Abstract
We describe a suite of SPACE tools for analysis and prediction of structures of biomolecules and their complexes. LPC/CSU software provides a common definition of inter-atomic contacts and complementarity of contacting surfaces to analyze protein structure and complexes. In the current version of LPC/CSU, analyses of water molecules and nucleic acids have been added, together with improved and expanded visualization options using Chime or Java based Jmol. The SPACE suite includes servers and programs for: structural analysis of point mutations (MutaProt); side chain modeling based on surface complementarity (SCCOMP); building a crystal environment and analysis of crystal contacts (CryCo); construction and analysis of protein contact maps (CMA) and molecular docking software (LIGIN). The SPACE suite is accessed at http://ligin.weizmann.ac.il/space.
INTRODUCTION
The Protein Data Base (PDB) (1,2) has become a major source for the analysis of biological processes at the molecular level, and allows analysis of interactions in proteins and their complexes. A number of web-based servers, including our own [LPC/CSU (3)], provide information on inter- and intra-molecular contacts in proteins [PDBsum (4,5), GRASS (6), Relibase (7), RankViaContact (8), STING CONTACTS (9) and Monster (10)]. The LPC/CSU approach differs from others mainly in the definition of contacting atoms (11) and in the provision of a more detailed description of contacts founded on an atom classification (12). Atoms are considered to be in contact with one another based on inter-atomic distances and the extent of crowding in the environment. For example, in non-packed regions two atoms could be listed as hydrogen bonded at distances up to 5 Å (assuming water mediation), while in packed regions they would not. In addition, a measure of contact surface area is provided. As a result, the LPC/CSU approach was applied not only for detailed structure analysis (13–15) but also for the derivation and application of knowledge-based functions to the protein folding problem (16–19) and to molecular docking (20,21).
In this communication, we describe a suite of SPACE tools designed to assist in the analysis and prediction of biomolecular structures and their complexes. A shared feature of all SPACE tools is the application of the LPC/CSU definition for inter-atomic contacts and surface complementarity. Inter-atomic contacts are calculated either numerically (11) or analytically (22). Complementarity is estimated based on the deviation of atoms into eight classes according to their physicochemical properties (12).
SPACE WEB TOOLS
LPC/CSU: contact analysis of biomolecules
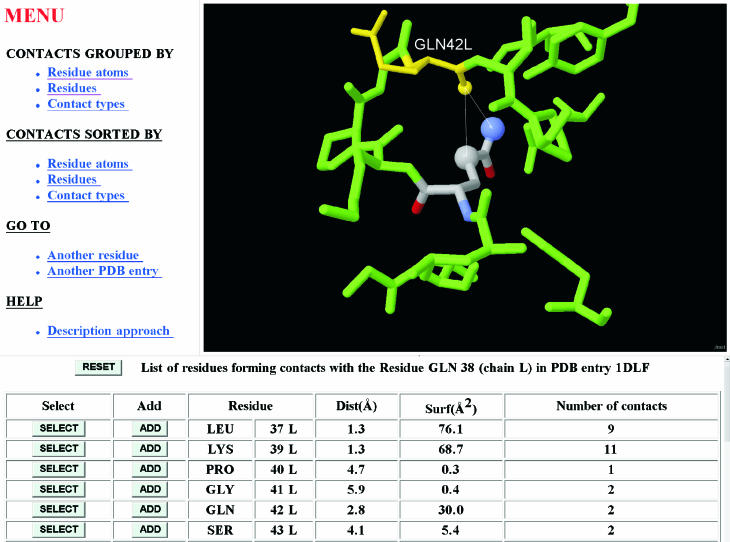
The LPC/CSU server in its current version analyzes and visualizes (either with Chime plug-in or Java based Jmol) atomic interactions within a protein or protein complex, including resolved water molecules and attached ligands, and nucleic acids. Different levels of analysis can be chosen: contacts can be grouped and sorted by atom, residue or contact type (H-bond, hydrophobic–hydrophobic and aromatic–aromatic). The output provides characteristics for every atom–atom contact (atom properties, distance and contact area). A typical output is illustrated in Figure 1.
Figure 1.
Graphical output of the CSU program. Interactions of residue Gln38L from pdb entry 1DLF (anti-dansyl Fv fragment) are illustrated. All residues and water oxygens in contact with Gln38L are evoked by clicking on ‘contacts grouped by residues’. Pressing ‘Select’ for Gln42L in the bottom frame results in labeling and highlighting (in yellow) of the residue selected. The visualization screen shows that only two contacts are formed and one of them is a H-bond to the backbone oxygen of Gln42L.
