Abstract
Similar to T cells and B cells, mast cell surfaces are dominated by microvilli, and like these other immune cells we showed with microvillar cartography (MC) that key signaling proteins for RBL mast cells localize to these topographical features. Although stabilization of ordered lipid nanodomains around antigen-crosslinked IgE-FcεRI is known to facilitate necessary coupling with Lyn tyrosine kinase to initiate transmembrane signaling in these mast cells, the relationship of ordered-lipid nanodomains to membrane topography had not been determined. With nanoscale resolution provided by MC, SEM and co-localization probability (CP) analysis, we found that FcεRI and Lyn kinase are positioned exclusively on the microvilli of resting mast cells in separate nano-assemblies, and upon antigen-activation they merge into overlapping populations together with the LAT scaffold protein, accompanied by elongation and merger of microvilli into ridge-like ruffles. With selective lipid probes, we further found that ordered-lipid nanodomains preferentially occupy microvillar membranes, contrasting with localization of disordered lipids to flatter regions. With this proximity of signaling proteins and ordered lipid nanodomains in microvilli, the mast cells are poised to respond sensitively and efficiently to antigen but only in the presence of this stimulus. Use of a short chain ceramide to disrupt ordered-lipid regions of the plasma membrane and evaluation with MC, CP, and flow cytometry provided strong evidence that the microvillar selective localization of signaling proteins and lipid environments is facilitated by the interplay between ordered-lipid nanodomains and actin attachment proteins, ERM (ezrin, radixin, moesin) and cofilin.
Keywords: Mast cell topography, plasma membrane domains, transmembrane signaling, super-resolution imaging-based Microvillar Cartography, FcεRI, ERM and Cofilin.
Significance Statement
Participation of ordered-lipid nanodomains (colloquially dubbed “rafts”) to target and regulate immune signaling in the plasma membrane is well documented. Recent studies also demonstrated the role of membrane topography, specifically microvilli, in T-cell and B-cell immune signaling. Here, we show how these features are coordinated in RBL mast cells, a well-established model for mast cells involved in multiple antigen-activated immune responses that include allergies and inflammation mediated by IgE-receptors (IgE-FcεRI). We found that the receptors and a key signaling kinase, together with ordered-lipid nanodomains localize to microvilli in resting cells, forming separated nano-assemblies. Antigen-activation causes elongation and merger of microvilli into ruffles where receptors and kinase coalesce to initiate transmembrane signaling. Selective pre-organization of signaling proteins and targeting lipid domains in microvilli facilitates sensitive and efficient responses to antigenic stimulation. Overall, our results demonstrate complex interplay between membrane topography, ordered-lipid nanodomains, and cytoskeleton attachment proteins in controlling mast cell activation.
Introduction
Although cartoons typically depict cells as smooth spheres, microscopic images reveal that the membrane surface of live mammalian cells is irregular, frequently dominated by different types of protrusions, including finger-like, actin-dependent microvilli. The mechanistic role of irregular and dynamic topography of plasma membranes remains a subject of investigation, with recent studies demonstrating its importance in different fields of cellular physiology (1), including immunology(2–8). In particular, microvilli have been found to be the predominant protruding structures on T cells and B cells involved in adaptive immune responses (2–6, 8–10). To fully elucidate participation of microvilli in immune cell function the three-dimensional (3D) topographical distribution of proteins and lipids within this milieu must be quantitatively assessed. However, this is technically challenging because the width of microvilli lies within ~100 nm, which is well below the diffraction limit of the optical microscope.
Recently, Haran and co-workers designed a super-resolution microscopy-based methodology called ‘Microvillar Cartography’ (MC) to examine the localization of membrane proteins with respect to the 3D plasma membrane topography in fixed samples (2, 4, 5, 11). They found strong evidence that the microvilli projections of Jurkat and human effector T cells serve as signaling hubs for T cell receptor (TCR)-mediated responses to antigen in cellular immune responses. In particular, they observed that TCR, co-receptor CD4, tyrosine kinase Lck, and adaptor LAT are substantially enriched in the microvilli of resting cells. This pre-assembly can thereby facilitate initial antigen sensing and the TCR complex to couple effectively with its proximal signaling molecules. This leads to effective cell activation, which then modulates membrane linkages with filamentous actin mediated by ezrin-radixin-moesin (ERM) proteins(5). Jun and co-workers employed scanning electron microscopy (SEM), total internal reflection fluorescence (TIRF) microscopy, and high-resolution confocal microscopy to show that microvilli-derived particles can form “immunological synaptosomes” with antigen-presenting cells (APCs) (6). This group also used high-resolution confocal microscopy to demonstrate TCR on the tips of microvilli, which were shed distally when these T cells formed synapses with activating surfaces (12). Utilizing dynamic lattice light sheet microscopy (LLSM), Krummel and coworkers demonstrated that microvilli serve as sensors for T-cells for finding specific APCs (3). Klotzsch and coworkers used SEM, super-resolution microscopy, and high-resolution confocal microscopy to demonstrate that the nanoconfinement of key signaling proteins in the microvilli is responsible for altering gene expression, leading to enhanced T-cell activation (7). Together, these studies provide a foundation for understanding the role of microvilli and 3D membrane topography more generally in targeting and regulating T cell activation.
Related studies have been carried out in B cells, another type of lymphocyte that plays complementary roles in immune responses. Employing dynamic LLSM, Reth and coworkers demonstrated that localization of antigen-sensing B cell receptors (BCRs) is similarly dictated by the microvillar topography of B cells (8). All these studies point strongly to topography-mediated signal initiation as a general mechanism for immune cells with microvillar dominated membrane surfaces. Mast cells are not lymphocytes but play diverse roles in innate and adaptive immunity, probably most notably in allergic and inflammatory responses (13). For mast cells, the high affinity receptor (FcεRI) for immunoglobulin E (IgE) is the dominant antigen-sensing receptor that is mediated by tightly bound, antigen-specific IgE (14). A microvillar topography like T cells and B cells has been characterized for RBL mast cells by SEM (15). However, the relationship between FcεRI, signaling partners, and the 3D membrane topography has not been defined.
Extensive investigations of the spatial relationship of antigen receptors and their signaling partners in the flattened ventral membrane of attached immune cells has been carried out using total internal reflection fluorescence (TIRF) microscopy and quantification of super resolution images. For all of T cells (16), B cells (17), and mast cells (18), antigen engagement of their respective receptors causes physical and functional coupling with key tyrosine kinases resulting in phosphorylation of the immune receptors, assembly of additional proteins and consequent downstream signaling. In general, the role of lateral membrane organization in receptor-mediated immune cell signaling has been the subject of intense interest, including the targeting role played by membrane lipid heterogeneity (19). With lipid compositions of model membranes representing those in the plasma membrane, nanoscopic regions of ordered lipids (characterized by saturated acyl chains) segregate from regions of disordered lipids, described as Lo-like and Ld-like, respectively. Although ordered-lipid nanodomains in cellular plasma membranes are almost certainly diverse in lipid and protein composition, these are often lumped loosely together and colloquially dubbed “rafts.” Many different studies have examined participation of “rafts” or Lo-like ordered-lipid nanodomains to initiate signaling in mast cells, T cells and B cells (17, 20–23). Compelling evidence supports the view that this phase-like behavior of lipids is important for regulating protein interactions in the membrane, and these may also couple to phase-based protein condensates in the cytoplasm (24–26).
Super-resolution microscopy has been applied extensively for detailed characterization of ordered-lipid nanodomains that are stabilized by immune-clustering of BCR or FcεRI to initiate functional coupling in B cells (17) or mast cells (16, 26), respectively. Imaging Fluorescence Correlation Spectroscopy (ImFCS) further demonstrated in mast cells that co-localization of Lyn tyrosine kinase with antigen-clustered FcεRI and consequent phosphorylation of FcεRI cytoplasmic segments is primarily mediated by Lyn’s saturated acyl chains that anchor it to the membrane inner leaflet and prefer an ordered lipid environment. Transmembrane phosphatases, which in the resting state suppress net phosphorylation, prefer a disordered environment and are excluded from the ordered regions surrounding the clustered FcεRI (23). Phosphorylated FcεRI provides a protein binding site for Lyn, and consequent recruitment of cytoplasmic Syk tyrosine kinase leads to phosphorylation of transmembrane LAT, which serves as a cytoplasmic scaffold for downstream signaling proteins.
Although the role of ordered-lipid nanodomains in regulating protein distributions during antigen-activation of mast cells has been amply investigated in the ventral surface of attached cells, these spatial/functional interactions as related to plasma membrane topography on suspended cells has received little attention and remains largely unknown. For the present study, we adopted the MC methodology and utilized the RBL mast cells (27) as a model system to decipher the correlation between membrane nanodomains and membrane topography and their participation in targeting antigen-stimulated mast cell signaling. We quantified the distribution of FcεRI and key signaling proteins, as well as order- and disorder-preferring lipid probes with respect to the 3D plasma membrane topography of resting suspended cells, and we observed similarities to T cells as revealed in previous studies (2, 4, 5). We observed how these protein and lipid distributions in mast cells change after antigen is introduced to crosslink FcεRI thereby activate the RBL mast cells, and found that the stimulated morphological changes differ markedly from those of T cells. We also examined the role of cytoskeleton attachment proteins, Ezrin/Radixin/Moesin and cofilin, which have been reported to regulate localization of signaling molecules in immune cells (5, 28, 29). Our study uncovers the complex interplay between membrane topography, ordered-lipid nanodomains, and cytoskeletal attachment proteins in controlling the distribution of signaling molecules, which together sense the presence of antigen and initiates cell activation such as occurs during immune responses mediated by mast cells.
