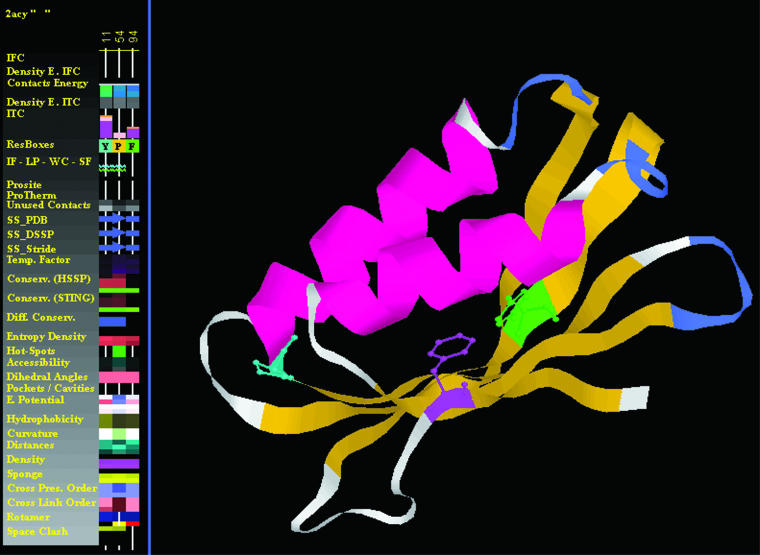
In the second exercise, we asked a similar question: is there a set of parameters (protein structure descriptors) that can define UNIQUELY an amino acid ensemble coinciding with the residues identified as essential in the folding process of a given protein?
We used the protein 2acy (see Figure 2) and again we were able to identify the very same amino acid triplet that Vendruscolo (16) and colleagues have identified in their experiment as crucial for the folding process. An important detail here is that one of the parameters used was the order of cross link. The cross links are defined in STING as the contacts [any type from a possible five classes: (i) hydrophobic interaction, (ii) hydrogen bonding, (iii) aromatic stacking, (iv) salt bridging and (v) cystein-bridging] established among residues that are far apart in the protein primary sequence, but are close in its 3D fold. The order of cross link is identified as a number of such cross links established among independent stretches of sequence (in STING we used three sizes for a sequence stretch: 15, 20 or 30). Only a single occurrence is counted for the order of cross link even though several could be registered (a central amino acid can make more than one contact and each one of them can be established with a different amino acid belonging to the same stretch of probing sequence size). The higher the order, the greater the importance that residue may have for the protein folding/stability. This specific STING parameter is calculated by varying three input parameters: (i) the size of the sequence stretch separating the residues in contact (15, 20 or 30); (ii) the size of the radius of the probing sphere within which the contacts are counted (3.5, 5 and 8.5 Å); (iii) the center of the probing sphere [Cα, Cβ or last heavy atom (LHA) in a side chain].
Figure 2.
The structure of acylphosphatase, AcP (2acy.pdb) and the key residues Tyr_11 (green), Pro_54 (cyan) and Phe_94 (magenta) found from the transition-state analysis to be crucial for the folding of this protein. While all three residues show cross presence ≥1, only the Tyr_11 and the Pro_54 have long-range contacts. Those three residues were selected by means of JPD and its select feature, using a set of parameters: (i) total unused contact energy; (ii) density; (iii) sponge; (iv) cross presence order; (v) secondary structure element; (vi) conservation Sh2Qs/evolutionary pressure; (vii) electrostatic potential @ Ca (viii) electrostatic potential @ surface. For the range of corresponding values see Table 3.
In Table 3 we show the set of parameters and values used to obtain a positive result in searching for the amino acid triplet in 2acy.pdb structure.
Table 3.
List of parameters and their numerical values/ranges used to filter out all amino acids from the 2acy.pdb except for the active site ones
| Parameter name | Parameter value (range) or description |
|---|---|
| Total unused contact energy | >72 Kcal/mol |
| Density | >1.08 for probing sphere 3A centered at Ca |
| Sponge | >0.75 for probing sphere 3A centered at Ca |
| Cross presence order | ≥1,1,2 (at Cα, Cβ and LHA) |
| Secondary structure element | β sheet |
| Conservation Sh2Qs/evolutionary pressure | <28 |
| Electrostatic potential at Ca | ≥7 |
| Electrostatic potential at surface | ≤0 |
CONCLUDING REMARKS
Diamond STING is described here as an improved interactive suite of tools for browsing the structure related STING_DB. The STING_DB has been expanded and the new parameters introduced contribute significantly to the understanding of phenomena such as protein stability. They also describe more fully the protein function. Furthermore, the integration of structural information with the thermodynamic data of proteins and mutants enables us to establish the relationship between structure and thermodynamics and also helps us decipher the function of proteins.