CryCo: analysis of crystal contacts
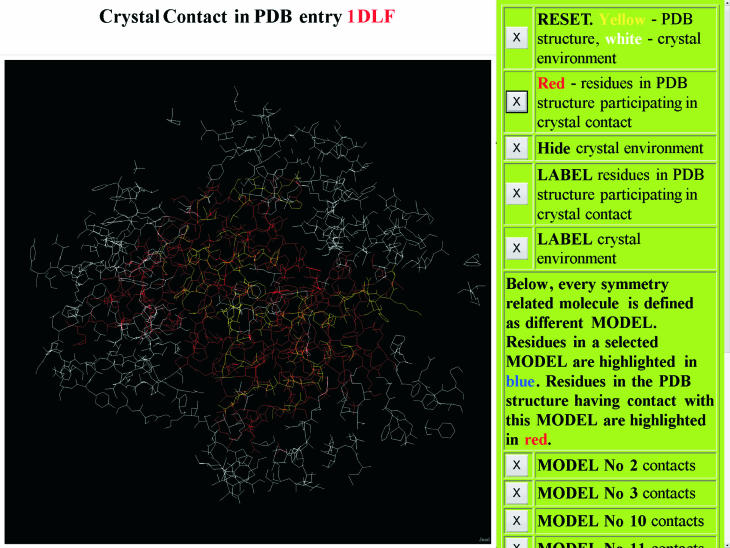
The CryCo server builds coordinate files in a PDB format for the unit cell as well as for the complete crystal environment of one molecule. The structural environment is built in several steps. First, symmetry related molecules are created using the PDBSET program from the CCP4 suite (23). When necessary, all molecules are translocated to one unit cell. The 26 adjacent cells in the crystal lattice are then constructed by translation and finally any atom farther than a chosen threshold from the closest atom of the central molecule is removed. Detailed analyses of atomic contacts are based on CSU software. Interactive visualization options and coordinate output files are provided, and also an option to submit a structural file for analysis. An example of the output from the CryCo server is provided in Figure 2. CryCo differs from existing tools such as the WHAT IF web server (24) and the xpack VRML-based program (25) in providing visualization options, detailed contact analyses and several files with new features for downloading.
Figure 2.
Graphical output of the CryCo server. The PDB structure under analysis is highlighted in yellow while its crystal environment is in white. Residues of the structure in crystal contact have been highlighted in red by clicking on the second panel. The atomic coordinates for every molecule in contact with the PDB structure are formatted in the output file as a different MODEL. Contacts formed by every such molecule can be visualized separately.
CMA: contact map analysis
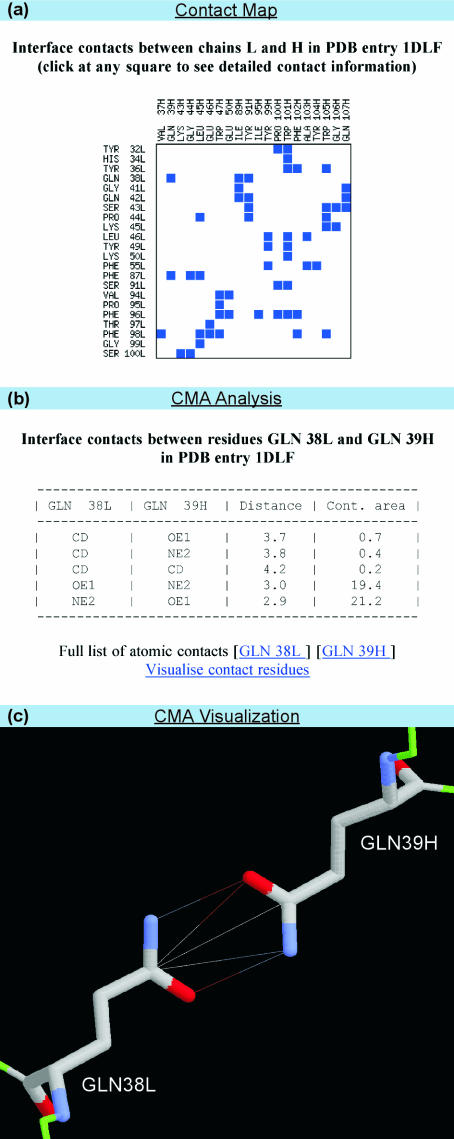
For a given PDB file, the ‘Contact Map Analysis’ server (CMA) evaluates residue–residue contacts between two chains or within a single one. In the example illustrated in Figure 3, the interface contacts between chains L and H in PDB file 1DLF are considered. Residue–residue contacts are represented as an interactive contact map, where a square at the crossing of two residues indicates a contact (Figure 3a). Positioning the cursor over the square highlights summary information about the contacting residues and their total contact area. Clicking on the square reveals a table with more detailed contact information based on LPC/CSU software, including names of the contacting atoms, distances and atom–atom contact areas (Figure 3b). Links for analysis and visualization of contact residues are likewise provided (Figure 3c). The CMA server was extensively used to analyze inter domain contacts in sandwich-like proteins (26). It differs from existing servers, such as WebMol (27), iMolTalk (28) and Stride (29), by providing detailed visualization and detailed contact analysis.