Results
Activation of mast cells is accompanied by changes in topographical features.
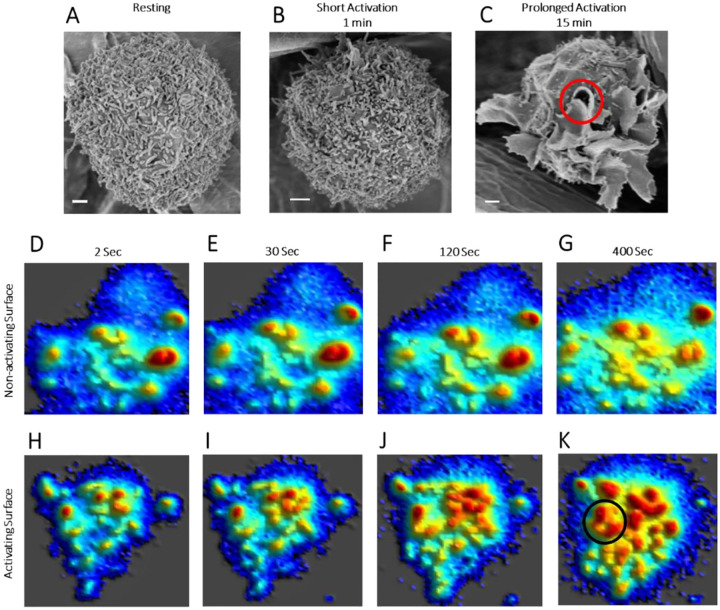
The topography of RBL mast cells resembles other mast cells (15, 30) and other white blood cells including lymphocytes involved in immune responses (T cells and B cells) (2, 4, 5, 9, 31, 32). We employed these RBL cells in the present study as a robust representative for mast cells more generally (27, 33). Like T cells (2), the surface of resting RBL cells is dominated by microvilli (Fig. 1A). However, unlike activation and synapse formation of T-cells (34) , SEM shows that activation of mast cells by soluble antigen does not cause immediate microvillar collapse. We found that short activation (1 min) with soluble antigen of suspended, IgE-sensitized RBL cells causes apparent elongation and merger of the microvilli (Fig. 1B). Prolonged activation (15 min) results in the formation of long and curvy ridges, commonly called “ruffles” (Fig. 1C). These observations are like those made previously with adherent RBL mast cells (15), and confocal microscopy of live RBL cells shows the ruffles to be dynamic (35, 36). As viewed with SEM, the antigen-activated ruffles sometimes include a donut-shape pit structure (Fig. 1C), which we speculate may be an opening through which release of fused secretory granules occurs in live cells.
Figure 1. Membrane Topography of RBL Mast Cells is Dominated by Microvilli which form Membrane Ruffles upon Activation:
A-C. Scanning electron microscopic (SEM) images of RBL cells. A. resting state, B. after 1 minute (short) and C. after 15 minutes (prolonged) of activation of suspended anti-DNP IgE-sensitized RBL cells using soluble antigen (DNP-BSA). The red circle in image C indicates the donut-shape pit-like structure formed after 15 min activation. Scale bar, 1μm.
D-K. Snapshots from movie of membrane topographical change of IgE-sensitized RBL cells while interacting with BSA coated non-activating glass surface (D-G) and 20 mol% DNP-BSA/BSA coated activating glass surface (H-K). The black circle in image K indicates the donut-shape pit-like structure formed on RBL cell membrane after interaction with activating surface. Complete movies are provided in SI Appendix Movies S1 and S2.
We adopted MC methodology (10) (SI Appendix Fig. S1A–M) to further delineate the changes in membrane topography and the redistribution of FcεRI and key signaling proteins that accompany activation by antigen. In one set of experiments, live cells were sensitized with anti-DNP IgE and stained with FM143 dye. Then suspended cells were dropped on glass surfaces coated with either BSA (non-activating) or BSA containing 20 mol% DNP-BSA (antigen-activating). We recorded a time series of variable-angle total internal reflection fluorescence microscope (VA-TIRFM) images of cells interacting with these activating or non-activating surfaces using MC methodology (10, 11), as detailed in the next section. Post-processing of this image time series allows the dynamics of membrane topography during the interactions to be observed. We found that for cells interacting with the non-activating surface the membrane topography of mast cells remains almost unaltered for up to ~ 400 seconds (Fig. 1D–G; SI Appendix Movies S1A and B). Within 30 min the membrane begins to spread on the glass surface as expected from previous studies showing integrin and cytoskeleton engagement (our data not shown; (36, 37)). In contrast, microvilli on cells interacting with the antigen-activating surface start to merge within 30 sec of interactions (Fig. 1H–K; Movies S2A and B). After a few minutes, the merging microvilli start to form ruffles (Fig. 1J–K). Sometimes, these ruffles coalesce in a circular fashion forming donut shape pit structures (Fig. 1K) like that observed in SEM images (Fig. 1C). At longer times the cells flatten and spread as observed previously in studies with activating surfaces (36, 37).
Membrane topography and localization of signaling molecules are correlated.
We used MC on fixed RBL cells to examine the spatial relationship between the membrane topography and membrane proteins that are involved in transmembrane signaling after engagement of antigen. FcεRI sensitized with IgE serves as the transmembrane receptor for multivalent antigen on mast cells. Crosslinking of these FcεRI complexes by antigen causes coupling with Lyn tyrosine kinase, which is anchored to the inner leaflet of the plasma membrane and phosphorylates cytoplasmic segments of FcεRI. Consequent recruitment of cytoplasmic Syk tyrosine kinase leads to phosphorylation of transmembrane LAT, which serves as a scaffold for downstream signaling proteins. To investigate distributions of FcεRI, Lyn, and LAT we fixed suspended, IgE(anti-DNP)-sensitized cells, either in a resting state or after activation for 1 min or 15 min with soluble antigen (DNP-BSA) at 37°C. For Lyn or LAT, cell membranes were homogeneously stained with FM143fx, followed by fixing and permeabilizing these cells, and labeling with the respective Alexa-647 tagged antibody. FcεRI was labeled by Alexa-647 tagged IgE during overnight cell culture, and this was followed by fixation and membrane staining with FM143fx. As we observe routinely, overnight sensitization of RBL cells with monomeric IgE does not cause cell activation (38, 39). The antibody-labeled, membrane-stained, fixed cells were attached to a poly-D-lysine (PDL) coated glass surface and imaged with TIRF microscopy. Because the cells are pre-fixed, this surface interaction causes no perturbation of the topography or activation of cells (5, 13). We observed similar cell topography in our MC imaging (microvillar density = 3.8±0.5/μm2) as in our SEM imaging (microvillar density = 4.4±0.5/μm2), confirming the robustness of the MC methodology. Similar microvillar density (2–4/μm2) was reported for T-cells (32).
MC of FM143 or FM143fx labeled cells utilizes VA-TIRFM imaging to quantify membrane topography. The relative distance of each point on a TIRFM image from the glass surface (δz) is determined from the measured fluorescence intensity at that point for each of several selected angles of incidence of the excitation beam (SI Appendix Fig. S1E–I) (5, 11, 40). The 3D-membrane topography map is then created from the set of δz values obtained by combining all the VA-TIRFM images (Fig. S1B, E–G), which increases the statistical precision. Because the typical width of microvilli (~100 nm) is well below the optical diffraction limit, the microvillar structure appears thicker (~ 250 nm diameter) in X-Y direction of VA-TIRFM images, which can achieve sub-diffraction resolution only in the Z-direction. That the microvillar density we observed in SEM images agrees well with the density calculated from the segmentation maps from VA-TIRFM images, provides confidence that the MC procedure does not underestimate or miss a significant fraction of microvilli. Previous studies on T cells showed that the membrane projections identified as microvilli by MC are true microvilli by co-localization of L-selectin (5).
In the next step, we determined the localization map of the fluorescent antibody-labeled membrane proteins in the X-Y direction with accuracy well below the diffraction limit using stochastic localization nanoscopy (SLN), in this case direct stochastic optical reconstruction microscopy (d-STORM) (Fig. S1C). This provided the nanoscale distribution pattern (in X-Y direction) of the specifically labeled membrane protein in the context of nanoscale membrane topography (in Z direction) by superimposing the SLN map of a membrane proteins on the VA-TIRFM membrane topography map (Fig. S1D). Because we are concerned only about the distribution of the membrane proteins (and not their absolute number), multiple fluorophores per labeling antibody do not lead to an artifactual conclusion. We recorded the SLN-based protein distribution map in two separate slices for each cell. One slice is at the glass surface (0 nm), typically visualizing mainly the microvillar region and a small fraction of cell body region (for shorter or bent-over microvilli). The other map is at a slice 400 nm away from the glass surface where both microvilli and cell body regions are included for most microvilli. We demonstrated previously that the TIRF evanescent field intensity, although decreasing with distance from the glass surface, is still high enough to detect fluorophores present in 0 nm and 400 nm slices, with equal probability (5).