There is an increased number of problems that STING tools can help tackle as they are maturing. With this maturation process we are closer to the general goal of the STING project: capture and describe in full detail nuances among amino acids flocking in regions with specific importance to the protein stability and function. Diamond STING is presented here as an intermediary but becomes a mature and powerful tool toward the final stage of its own development, having already the ability to identify with precision the flocks of residues with a very unique role in a protein: the active site amino acids and the folding essential amino acids. This step is the fundamental one toward the more interesting quest: predicting such residues. The discussion on the details of how general is the finding that the specific parameter set in a determined range of values can be used for the whole family of proteins in order to filter out all other amino acids but the ones belonging to the active site or are essential for the folding; and what this finding can achieve in terms of learning and predicting such important sites in the proteins is beyond the scope of this publication and will be discussed in a separate one.
It is clear that the STING Suite of programs may be used both in research and teaching. Diamond STING plots offer visual but powerful tools for detecting the constellations of amino acids with peculiar characteristics within a protein structure or in the structural alignment of two proteins. Such ability is crucially dependent on the Select feature in JPD and consequently is recommended as the filter for accepting and/or rejecting a hypothesis described in an experiment or in the literature.
Although we are adding many new parameters and features to the STING over the years, our initial goal of having most parameters at the same place represented in an easy to see and easy to interpret fashion has not yet been challenged. On the contrary, the more the number of features/parameters added, the greater the grasp of the capability of the STING in composing with ever higher resolution.
FUTURE DEVELOPMENTS
Diamond STING is already being expanded and the STING_DB being ported into relational database.
The new version—STAR STING—will be able to accept users' local files in a much larger quantity (we expect to double the CPU capacity dedicated exclusively to local file requests). By porting the STING_DB into relational database we will be able to ask questions that are now only answered within a single PDB file. Those questions can be formulated in a way similar to this, for example: which are the protein structures available in the PDB that do posses a cavity of the determined volume, and the residues forming the cavity are conserved to a determined value of the relative entropy/evolutionary pressure, and the cavity forming residues have the electrostatic potential at the surface which is in a negative range and those proteins also do have at least two residues in their structure that are identified as having the Order of Cross Links of at least {1, 1, 1}. What the above question has just defined is the search across the PDB for the binding site that can accommodate a ligand of a determined size/volume, formed by amino acids which are very conserved, with the negative EP at the pocket surface (providing for the adequate activity of the protein) and that protein probably has a pocket with the mouth too tight for the ligand penetration. Consequently, it has to open for the ligand binding by some gating mechanism that involves an amino acid which can make the cross links across the pocket mouth. Upon a ligand binding, that residue will be responsible for keeping the pocket closed until the chemical action is completed. This residue is definitely making at least one Cross Link of Order equal or higher than {1, 1, 1} (Cα, Cβ, LHA).
STAR STING should also present to the user an option for the graphical molecular viewer. STING is currently dependent on Chime. However, we would like to make it more OS independent and for that reason we plan to integrate STING with the JMOL (http://jmol.sourceforge.net). By making available JMOL as the STING molecular viewer, we also hope to get STING functional on Mac stations, an important achievement in light of widespread use of this platform.
With respect to new parameters that will be added to the STAR STING, we are working in complementing the current classes of the contacts with the important missing one: Protein–ligand contacts. In addition, we should be adding the moving protein parts identification (by identifying the residues involved in protein domain movements), as well as the preferred local environment reported in per-residue manner.
We have moved toward the integration between structural information and thermodynamic data of protein stability, by linking between STING and ProTherm. We plan to integrate thermodynamic data for molecular interactions, such as protein–nucleic acid and protein–ligand interactions, along this direction.
Finally, we are working to add more convenience to the STING Report in the STAR STING by adding a capability of creating the Reports in PDF format with a page break feature that allows for the figures being always integral and presented in a single page, as well as adding an important visual description [such as MolScript (17)] of the residue being explored by the STING Report.