Figure 3.
Server for construction and analysis of protein contact maps. (a) Contact map (b) CMA analysis and (c) CMA visualization.
MutaProt: structural rearrangements upon point mutations
MutaProt contains a database of pairs of PDB files whose members differ in one or two amino acids (30). The software examines the microenvironment of the mutated residues. The database is accessed by specifying a PDB file, keyword or a pair of amino acids. Accessibility and atomic contacts of the mutated residue are provided by CSU software. The current version of the server has a number of significant improvements. MutaProt now extracts pairs based on differences at the chain level. This dramatically increases the database to ∼200 000 pairs. Wild-type structures are distinguished from mutant ones where information is available. An option has been included for user submission of a structural file for pairing up with PDB entries and MutaProt analysis. The interactive graphics have been expanded to include the entire PDB structure and presentation of the protein sequence is included along with secondary structure assignment based on DSSP (31). In addition, superposition of the two pair members is now done analytically (32). A list of publicly available mutation databases is provided. MutaProt is unique in providing detailed on line analysis of atomic contacts and offering a superimposed 3D presentation of regions being compared.
SCCOMP: side chain modeling
SCCOMP is a server for side chain modeling. It uses a scoring function (33) that includes terms for complementarity (CSU definitions of geometric and chemical compatibility), excluded volume, internal energy based on rotamer probability and solvent accessible surface. The input for the program is a coordinate file in the PDB format with or without side chain coordinates. The output is the file with predicted coordinates for the side chains. The program has an accuracy of ∼93% for χ1 prediction (±40°) of buried residues, ∼71% for exposed residues, ∼83% for all residues and an overall RMSD of 1.7 Å (not including Cβ). A fast iterative search takes ∼1 min for a typical protein; the slower stochastic search takes ∼12 min and improves prediction by ∼2% and 0.1 Å RMSD. SCCOMP permits modeling a subset of residues, introducing any number of mutations, and using homologous structures as templates. It complements another publicly available server (http://www1.jcsg.org/scripts/prod/scwrl/serve.cgi) uses a less sophisticated scoring function (34). Although our program is slower, it more accurately predicts χ1+2 and returns a lower RMSD for the overall structure. Furthermore, SCCOMP is convenient for performing in silico mutagenesis.
SPACE PROGRAMS
The SPACE suite provides an option to download a number of programs. Source codes for the LIGIN (12) (molecular docking) and SCCOMP (33) (side chain modeling) programs are available at the SPACE website. To enable the analysis of a large number of PDB files, LPC and CSU programs with output as simple text files are also provided.
Acknowledgments
Funding to pay the Open Access publication charges for this article was waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bernstein F.C., Koetzle T.F., Williams G.J.B., Meyer E.F., Jr, Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 2.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobolev V., Sorokine A., Prilusky J., Abola E.E., Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 4.Laskowski R.A., Hutchinson E.G., Michie A.D., Wallace A.C., Jones M.L., Thornton J.M. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 5.Laskowski R.A., Chistyakov V.V., Thornton J.M. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayal M., Hitz B.C., Honig B. GRASS: A server for the graphical representation and analysis of structures. Protein Sci. 1999;8:676–679. doi: 10.1110/ps.8.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendlich M., Bergner A., Gunther J., Klebe G. Relibase: design and development of a database for comprehensive analysis of protein–ligand interactions. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 8.Shen B.R., Vihinen M. RankViaContact: ranking and visualization of amino acid contacts. Bioinformatics. 2003;19:2161–2162. doi: 10.1093/bioinformatics/btg293. [DOI] [PubMed] [Google Scholar]
- 9.Mancini A.L., Higa R.H., Oliveira A., Dominiquini F., Kuser P.R., Yamagishi M.E.B., Togawa R.C., Neshich G. STING Contacts: a web-based application for identification and analysis of amino acid contacts within protein structure and across protein interfaces. Bioinformatics. 2004;20:2145–2147. doi: 10.1093/bioinformatics/bth203. [DOI] [PubMed] [Google Scholar]