The membrane topography map of each cell was segmented into the microvillar region and the cell-body region following the established MC methodology (11). From our combined measurements we quantified three features in each cell: 1) the fraction of labeled proteins of each type residing in the microvilli in 0 nm and 400 nm imaging slices (SI Appendix Fig. S1J–K); 2) the fraction of microvilli occupied by each labeled protein; 3) the fluorophore density (δCount/δArea) as a function of radial distance (R) from the tips of microvillar structures in the 0 nm imaging slice (SI Appendix Fig. S1L–M). The central microvillar regions are defined as those regions within 20 nm in the Z-direction from the pixel with the minimum δz value (microvillar tip). Importantly, the topographical analysis relies only on the sub-diffraction resolution in the Z-direction, as determined by VA-TIRFM. MC-based superposition of topography and SLN allowed us to determine the distribution of endogenous membrane proteins involved in FcεRI-mediated signaling in RBL mast cells: FcεRI, Lyn kinase, and LAT scaffold. We also investigated ectopically expressed, transmembrane tyrosine phosphatase, PTPα, which counteracts Lyn kinase activity (41). We made these measurements for suspended cells in the resting state and after 1 min and 15 min activation with soluble antigen. A summary of our findings is presented in Fig. 2 and described below.
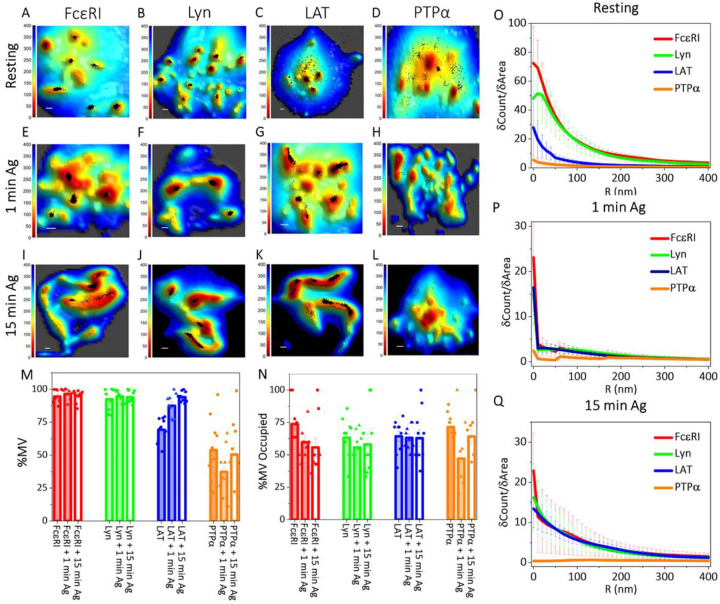
Figure 2. Membrane Topography and Localization of Signaling Proteins are Correlated in Resting and Antigen-Activated States of RBL Mast Cells:
A-L. Localization maps of RBL cell membrane signaling proteins with respect to the three-dimensional membrane topography in suspended resting cells (A-D), and after 1 min (E-H) or 15 min (I-L) activation with soluble antigen. Positions of proteins obtained from SLN (black dots) in the 0 nm imaging slice are superimposed on membrane topography maps obtained from VA-TIRFM. The color bars represent the distance from the glass in nanometers (nm). Scale bars, 0.5 μm.
M-Q. Quantified distributions of proteins on the RBL cell surface in resting, 1 min and 15 min antigen-activated states. The error bar represents the standard error of the mean.
M. Percentage of proteins on microvillar (MV) regions of the membrane in resting, 1 min, and 15 min activated states in the 0 nm slice. The values for individual cells are shown as dots in the plot.
N. Fraction of microvilli (MV) occupied by the respective proteins in resting, 1 min and 15 min activated states. The values for individual cells are shown as dots in the plot.
O-Q. Protein fluorophore density (δCount/δArea) as a function of radial distance (R) from the central microvilli region in the 0 nm imaging slice in resting (O), 1 min (P), and 15 min (Q) activated states.
FcεRI:
FcεRI is specifically labeled with monomeric IgE. Crosslinking of FcεRI complexes by soluble antigen causes the formation of nanoclusters and activates the cell (22, 23, 26).
Resting state.
We observed that 94±2% of FcεRI localize to microvilli in the 0 nm slice (Fig. 2A and M). The segregation pattern of FcεRI is very similar in the 400 nm slice, 92±3% localize to microvilli (SI Appendix Fig. S2A and M). 74±6% of microvilli are occupied by FcεRI (Fig. 2N). We found the density plot for FcεRI (δCount/δArea vs R) decays sharply in the 0 nm SLN slice as the radial distance (R) from the central microvillar region increases, indicating predominant localization nearer the microvillar tips (Fig. 2O).
Changes with activation.
Incubation with soluble antigen for 1 min or 15 min does not lead to significant changes in localization of FcεRI with respect to the microvilli although these structures change from separate microvilli to elongated/merged microvilli to ruffles during this time period (0 nm slice: Fig. 2A, E, I and M; 400 nm slice: Fig. S2E, I and M). The fraction of occupied microvilli appears to decrease slightly upon activation (Fig. 2N), although the decrease is not statistically significant according to one-way ANOVA analysis (P>0.05). The δCount/δArea vs R plot for FcεRI becomes significantly sharper upon activation for 1 min, consistent with the view that more FcεRI are accumulating towards the tips of the microvilli (Fig. 2P). By 15 min the δCount/δArea plot becomes more gradual, probably reflecting formation of ruffles rather than sharp microvilli (Fig. 2Q).
Lyn:
Cytoplasmic, membrane-anchored Lyn couples with and phosphorylates antigen-clustered FcεRI as the first step of transmembrane signaling (18, 23). This coupling involves partitioning of Lyn, via saturated palmitate and myristoylate chains, into ordered-lipid nanodomains that coalesce around the clustered FcεRI (23, 42, 43).
Resting state.
We observed that, like FcεRI, most Lyn localizes in the microvilli of RBL cells in imaging slices at 0 nm (92±3 %; Fig. 2B and M) and 400 nm (91 ±2 %; Fig. S2B and M). 63±5 % of microvilli are occupied by Lyn (Fig. 2N). Interestingly, the δCount/δArea vs R plot for Lyn shows a peak at ~20 nm away from the central microvillar region then decays sharply (Fig. 2O). This is a distinctive difference from the same plot for FcεRI, which decays sharply directly from the central microvillar region (Fig. 2O), suggesting an inherent separation of FcεRI and Lyn within the same microvillus for resting RBL mast cells. In a subsequent section, we describe co-localization probability (CP) analysis to quantify further the separation of these two proteins within microvilli.
Changes with activation.
Antigen activation for 1 min or 15 min causes little or no change in the percentage of Lyn on the microvilli (Fig. 2M, and Fig. S2M) or the percentage of microvillar structures occupied by Lyn (Fig. 2N). However, antigen-activation for 1 min causes the δCount/δArea plot for Lyn kinase to decay sharply from the central microvillar region (Fig. 2P), and this decay appears to become more gradual after 15 min activation (Fig. 2Q). The δCount/δArea plots for Lyn after both 1 min and 15 min activation have nearly the same shape as the corresponding plots for FcεRI, consistent with the view that antigen-clustering of FcεRI causes coupling of these signaling molecules on the microvilli.
LAT:
LAT is primarily cytoplasmic and anchored to the membrane by a short transmembrane segment; reversible palmitoylation enhances partitioning into ordered-lipid nanodomains (44). After antigen-clustering of FcεRI, palmitoylated LAT serves as a scaffold for the assembly of proteins necessary for downstream signaling (23, 45). LAT plays a similar role in T cells (16), and previous MC studies showed that ~70% of LAT molecules reside in the microvilli of resting T cells (5)
Resting state.
Compared to FcεRI and Lyn, LAT localizes somewhat less prominently in the microvilli of RBL cells in imaging slices at 0 nm (69.3±2.9%; Fig. 2C and M) and 400 nm (67.7±3.0%; Fig. S2C and M). 64.3±4.0% of the microvilli of the cells are occupied with LAT molecules (Fig. 2N). The δCount/δArea vs R plot for LAT exhibits a decay shape that is shallower compared to those of FcεRI and Lyn indicating that although LAT proteins tend to localize to microvilli, they also distribute across the cell body (Fig. 2O).
Changes with activation.
The percentage of LAT localized in the microvilli in the 0 nm slice increases from 1 min (87.4±3.0%) to 15 min (94.0±1.3%) after antigen-clustering of FcεRI (Fig. 2G, K and M), and a similar increase is observed in the 400 nm slice (Fig. S2G, K and M). This increase is statistically significant (P value, 3.82726×10−7<<0.05). The fraction of microvilli occupied by LAT remains almost unchanged after antigen activation (Fig. 2N). By 1 min the δCount/δArea plot for LAT increases in magnitude at the microvillar central region with a steeper decay and a shape that is very similar to those of FcεRI and Lyn (Fig. 2P). These three plots continue to be very similar after 15 min (Fig. 2Q), consistent with coalescence of these three signaling proteins on microvilli upon antigen activation.
PTPα:
By counterbalancing tyrosine kinase activities, transmembrane tyrosine phosphatases are known to regulate the activation of immune cells including mast cells (46), such that the spatial relationship of these proteins to microvilli is of considerable interest. CD45 has been established as a negative regulator in T cells, and previous studies showed that this protein distributes uniformly between microvilli and flatter cell regions (5, 47). CD45 appears to have positive and negative regulatory roles in mast cells (48). Moreover, this protein is expressed in low levels in normal mast cells, including RBL cells (41, 49), which precluded a MC-based distribution map of CD45 in these cells with high confidence. Instead, we chose to investigate ectopically expressed PTPα, a transmembrane tyrosine phosphatase that we found previously to negatively regulate Lyn kinase activity in a reconstituted system (41). Our previous studies showed that PTPα localizes preferentially in disordered-lipid regions, away from antigen-clustered FcεRI which stabilizes ordered-lipid nanodomains domains proximally (23).