Acknowledgments
This work was supported in part by the following grants: FAPESP 01/08895-0, FINEP 1945/01 and CNPq 401695/2003–2004. The authors wish to express special thanks to Phil Bourne and Wolfgang Bluhm from RCSB/PDB-San Diego, Jose Valverde from CNB-Madrid-Spain, Oscar Grau from LaPlata University in Argentina and Megan Restuccia from Columbia University at New York City for their collaboration in testing and maintaining STING at the respective STING mirror sites. Funding to pay the Open Access publication charges for this article was provided by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), FINEP (Financiadora de Estudos e Projetos) and CNPq (Conselho Nacional de Desenvolvimento Científico e Techológico).
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H.M., Bourne P.E., Westbrook J. The Protein Data Bank: a case study in management of community data. Current Proteomics. 2004;1:49–57. [Google Scholar]
- 2.Neshich G., Togawa R.C., Mancini A.L., Kuser P.R., Yamagishi M.E.B., Pappas G., Jr, Torres W.V., Campos T.F., Ferreira L.L., Luna F.M., et al. STING Millennium: a web based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res. 2003;31:3386–3392. doi: 10.1093/nar/gkg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higa R.H., Togawa R.C., Montagner A.J., Palandrani J.C., Okimoto I.K., Kuser P.R., Yamagishi M.E., Mancini A.L., Neshich G. STING Millennium Suite: integrated software for extensive analyses of 3d structures of proteins and their complexes. BMC Bioinformatics. 2004;5:107–115. doi: 10.1186/1471-2105-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neshich G., Rocchia W., Mancini A.L., Yamagishi M.E., Kuser P.R., Fileto R., Baudet C., Pinto I.P., Montagner A.J., Palandrani J.F., et al. JavaProtein Dossier: a novel web-based data visualization tool for comprehensive analysis of protein structure. Nucleic Acids Res. 2004;32:W595–W601. doi: 10.1093/nar/gkh480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higa R.H., Montagner A.J., Togawa R.C., Kuser P.R., Yamagishi M.E., Mancini A.L., Pappas G., Jr, Miura R.T., Horita L.G., Neshich G. ConSSeq: a web-based application for analysis of amino acid conservation based on HSSP database and within context of structure. Bioinformatics. 2004;20:1983–1985. doi: 10.1093/bioinformatics/bth185. [DOI] [PubMed] [Google Scholar]
- 6.Higa R.H., Oliveira A.G., Horita L.G., Miura R.T., Inoue M.K., Kuser P.R., Mancini A.L., Yamagishi M.E., Togawa R.C., Neshich G. Defining 3D residue environment in protein structures using SCORPION and FORMIGA. Bioinformatics. 2004;20:1989–1991. doi: 10.1093/bioinformatics/bth190. [DOI] [PubMed] [Google Scholar]
- 7.Mancini A.L., Higa R.H., Oliveira A., Dominiquini F., Kuser P.R., Yamagishi M.E., Togawa R.C., Neshich G. STING Contacts: a web-based application for identification and analysis of amino acid contacts within protein structure and across protein interfaces. Bioinformatics. 2004;20:2145–2147. doi: 10.1093/bioinformatics/bth203. [DOI] [PubMed] [Google Scholar]
- 8.Neshich G., Mancini A.L., Yamagishi M.E., Kuser P.R., Fileto R., Pinto I.P., Palandrani J.F., Krauchenco J.N., Baudet C., Montagner A.J., et al. STING Report: convenient web-based application for graphic and tabular presentations of protein sequence, structure and function descriptors from the STING database. Nucleic Acids Res. 2005;33:D269–D274. doi: 10.1093/nar/gki111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin M.Y. The molecular biology database collection: 2005 update. Nucleic Acids Res. 2005;33:D5–D24. doi: 10.1093/nar/gki139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 11.Schneider R., de Daruvar A., Sander C. The HSSP database of protein structure-sequence alignments. Nucleic Acids Res. 1997;25:226–230. doi: 10.1093/nar/25.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider R., Sander C. The HSSP database of protein structure-sequence alignments. Nucleic Acids Res. 1996;24:201–205. doi: 10.1093/nar/24.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasteiger E., Jung E., Bairoch A. Swiss-Prot: connecting biological knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 14.Bava K.A., Gromiha M.M., Uedaira H., Kitajima K., Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32:D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgur Y., Dyda F., Hickman A.B., Jenkins T.M., Craigie R., Davies D.R. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vendruscolo M., Paci E., Dobson C.M., Karplus M. Three key residues form a critical contact network in a protein folding transition state. Nature. 2001;409:641–645. doi: 10.1038/35054591. [DOI] [PubMed] [Google Scholar]
- 17.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 18.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19.Heinig M., Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–W502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]