- 10.Salerno W.J., Seaver S.M., Armstrong B.R., Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–W568. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobolev V., Edelman M. Modeling the quinone-B binding site of the photosystem-II reaction center using notions of complementarity and contact-surface between atoms. Proteins. 1995;21:214–225. doi: 10.1002/prot.340210304. [DOI] [PubMed] [Google Scholar]
- 12.Sobolev V., Wade R.C., Vriend G., Edelman M. Molecular docking using surface complementarity. Proteins. 1996;25:120–129. doi: 10.1002/(SICI)1097-0134(199605)25:1<120::AID-PROT10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 13.Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., Pietrokovski S. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004;344:1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 14.Swint-Kruse L. Using networks to identify fine structural differences between functionally distinct protein states. Biochemistry. 2004;43:10886–10895. doi: 10.1021/bi049450k. [DOI] [PubMed] [Google Scholar]
- 15.Reichmann D., Rahat O., Albeck S., Meged R., Dym O., Schreiber G. The modular architecture of protein–protein binding interfaces. Proc. Natl Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConkey B.J., Sobolev V., Edelman M. Discrimination of native protein structures using atom-atom contact scoring. Proc. Natl Acad. Sci. USA. 2003;100:3215–3220. doi: 10.1073/pnas.0535768100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaya H., Chan H.S. Solvation effects and driving forces for protein thermodynamics and kinetic cooperativity: how adequate is native-centric topological modeling? J. Mol. Biol. 2003;326:911–931. doi: 10.1016/s0022-2836(02)01434-1. [DOI] [PubMed] [Google Scholar]
- 18.Yang S.C., Cho S.S., Levy Y., Cheung M.S., Levine H., Wolynes P.G., Onuchic J.N. Domain swapping is a consequence of minimal frustration. Proc. Natl Acad. Sci. USA. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ollerenshaw J.E., Kaya H., Chan H.S., Kay L.E. Sparsely populated folding intermediates of the Fyn SH3 domain: matching native-centric essential dynamics and experiment. Proc. Natl Acad. Sci. USA. 2004;101:14748–14753. doi: 10.1073/pnas.0404436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobolev V., Niztaev A., Pick U., Avni A., Edelman M. A case study in applying docking prediction: modeling the tentoxin binding sites of chloroplast F1-ATPase. Curr. Sci. 2002;83:857–867. [Google Scholar]
- 21.Lloyd D.G., Hughes R.B., Zisterer D.M., Williams D.C., Fattorusso C., Catalonotti B., Campiani G., Meegan M.J. Benzoxepin-derived estrogen receptor modulators: a novel molecular scaffold for the estrogen receptor. J. Med. Chem. 2004;47:5612–5615. doi: 10.1021/jm0495834. [DOI] [PubMed] [Google Scholar]
- 22.McConkey B.J., Sobolev V., Edelman M. Quantification of protein surface, volumes and atom–atom contacts using a constrained Voronoi procedure. Bioinformatics. 2002;18:1365–1373. doi: 10.1093/bioinformatics/18.10.1365. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez R., Chinea G., Lopez N., Pons T., Vriend G. Homology modeling, model and software evaluation: three related resources. Bioinformatics. 1998;14:523–528. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 25.Fu T.Y., Chen Y.W. Visualization of macromolecular crystal packing using Virtual Reality Modelling Language (VRML) J. Appl. Crystallogr. 1996;29:594–597. [Google Scholar]
- 26.Potapov V., Sobolev V., Edelman M., Kister A., Gelfand I. Protein–protein recognition: juxtaposition of domain and interface cores in immunoglobulins and other sandwich-like proteins. J. Mol. Biol. 2004;342:665–679. doi: 10.1016/j.jmb.2004.06.072. [DOI] [PubMed] [Google Scholar]
- 27.Walther D. WebMol—a Java based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 28.Diemand A.V., Scheib H. iMolTalk: an interactive, internet-based protein structure analysis server. Nucleic Acids Res. 2004;32:W512–W516. doi: 10.1093/nar/gkh403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 30.Eyal E., Najmanovich R., Sobolev V., Edelman M. MutaProt: a web interface for structural analysis of point mutations. Bioinformatics. 2001;17:381–382. doi: 10.1093/bioinformatics/17.4.381. [DOI] [PubMed] [Google Scholar]
- 31.Kabasch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Arun K.S., Huang T.S., Blostein S.D. Least-squares fitting of two 3-D point sets. IEEE Trans. Pattern Anal. Mach. Intel. 1987;9:698–700. doi: 10.1109/tpami.1987.4767965. [DOI] [PubMed] [Google Scholar]
- 33.Eyal E., Najmanovich R., McConkey B.J., Edelman M., Sobolev V. Importance of solvent accessibility and contact surfaces in modeling side-chain conformations in proteins. J. Comput. Chem. 2004;25:712–724. doi: 10.1002/jcc.10420. [DOI] [PubMed] [Google Scholar]
- 34.Canutescu A.A., Shelenkov A.A., Dunbrack R.L., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]