For our MC studies, we transiently transfected human influenza hemagglutinin (HA)-tagged PTPα. Our transfection efficiency for HA-PTPα in RBL cells was ~30%, and with Alexa-647 labeled anti-HA antibodies we could identify the transfected population. For MC imaging, we fixed the transfected cells, stained with FM143fx, and labeled with fluorescent antibodies after permeabilization. Interestingly, we found that microvilli observed in these samples depended on the level of transfection as visualized with fluorescence. Although cells with low levels of PTPα expression looked somewhat like the samples stained for FcεRI, Lyn, or LAT, cells with high expression levels of PTPα exhibited much fewer microvilli. For MC analysis we selected transfected cells that had moderate levels of PTPα expression. For those cells we found 54±7% and 58±6% of PTPα on the microvilli in 0 nm and 400 nm slices, respectively (Fig. 2D and M; Fig. S2D and M). We found that 72±3% of microvilli of those transfected cells have some PTPα (Fig. 2N). However, the slope of δCount/δArea vs R plot for PTPα molecules is very shallow (almost flat) compared to FcεRI, Lyn, and LAT, indicating an almost uniform distribution of PTPα across the membrane microvilli and cell body (Fig. 2O). When these cells transfected with PTPα are treated with antigen to cluster FcεRI, we found that their membrane topography changes are disrupted compared to those observed for FcεRI, Lyn, and LAT described above: Elongation/merger of microvilli and formation of ruffles do not occur in the same manner (Fig. 2D, H and L).
To evaluate how PTPα transfection of RBL cells affects their function, we performed an assay of the ultimate cell response to effective stimuli: degranulation. We found that the percentage of degranulation stimulated by antigen is decreased by ~25% for PTPα transfected cell batches compared to cells that had not been transfected (Fig. S2N). Comparing this result to a transfection efficiency of ~30% suggests that our transfection of PTPα substantially inhibits cellular degranulation. Thus, although we imaged the change in the distribution of moderately expressed PTPα before and after antigen-activation of transfected RBL mast cells (Fig. 2D – Q), interpretation is complicated by the evidence for dysfunction. Nonetheless, these experiments demonstrate a correlation between disrupted antigen-activation and disrupted changes in membrane topography.
Membrane topography correlates localization of ordered-lipid nanodomains and signaling proteins.
Previous studies established that antigen-clustering of FcεRI in RBL mast cells causes proximal stabilization of ordered-lipid nanodomains, which facilitate functional coupling with Lyn kinase to initiate transmembrane signaling (18, 23, 42). A similar mechanism has been described for immune receptors in B cells and T cells (17, 20, 21). Localization of ordered-lipid nanodomains with microvilli was recently indicated for T cells (12). However, how these lipid domains and microvilli coordinate to localize signaling proteins and modulate their distribution after antigen activation for any immune cell has not been described. Several genetically encoded constructs have been developed to serve as lipid probes, including fluorescent proteins fused with fatty acylated peptides that anchor these probes to the inner leaflet of the plasma membrane (18, 23, 50). The PM lipid probe incorporates the short N-terminal segment of Lyn, including its palmitoylation and myristoylation sites that cause preferential localization to ordered-lipid regions. The GG lipid probe, commonly used for partitioning preferentially into disordered-lipid regions, contains the membrane anchorage motif likr that of K-Ras consisting of a geranylgeranyl modification and a juxta-membrane polybasic sequence.
We employed the PM lipid probe to identify the distribution of ordered-lipid nanodomains with respect to the membrane topography of RBL mast cells. We prepared a 3xHA-tagged construct of PM, which could be labeled with fluorescent anti-HA antibodies. After transfection of 3HA-PM plasmid, the cells were stained with FM143fx, permeabilized, and labeled with anti-HA conjugated to Alexa 647. Results after MC imaging are shown in Fig. 3. We found that 93±1% and 90±2% of PM probes localize to the microvilli in the 0 nm and 400 nm slice, respectively (Fig. 3A and E; Fig. S3A and E). PM probes occupy 81 ± 4% of the microvilli (Fig. 3F). Moreover, the δCount/δArea vs R plot for PM molecules decreases sharply as the distance from the central microvillar region increases (Fig. 3G). This strong localization of PM probes indicates that the ordered-lipid nanodomains are found predominately in microvilli of suspended RBL mast cells.
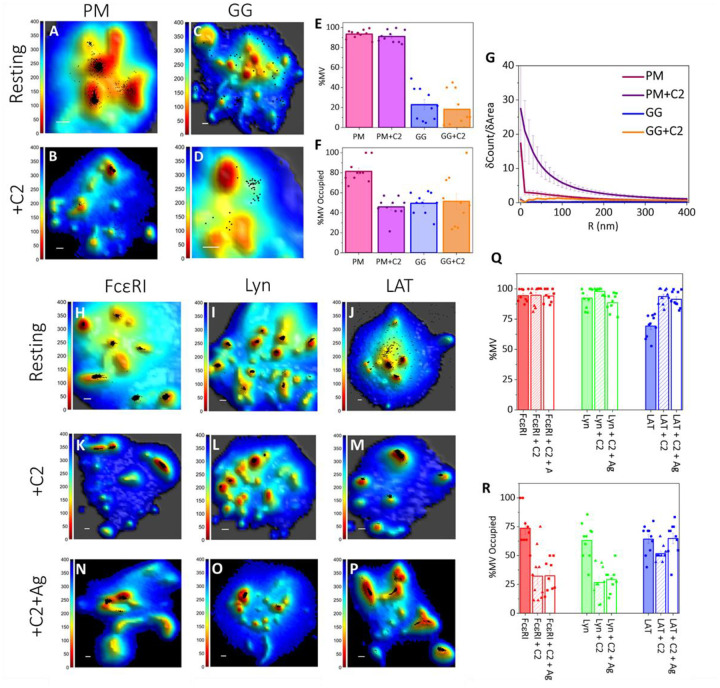
Figure 3: Synergy between Membrane Topography and Membrane Domains Controls the Localization of Signaling Proteins in Resting and Antigen-Activated States of RBL Mast Cells:
A-D. Localization maps of probes preferring either ordered lipid (PM-3HA; A-B) or disordered lipid (3HA-GG; C-D) regions with respect to three-dimensional membrane topography in resting untreated (A, C), and C2-ceramide treated (B, D) cells. Positions of probes obtained from SLN (black dots) in the 0 nm imaging slice are superimposed on membrane topography maps obtained from VA-TIRFM. The color bars represent the distance from the glass in nanometers (nm). Scale bars, 0.5 μm.
E-G. Quantified distributions of lipid probes on the RBL cell surface in resting untreated and C2-ceramide treated cells. The error bar represents the standard error of the mean.
E. Percentage of molecules on microvillar (MV) regions of the membrane in resting untreated and C2 ceramide treated cells at 0 nm imaging plane. The values for individual cells are shown as dots in the plot.
F. Fraction of microvilli (MV) occupied by the respective lipid probes in resting untreated and C2-ceramide treated cells. The values for individual cells are shown as dots in the plot.
G. Probe density (δCount/δArea) as a function of radial distance (R) from the central microvilli region in the 0 nm imaging slice for resting untreated and C2-ceramide treated cells.
H-P. Localization maps of signaling proteins with respect to three-dimensional membrane topography in resting untreated (H-J), C2-ceramide treated (K-M) cells and after 15 min antigen-activation in continued presence of C2-ceramide (N-P). Positions of proteins obtained from SLN (black dots) in the 0 nm imaging plane are superimposed on membrane topography maps obtained from VA-TIRFM. The color bars represent the distance from the glass in nanometers (nm). Scale bars, 0.5 μm.
Q-R. Quantified distributions of proteins in untreated, C2-ceramide treated resting cells, and after 15 min antigen-activation in continued presence of C2-ceramide. The error bar represents the standard error of the mean.
Q. Percentage of probes on membrane microvilli (MV) in untreated and C2-ceramide treated resting cells, and after 15 min antigen-activation in continued presence of C2-ceramide in the 0 nm imaging slice. The values for individual cells are shown as dots in the plot.
R. Fraction of microvilli (MV) occupied by the respective proteins in untreated and C2-ceramide treated resting cells, and after 15 min antigen-activation in continued presence of C2-ceramide. The values for individual cells are shown as dots in the plot.
Using a 3xHA derivative of the disorder-preferring GG lipid probe, we found that the distribution of GG-3HA differs markedly from that of 3HA-PM. Only 23 ± 5% and 18 ± 5% of GG probes localize to the microvilli of suspended cells in 0nm and 400 nm slices, respectively (Fig. 3B and E; Fig. S3C and E). The fraction of microvilli occupied by the GG probes is 49 ± 3% compared to 81 ± 4% for PM (Fig. 3F). These partitioning values suggest that disordered-lipid regions preferentially occupy the flatter membranes of the cell body. The flat δCount/δArea vs R plot for GG probes also indicates that GG is spread more evenly across the cell membrane (Fig. 3G).
Short chain ceramides, including N-acetyl-D-sphingosine (C2-ceramide) have been shown to disrupt ordered-lipid nanodomains (60–62) with functional consequences in RBL mast cells. For example, antigen-stimulated phosphorylation of FcεRI by Lyn is inhibited by this treatment (27). To investigate how microvillar structure or density might depend on ordered-lipid nanodomains, we treated suspended RBL cells with 32μM C2-ceramide (27, 51). MC imaging shows no substantial change in the microvilli or their density after this treatment for resting cells (Fig. 3A–D and H–M). We found the percentage of PM probes localizing to microvilli of C2-ceramide treated cells is also like that of untreated cells (~90%; Fig. 3E; Fig. S3B and E). However, the fraction of microvilli occupied by PM probes decreases by 45% (from 81±4% to 46±3%) for C2-ceramide treated cells compared to untreated cells (Fig. 3F). In other words, C2-ceramide apparently causes PM probes to become more concentrated on fewer microvilli. Notably, the distribution of the GG probe does not change significantly upon C2-ceramide treatments (Fig. 3B and D–H).
Perturbation of ordered-lipid nanodomains affects topographical localization of signaling proteins and degranulation.
Disruption of the ordered-lipid nanodomains has proven useful for investigating their functional relevance (23, 50, 52, 53). We treated the cells with C2-ceramide to determine effects on the topographical localization of FcεRI, Lyn and LAT, before and after activation with antigen. We found that, compared to untreated cells (Fig. 2 I–K), antigen addition to C2-ceramide-treated cells for 15 min does not cause a dramatic topographical change, MC imaging showing only limited elongation/merger of microvilli (Fig. 3 N–P). This observation suggests that C2-ceramide-treated mast cells do not activate normally after engagement with antigen, which is consistent with our previous findings that C2-ceramide inhibits antigen-stimulated signaling events including Ca2+ mobilization and phospholipase D activity (50, 53). To evaluate further, we measured antigen-stimulated degranulation of cells with and without C2-ceramide treatment. We found C2-ceramide essentially prevents stimulated degranulation (Fig. S3 M), consistent with the disruption in treated cells of ordered-lipid nanodomains necessary to initiate signaling. We also tested localization of signaling proteins as follows.
FcεRI:
For cells treated with C2-ceramide, FcεRI still localize preferentially to the microvilli (95 ± 2% and 95 ± 2% on the 0 and 400 nm slices, respectively) like untreated cells (Fig. 3K and R; Fig. S3F and L). However, the fraction of microvilli occupied by FcεRI decreases by 60% from 74±6% to 32±7%, after C2 ceramide-treatment (Fig. 3K and S). Consistent with the substantial topographical and functional disruption by C2ceramide, we found that 15 min activation of C2 ceramide-treated cells does not significantly change the percentage of FcεRI on the microvilli (94 ± 2% and 89 ± 3% on 0 and 400 nm slices, respectively (Fig. 3N and R; Fig. S3I and L). Also, the percentage of microvilli occupied by FcεRI after activation does not change much from resting, C2-ceramide treated cells (both 32%; Fig. 3K, N and S).
Lyn:
The distributional responses of Lyn to C2-ceramide treatment are very similar to those of FcεRI. For resting cells, there is very little effect on Lyn’s predominant localization to microvilli (98 ± 1% and 94 ± 2%in the 0 nm and 400 nm slices, respectively, (Fig. 3L and R; Fig. S3G and L). Further, the percentage of microvilli occupied by Lyn is markedly reduced to less than half that of untreated cells (63 ± 5% to 27 ± 5%, Fig. 3S). Notably, the C2-ceramide-induced decrease in the fraction of microvilli occupied by Lyn parallels the decrease in the fraction of microvilli occupied by the ordered-lipid preferring PM probes in resting cells (Fig. 3F). Lyn still localizes primarily to microvilli after 15 min antigen-activation (89 ± 3% and 87 ± 3% in 0 nm and 400 nm slices, respectively (Fig. 3O and R; Fig. S3J and R), and the fraction of microvilli occupied by Lyn kinase (29 ± 4 %, Fig. 3S) also remains the same as the C2-ceramide-treated, resting cells.
LAT:
In contrast to FcεRI and Lyn, LAT shows ~1.5-fold increased (P value, 5.8212×10−8 <<0.05) localization on the microvilli in C2-ceramide-treated, resting cells compared to untreated cells: 69 ± 3% to 94 ± 2% (Fig. 3 J, M and R) and 68 ± 3% to 91 ± 3% (Fig. S3H and L) in 0 nm and 400 nm slices, respectively. The percentage of microvilli occupied by LAT molecules decreases from 64±4% to 52±2% in C2-ceramide-treated resting cells compared to untreated resting cells ((P value, 0.02883<0.05) Fig. 3S), also differing from FcεRI and Lyn. 15 min after antigen addition to C2-ceramide-treated cells, LAT continues to localize predominantly to microvilli, 91±2% Fig. 3P and R) and 91±2% (Fig. S3K and L) in 0 nm and 400 nm slices, respectively. The fraction of microvilli occupied by LAT appears to increase slightly after antigen-activation of C2-ceramide-treated cells (to 65±5%, Fig. 3S). This variable behavior of LAT after C2-ceramide treatment is not readily interpreted but may reflect previous observations that the order-preference of this scaffold protein is related to its phosphorylation state (44), which is unknown under these conditions.
Lyn and FcεRI localize to mutually exclusive microvilli after C2-ceramide treatment.
Our observation that both FcεRI and Lyn are restricted to roughly half the number of microvilli after C2-ceramide treatment raises the question whether both localize to the same microvillus, or they localize to different microvilli. If they localize to the same microvillus, then we might expect increased interactions and thereby enhanced cell activation, which we do not observe. To investigate we performed co-localization probability (CP) analysis (4, 5, 11) for FcεRI and Lyn in resting RBL cells that were treated or not with C2-ceramide. For that purpose, we labeled FcεRI with Alexa-647 tagged IgE and Lyn with Alexa-488 tagged anti-Lyn antibody. After our usual procedure to fix and label the suspended cells, we performed sequential dual-color SLN imaging to assess the CP values for these molecules in pairs on cells with nanometer resolution (Fig. S4). The distance-dependent CP is the probability that a probe of one type will have at least one partner of the other type within a specified interaction distance. The CP for probes a and b within distance R is given by CP = Nab(R)/Na, where Na is the total number of detected points of probe a, and Nab(R) is the number of points of probe a that have at least one partner of molecule b within R. We found that for resting cells at least ~50% of FcεRI have one Lyn within 100 nm diameter, i.e., within the radius of the microvilli (Fig. S4A–C and G). However, the fraction of IgE-FcεRI that have one Lyn within 20 nm diameter is below 5% for resting cells (Fig. S4G). This indicates that although nearly half of the IgE-FcεRI localize with Lyn kinase molecules in the same microvillus, they are in separate assemblies. This might play a crucial role in controlling the auto-activation of mast cells in the resting state. Notably, in C2-ceramide treated cells, only ~ 5% of FcεRI receptors have one Lyn kinase molecule within 100 nm diameter, i.e., within the same microvillus (Fig. S4D–F and G). This indicates that the majority of IgE-FcεRI and Lyn kinase molecules localize on mutually exclusive microvilli after C2-ceramide treatment.
Cytoskeletal attachment proteins are involved in topographical changes accompanying RBL mast cell activation.
Previous reports suggest that the interplay between the ERM proteins and cofilin controls the membrane topographical changes upon activation of T cells and B cells (5, 28, 29). Activated forms of ERM proteins, i.e. phosphorylated ERM, are shown to stabilize the actin cytoskeleton and thereby support the formation of microvilli (5, 28, 29). The effect of ERM proteins is counterbalanced by cofilin proteins which when dephosphorylated increase actin severing thereby reversing the formation of these membrane protrusions (29). We used antibodies specific for either phosphorylated forms or all forms of ERM and cofilin to label the endogenous ERM and cofilin proteins in RBL mast cells. After tagging with fluorescent secondary antibodies, we used flow cytometry to measure the amount of phosphorylated and total ERM and cofilin proteins in RBL cells in the presence and absence of C2-ceramide under four cell conditions: 1) resting cells not sensitized with IgE (NS), 2) IgE-sensitized cells with no (resting, S), or 3) 1 min, or 4) 15 min antigen activation (Fig. S5A–H). Using the NS cells as a normalizing baseline we calculated the percentage increase shift in the Mean Fluorescent Intensity (MFI) for ERM, P-ERM, cofilin and P-cofilin in the presence and absence of C2-ceramides after antigen activation (Fig. S5I–J). In the absence of C2-ceramide, P-ERM content of the cells appears to increase substantially after antigen-activation for 1 min (57±16%) and 15 min (75±25%). An increasing trend is also seen with total ERM content following antigen-activation for 1 min (32±11%), and 15 min (62±7%).
Visualization of immunoblots (Fig. S5K–L) support the flow cytometry results. The presence of two ERM bands in these blots is consistent with expression of Moesin and Ezrin but not Radixin in mast cells (54); we cannot rule out two bands for P-ERM. The lower values for ERM compared to P-ERM (Fig. S5I) after 1 min indicates that the ratio of P-ERM vs total ERM increases significantly upon antigen activation of mast cells, and this may be related to the initial elongation/merger of microvilli. The ratio of P-ERM to ERM decreases somewhat after 15 min when most of the microvilli have merged into ruffles. Notably, in presence of C2-ceramide neither the P-ERM nor the ERM content increases significantly, after 1 min or 15 min of antigen addition. Considering that C2-ceramide treatment causes FcεRI and Lyn to relocate to mutually exclusive microvilli, these results are consistent with the view that effective coupling of FcεRI with Lyn stimulates new ERM expression, enhanced ERM phosphorylation and merging of microvilli.
In the case of cofilin, the phosphorylated cofilin and total cofilin do not appear to significantly increase after 1 min antigen activation (Fig. S5J). Both forms of the protein increase after 15 min antigen activation, with the increase of P-cofilin (26±8%) about half that of total cofilin (50±15%) (Fig. S5J). Overall, the changes of cofilin after antigen-activation are considerably less compared to ERM proteins as quantified by flow cytometry (Fig. S5I) and visualized in immunoblots (Fig. S5M and N). Because phosphorylation of cofilin inactivates this enzyme, the ratio of active cofilin vs total cofilin appears to increase after longer activation, corresponding to more severing of the actin cytoskeleton by cofilin. This may play a role in transforming microvilli by severing initial cytoskeletal attachments and merging these structures into ruffles. C2-ceramide treatment does not appear to have a significant effect on the expression of P-cofilin or total cofilin. Together our results indicate that FcεRI-mediated stimulation modulate the phosphorylation state of ERM proteins and less so for cofilin proteins.
Discussion
FcεRI concentrates on RBL mast cell microvilli which merge to form ruffles upon antigen activation.
Like B cells, T cells and lymphocytes more generally (38), the plasma membrane of resting mast cells is dominated by dynamic, actin-mediated, finger-like protrusions known as microvilli (3, 35). Previous studies demonstrated concentration of immune receptors in microvilli of B cells and T cells could be directly related to antigen sensing (2, 3, 5, 6, 8, 9, 12). The present study investigated whether the same is true in mast cells which play multiple roles in immune responses, including allergic reactions. RBL mast cells have proved to be an experimentally robust model for examining mechanisms of mast cells more generally (33). For these mast cells, the presence of microvilli that transform to membrane ruffles after antigen-activation has been observed with SEM and fluorescence microscopy (15, 36), but participation of membrane topography in antigen sensing and FcεRI-mediated signaling has been largely unexamined. Recent studies using confocal microscopy, microcartography (MC) and lattice light-sheet microscopy (LLSM) highlighted the role of microvilli to present immune receptors for optimal antigen surveillance and to facilitate stimulated signaling pathways in T-cells and B-cells (2, 3, 5, 6, 8, 9, 12). Our present study extends this investigation by building on extensive studies that have been carried out on signaling in RBL mast cells initiated by FcεRI. We now advance this characterization as enabled by MC, which with VA-TIRFM can nanoscopically image the three-dimensional structure of microvilli and with Stochastic Localization Nanoscopy (SLN) can identify labeled signaling molecules with respect to these topographically defined microvilli. Thus, we can correlate changes in plasma membrane topography of mast cells with distributions of signaling proteins as well as participation of ordered-lipid (Lo-like) nanodomains (colloquially known as “rafts”), which coordinately regulate and target antigen-induced signal initiation. Our experimental results further provide new insight into the role of cytoskeleton attachment proteins, ERM and cofilin, in controlling membrane domains and topography in these cells.
Our present study examined suspended RBL mast cells and the hypothesis that their microvilli pre-organize FcεRI and other membrane components to facilitate transmembrane signaling after the addition of antigen. Previous studies using this MC approach with human effector and Jurkat T cells showed that TCR, co-receptor CD4, tyrosine kinase Lck, and adaptor LAT are substantially enriched in the microvilli on resting cells (2, 5), providing strong evidence that this level of localization yields a signaling hub that is key to T cell capacity to surveil juxtaposed antigen presenting cells (31, 55). Confirming a tight spatial correlation between microvilli and P-ERM (56, 57), Ghosh et al showed that these actin attachment proteins are necessary for localization of TCR to microvilli on resting cells (5). We found that the microvillar structure as defined by MC is similar in the resting state for both T cells and RBL mast cells, and moreover that FcεRI and signaling partners concentrate in these structures. Like T cells and B cells, this level of organization may serve to enhance the capacity of resting mast cells to respond quickly and efficiently to environmental antigens.
Ghosh et al (5) further showed with MC that addition of anti-CD3 to activate TCR on Jurkat cells as they interacted with an ICAM coated surface caused cells to spread quickly on those surfaces. By 3 min, the microvilli largely collapsed and TCRαβ were distributed across the interfacing membrane (5). In contrast, our MC analysis revealed that RBL cell microvilli do not quickly collapse on an antigen-presenting surface or in the presence of soluble antigen, but rather transform within a few minutes to ridge-like ruffles. In comparison, Cai et al used diffraction-limited LLSM images of T cells to identify elongated membrane protrusions as microvilli and to sensitively monitor their dynamics (3). These studies showed TCR clusters on the microvillar structures of resting cells and on actin-independent membrane protrusions within synapses. Park et al used high resolution confocal microscopy to demonstrate TCR on the tips of microvilli that were shed distally when these T cells formed synapses with activating surfaces (12). LLSM used to evaluate the structure and dynamics of membrane topography of Ramos B cells revealed dynamic microvilli interconnected by ridge-like structures (8). IgM-BCRs localize to the tops of these microvilli, evidently by an actin-driven mechanism, and antigen-induced clustering causes their immobilization on the ridges. Thus, antigen activation of RBL mast cells induces microvillar dynamics that differ significantly in comparison to those occurring with their immune cell counterparts, T cells or B cells.
FcεRI and Lyn kinase localize to microvilli in separate protein assemblies that come together with LAT upon antigen activation.
Using MC to examine the distribution of endogenous signaling partners with respect to the 3D membrane topography of RBL mast cells, we found that IgE-FcεRI and Lyn, localize almost entirely in the microvilli on resting cells (Fig. 2). This specific localization resembles that of T cells for which TCR (analogous to FcεRI), Lck (analogous to Lyn), CD2, and CD4 are >80% localized to microvilli (5). We found that antigen engagement causes the microvilli structures to merge into ruffles (Fig. 1) and that both FcεRI and Lyn continue to localize almost exclusively on these protrusions (Fig. 2). Our results are summarized in the schematic of Fig. S6 Microvillar localization of both FcεRI and Lyn on resting mast cells raises the question how auto-activation in the absence of antigen is avoided. Our δCount/δArea vs R plot from MC analysis shows slightly shifted peaks (Fig. 2) indicating that although occupying the same microvillus (~100 nm diameter), FcεRI and Lyn maintain an inherent separation of a few tens of nanometers in the resting state. Further assessment with co-localization probability (CP) analysis clearly demonstrates that, although at least 50% of FcεRI have at least one Lyn partner within 100 nm (i.e. within a single microvillous) in the resting state, the percentage of FcεRI having a Lyn partner within very close proximity, i.e. within 20 nm, is negligible (<5%) (Fig. S4). Thus, despite co-localization on the same microvilli in resting cells, this distinctive separation of FcεRI and Lyn is likely important for preventing spontaneous activation of mast cells. Upon antigen-crosslinking of FcεRI, these receptors are known to co-cluster with Lyn (18, 23, 26, 58). Consistent with those established results, MC analysis shows overlapping δCount/δArea curves, indicating their merger (Fig. 2).
As for T cells, LAT in activated RBL mast cells serves as a scaffold for multiple signaling components, including Grb2, Gads, and SLP76 which connect to phosphorylated LAT via their SH2 motifs and connect to phospholipase-Cγ and other signaling enzymes (37, 59, 60). We determined for resting RBL cells that the larger fraction of LAT proteins localize to microvilli, but a significant smaller fraction (~30%) are found on the flatter cell body surface (Fig. 2). This distribution of LAT molecules in resting RBL mast cells is very similar to that in resting T-cells (5). By a minute after antigen-activation of RBL cells, almost all the LAT molecules localize to the merging microvillar structures (>90%), and the population in non-microvillar regions drastically decreases. Correspondingly, the steepness of δCount/δArea vs R plot increases dramatically (Fig. 2). Remarkably, the shapes of δCount/δArea plots for IgE-FcεRI, Lyn, and LAT are almost the same, indicating that these three proteins assemble together upon activation. These results are consistent with our previous backscatter detection of immuno-gold particles in SEM images of adherent RBL cells. These dorsal surface images showed separation of FcεRI, Lyn, and LAT in the resting state of mast cells and their co-clustering after antigen-crosslinking of FcεRI (58).
Because exceeding the phosphorylation threshold for antigen-crosslinked FcεRI to initiate cytoplasmic signaling involves exclusion of transmembrane phosphatases from these clusters (23, 61), we attempted to investigate their localization in resting and activated cells. For this purpose, we employed genetically encoded PTPα as a good proxy for these phosphatases in RBL mast cells (41), analogous to CD45 which operates in T cells. As monitored with fluorescent anti-HA antibodies, we found that PTPα-HA transiently transfected into resting RBL cells distribute randomly on the membrane surface without apparent selectivity for membrane topography (Fig. 2). We further observed that those cells with a high level of HA-PTPα expression do not undergo the signature membrane topographical changes to ruffles when exposed to antigen and that HA-PTPα transfected cells exhibit significantly reduced degranulation (Fig. S2). Although our experimental results with HA-PTPα may not represent the topographical distribution of endogenous transmembrane phosphatases, they are consistent with the requirement of FcεRI-mediated activation for changes in membrane topography that accompany downstream signaling in suspended cells.
Plasma membrane topography coordinates with ordered-lipid nanodomains to regulate localization in resting and activated cells.
Correlation of microvilli and ordered-lipid nanodomains was recently indicated for T cells by Park et al who found that microvilli bearing TCR are also decorated by added cholera toxin B, which binds to order-preferring ganglioside GM1(12). For a more thorough investigation of nanodomains in RBL mast cells, we employed genetically encoded probes that have been established to partition preferentially into ordered-lipid (palmitate/myristoylate: PM probe) or disordered-lipid (geranylgeranyl: GG probe) regions of the plasma membrane (49). With MC analysis we found that 3HA-PM and GG-3HA localize differentially with respect to membrane topography in resting cells (Fig. 3). The ordered-lipid probe localizes predominantly with the microvilli, like Lyn and FcεRI. In contrast, the disordered-lipid probe prefers the flat, cell-body regions (Fig. 3 and as depicted in Fig. S6). Thus, these results demonstrate that ordered-lipid nanodomains tend to form on plasma membrane microvilli, whereas the membrane in the flat regions is more disordered in character. C2-ceramide treatment to disrupt ordered-lipid nanodomains (50, 52) allowed us to evaluate possible coordination between these domains and actin-mediated membrane microvilli to control localization of FcεRI and Lyn. Although this perturbation does not significantly alter the microvillar-specific localization of FcεRI and Lyn, the fraction of microvilli occupied by each of those proteins decreases dramatically (by 50%) (Fig. 3). Remarkably, CP analysis revealed that those two proteins occupy mutually exclusive microvilli after C2-ceramide treatment (Fig. S4 and as depicted in Fig. S6). These results point to distinctive structural underpinnings for microvilli-selective localization of FcεRI and Lyn.
Lyn is known to partition into ordered-lipid regions as driven by dual palmitoylation and myristolyation of N-terminal amino acids (62), and 3HA-PM retains this segment of Lyn (49). Interestingly, the PM probe also reports that C2-ceramide mediated perturbation causes redistribution of the ordered-lipid domains to ~50% of the microvilli, and this is likely to be the same set of microvilli as the 50% occupied by Lyn. Thus, for resting cells, the localization of Lyn and PM to microvilli is consistent with ordered-lipid nanodomains localized to these membranes. However, this is not a simple explanation for monomeric FcεRI, which exhibit little detectable preference for ordered-lipid regions before crosslinking by multivalent antigen leads to proximal stabilization of ordered-lipids (18, 42). This question is considered further in a subsequent section discussing actin-attachment proteins.
Organization of membrane proteins and lipids in microvilli is mediated by the actin-cytoskeleton.
It is known that parallel actin filaments create microvilli, and their stability is scaffolded by active (phosphorylated) ERM proteins (P-ERM), which locate almost exclusively with the microvillar structures (5, 57). Serving as a counterbalance is active cofilin, which severs the actin cytoskeleton upon dephosphorylation (29, 63, 64). We evaluated possible antigen-induced changes in these actin-attaching proteins that accompany changes in RBL mast cell signaling and membrane topography (Fig. S5). Our flow cytometry results show that by 1 min after antigen stimulation the phosphorylation of ERMs clearly outweighs dephosphorylation of cofilin (P-ERM/ERM > cofilin/P-cofilin), consistent with further enhancement of microvillar structures that are elongating and merging. By 15 min of stimulation the ratio of P-ERM/ERM compared to cofilin/P-cofilin decreases somewhat suggesting that the membrane ruffling occurring at this stage involves a balance of PERM/cofilin differing from that optimal for maintaining individual microvilli. C2-ceramide treatment prevents the increase of both the phosphorylated and total ERM after antigen exposure, consistent with preventing FcεRI-mediated cell activation by disrupting the coupling with Lyn. Our results with RBL cells can be compared to those for DT40 B cells which also showed that a consequence of antigenic stimulation is modulating the phosphorylation pattern of ERM proteins and cofilin. However, different from the RBL cells, simultaneous dephosphorylation of ERM and cofilin in stimulated DT40 B cells leads to local depletion of actin, diminished microvillar structures, and enhanced BCR clustering (29).
We speculate that actin attachment proteins participate in the exclusive localization of Lyn and FcεRI to microvilli in resting mast cells. We previously evaluated the segregation of ordered-lipid vs disordered-lipid regions of the plasma membrane as may be mediated by actin attachments (20). Our SLN measurements in those RBL cell studies are generally consistent with the findings of others that order-preferring attachment proteins can mediate actin contacts with order-preferring membrane proteins in lymphocyte signaling (65, 66). A relevant example is Cbp/PAG, which is strongly order-preferring, a known interaction partner of Lyn, and coupled to the actin cytoskeleton via interactions with EBP-50 and ezrin (67). In this view, actin aligned in the microvilli might stabilize ordered-lipid nanodomains in the adjoining plasma membrane, as mediated in part by P-ERM and PAG-like associations. These nanodomains would also then preferentially include palmitoylated/myristoylated Lyn and other saturated fatty acid anchors (e.g., PM-3HA). That Lyn and FcεRI segregate to separate microvilli after C2-ceramide underscores differential actin association by FcεRI. Crosstalk between the FcεRI and ERM proteins has been observed experimentally in mast cells (68), but particular structural associations have not been identified. Separate studies have demonstrated that Moesin is the dominant ERM protein in mast cells with less Ezrin and little or no Radixin (54).
Microvilli facilitate antigen-sensitive transmembrane signaling in mast cells to cause functional morphological changes.
Our nanoscopic MC examination of RBL mast cells showed that, like T cells, the antigen receptor (FcεRI) concentrates on microvilli together with its key signaling partner (Lyn kinase), providing strong evidence that this assembly serves as an efficient sense-and-response mechanism for antigenic stimuli (5, 31, 55). Antigen activation of T cells, B cells, and mast cells is followed by topographical changes such as those occurring on antigen presenting cells or other activating surfaces. As shown for B cells (29) and RBL mast cells (present study), one consequence of cell activation is modulating the phosphorylation pattern of ERM proteins, which in turn modulates actin attachment in the microvilli (5). Current studies indicate differences among these three types of immune cells regarding the nature of the microvilli collapse (or distal molting) at an activating surface, although these apparent differences may be derived in part to the imaging approach employed (e.g., MC vs LLSM vs confocal microscopy) (5, 10, 12). Our present MC studies on RBL mast cells can be compared directly to those carried out on T cells (5) , which showed rapid microvillar collapse in a T cell synapse. In contrast, mast cell microvilli elongate and merge into ruffles within a minute after engaging a surface-bound or soluble antigen.
We note that MC shows that flattening of RBL mast cells on surfaces is delayed compared to T cells, but that flattening and adherence does occur after about 30 min, as described previously (36, 37) and is observed routinely with these cells in culture or dropped on glass or plastic surfaces. The adherence process involves integrins but FcεRI-mediated signaling does not occur in the absence of antigen (36, 37). Antigen addition causes the adherent cells to spread out farther and ruffles appear on the periphery (36, 37). Thus, although antigen causes microvillar collapse of suspended cells, cell flattening during adherence does not by itself initiate signaling. Subject to further investigation is if and how the spatial relationships and structural associations of FcεRI, Lyn, Lat and ordered-lipid nanodomains change when microvilli flatten on the ventral surface of adherent cells. Overall, our MC results are consistent with previous extensive SLN and ImFCS imaging carried out in TIRF mode on the ventral surface and SEM imaging on the dorsal surface, showing that antigen crosslinking of FcεRI stabilizes an ordered-lipid environment wherein coupling with Lyn and Lat occurs.
Although antigen-activated ruffling has long been observed in mast cells, its physiological role has not been clearly defined. Oliver and co-workers demonstrated previously that ruffling can be separated from cell activation pathways necessary for degranulation (15, 69): Membrane ruffling can be stimulated by phorbol esters, which mimic the PIP2 metabolite diacyl glycerol and act as an agonist for protein kinase C. This is interesting because PKC causes exposure of PIP2, implicated in antigen-stimulated phosphorylation of ERM proteins in B cells (29). However, phorbol ester-stimulated ruffling in mast cells is Ca2+ independent, whereas antigen-stimulated signaling leading to PKC activation and degranulation also requires Ca2+ mobilization (15, 69). After sensitive antigen recognition via the FcεRI-bearing microvilli, stimulated ruffling may be productively involved in mast cell motility. This possibility is consistent with our previous characterization of RBL mast cells moving up a chemical gradient of antigen (70). At low antigenic concentrations within the gradient, the motile cells move toward the source, but at a threshold concentration the cells adhere, spread, ruffle, and begin to degranulate. Mast cells are known to migrate toward source of infection, making physiological sense that membrane dynamics facilitate motion until the stimulus becomes sufficiently strong to cause proximal degranulation to release of chemical mediators of inflammation.
In conclusion, using MC and SEM, we have confirmed with RBL mast cells that microvilli abundantly cover the mast cell surface and that antigen-stimulation causes microvillar elongation and merger into ruffles (Fig. S6). MC analysis has now shown for the first time that the high affinity receptor for IgE, FcεRI, and its initial signaling partner, Lyn tyrosine kinase, concentrate in microvilli of the resting cells. The two proteins group separately but closely within 100 nm dimension of these topographical features. This microvillar-specific pre-organization of the mast cell signaling proteins appears to be maintained by synergy between ordered-lipid nanodomains and ERM proteins that attach the plasma membrane to the actin cytoskeleton. With Lyn and its isolated palmitoylated/myristoylated segment (PM) as order-preferring probes, we found that ordered-lipid domains are almost exclusively localized on the microvilli of the cells, whereas the disorder-preferring probes (GG) distribute preferentially to the flatter cell body regions. This resting state organization is poised for sensitive recognition and response to environmental antigens. Upon engaging the antigen, FcεRI, Lyn, and LAT coalesce near the tips of the microvilli which then elongate and form ruffles within minutes. Simultaneously the phosphorylation pattern of ERMs and cofilin changes, consistent with topographical transformation of the microvilli. Our studies on RBL mast cells found some possibly significant differences from the T cells and B cells that have been investigated, but together they affirm the importance of microvilli for organizing immune receptors, signaling partners, and ordered-lipid nanodomains on microvillar tips. With such pre-organization these immune cells are poised for exquisite antigen sensing, and consequent antigen-stimulated microvillar transformation facilitates the consequent cellular response.
Materials and Methods
MC methodology images and dual color super-resolution images were recorded using a homebuilt TIRF microscope equipped with an electron multiplying charge-coupled device (EMCCD) camera (5, 11, 23). The raw images were further processed using custom-made MATLAB codes (11) to determine the quantitative distribution of membrane protein concerning membrane topography and co-localization probability. The flow cytometry experiments were conducted on a Thermo Fisher Attune NxT flow cytometer. The SEM experiments were carried out on a Keck Leo 1550 SEM. Thorough descriptions of the source of all materials including chemicals and plasmids, instrumental setups, sample preparation, image processing, and quantitative analyses are provided in the SI Appendix.
Supplementary Material
Classification:
Biological Sciences: Biophysics and Computational Biology + Immunology and Inflammation.
Acknowledgments
This work is supported by the National Institute of General Medical Sciences (NIGMS) Grant R01GM117552. SG acknowledges Start-up Research Grant (SRG/2023/000147), SERB-India for financial support during manuscript preparation after experimental work was completed. This work made use of the shared instrumentation facility at the Cornell Center for Materials Research and the BRC Flow Cytometry Facility (RRID:SCR_021740) at the Cornell Institute of Biotechnology. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or NIH.
Footnotes
Competing Interest Statement: The authors declare no competing interest.
Data sharing plans:
The code used in the analysis is available with the previous publication (13). Data and documentation are available upon reasonable request from the corresponding authors.
References
- 1.Cail R. C., Drubin D. G., Membrane curvature as a signal to ensure robustness of diverse cellular processes. Trends Cell Biol (2022). 10.1016/j.tcb.2022.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Jung Y., et al. , Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proceedings of the National Academy of Sciences 113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai E., et al. , Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S., et al. , CCR7 signalosomes are preassembled on tips of lymphocyte microvilli in proximity to LFA-1. Biophys J 120, 4002–4012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh S., et al. , ERM-Dependent Assembly of T Cell Receptor Signaling and Co-stimulatory Molecules on Microvilli prior to Activation. Cell Rep 30, 3434–3447.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kim H.-R., et al. , T cell microvilli constitute immunological synaptosomes that carry messages to antigen-presenting cells. Nat Commun 9, 3630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aramesh M., et al. , Nanoconfinement of microvilli alters gene expression and boosts T cell activation. Proceedings of the National Academy of Sciences 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saltukoglu D., et al. , Plasma membrane topography governs the 3D dynamic localization of IgM B cell antigen receptor clusters. EMBO J 42 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung Y., Wen L., Altman A., Ley K., CD45 pre-exclusion from the tips of T cell microvilli prior to antigen recognition. Nat Commun 12, 3872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben Sahel Y., Dardikman-Yoffe G., Eldar Y. C., Gosh S., Haran G., Super-Resolved Imaging of Early-Stage Dynamics in the Immune Response in 2021 IEEE International Conference on Image Processing (ICIP), (IEEE, 2021), pp. 3468–3472. [Google Scholar]
- 11.Ghosh S., Alcover A., Haran G., “Microvillar Cartography: A Super-Resolution Single-Molecule Imaging Method to Map the Positions of Membrane Proteins with Respect to Cellular Surface Topography” in The Immune Synapse: Methods and Protocols, Baldari C. T., Dustin M. L., Eds. (2023). [DOI] [PubMed] [Google Scholar]
- 12.Park J.-S., et al. , Trogocytic molting of T cell microvilli upregulates T cell receptor surface expression and promotes clonal expansion. Nat Commun 14, 2980 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metcalfe D. D., Baram D., Mekori Y. A., Mast cells. Physiol Rev 77, 1033–1079 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Metzger H., The Receptor with High Affinity for IgE. Immunol Rev 125, 37–48 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Pfeiffer J. R., Seagrave J. C., Davis B. H., Deanin G. G., Oliver J. M., Membrane and cytoskeletal changes associated with IgE-mediated serotonin release from rat basophilic leukemia cells. J Cell Biol 101, 2145–55 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman E., et al. , Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity 35, 705–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone M. B., Shelby S. A., Núñez M. F., Wisser K., Veatch S. L., Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelby S. A., Veatch S. L., Holowka D. A., Baird B. A., Functional nanoscale coupling of Lyn kinase with IgE-FcεRI is restricted by the actin cytoskeleton in early antigen-stimulated signaling. Mol Biol Cell 27, 3645–3658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levental I., Veatch S. L., The Continuing Mystery of Lipid Rafts. J Mol Biol 428, 4749–4764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harder T., Lipid raft domains and protein networks in T-cell receptor signal transduction. Curr Opin Immunol 16, 353–359 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Dinic J., Riehl A., Adler J., Parmryd I., The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci Rep 5, 10082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bag N., London E., Holowka D. A., Baird B. A., Transbilayer Coupling of Lipids in Cells Investigated by Imaging Fluorescence Correlation Spectroscopy. J Phys Chem B 126, 2325–2336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bag N., et al. , Lipid-based and protein-based interactions synergize transmembrane signaling stimulated by antigen clustering of IgE receptors. Proceedings of the National Academy of Sciences 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouches M., Veatch S. L., Machta B. B., Surface densities prewet a near-critical membrane. Proceedings of the National Academy of Sciences 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H.-Y., et al. , Coupling of protein condensates to ordered lipid domains determines functional membrane organization. Sci Adv 9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelby S. A., Holowka D., Baird B., Veatch S. L., Distinct stages of stimulated FcεRI receptor clustering and immobilization are identified through superresolution imaging. Biophys J 105, 2343–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seldin D. C., et al. , Homology of the rat basophilic leukemia cell and the rat mucosal mast cell. Proceedings of the National Academy of Sciences 82, 3871–3875 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faure S., et al. , ERM proteins regulate cytoskeleton relaxation promoting T cell–APC conjugation. Nat Immunol 5, 272–279 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Droubi A., et al. , The inositol 5-phosphatase INPP5B regulates B cell receptor clustering and signaling. Journal of Cell Biology 221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knutton S., Sumner M. C., Pasternak C. A., Role of microvilli in surface changes of synchronized P815Y mastocytoma cells. J Cell Biol 66, 568–576 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orbach R., Su X., Surfing on Membrane Waves: Microvilli, Curved Membranes, and Immune Signaling. Front Immunol 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majstoravich S., et al. , Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood 104, 1396–403 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Falcone F. H., Wan D., Barwary N., Sagi-Eisenberg R., RBL cells as models for in vitro studies of mast cells and basophils. Immunol Rev 282, 47–57 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Govendir M. A., et al. , T cell cytoskeletal forces shape synapse topography for targeted lysis via membrane curvature bias of perforin. Dev Cell 57, 2237–2247.e8 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Oliver J. M., Seagrave J., Stump R. F., Pfeiffer J. R., Deanin G. G., Signal transduction and cellular response in RBL-2H3 mast cells. Prog Allergy 42, 185–245 (1988). [PubMed] [Google Scholar]
- 36.Orth R. N., Wu M., Holowka D. A., Craighead H. G., Baird B. A., Mast Cell Activation on Patterned Lipid Bilayers of Subcellular Dimensions. Langmuir 19, 1599–1605 (2003). [Google Scholar]
- 37.Wakefield D. L., Holowka D., Baird B., The FcεRI signaling cascade and integrin trafficking converge at patterned ligand surfaces. Mol Biol Cell 28, 3383–3396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill H. C., Parish C. R., A rapid, automated colorimetric assay for measuring antibody binding to cell surface antigens. J Immunol Methods 64, 257–268 (1983). [DOI] [PubMed] [Google Scholar]
- 39.Baird B., Sajewski D., Mazlin S., A microtiter plate assay using cellulose acetate filters for measuring cellular [3H]serotonin release. J Immunol Methods 64, 365–375 (1983). [DOI] [PubMed] [Google Scholar]
- 40.Sundd P., et al. , Quantitative dynamic footprinting microscopy reveals mechanisms of neutrophil rolling. Nat Methods 7, 821–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young R. M., Zheng X., Holowka D., Baird B., Reconstitution of regulated phosphorylation of FcepsilonRI by a lipid raft-excluded protein-tyrosine phosphatase. J Biol Chem 280, 1230–5 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Field K. A., Holowka D., Baird B., Fc epsilon RI-mediated recruitment of p53/56lyn to detergent-resistant membrane domains accompanies cellular signaling. Proc Natl Acad Sci U S A 92, 9201–5 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davey A. M., Walvick R. P., Liu Y., Heikal A. A., Sheets E. D., Membrane order and molecular dynamics associated with IgE receptor cross-linking in mast cells. Biophys J 92, 343–55 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levental I., Lingwood D., Grzybek M., Coskun Ü., Simons K., Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proceedings of the National Academy of Sciences 107, 22050–22054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitoh S., et al. , LAT is essential for Fc(epsilon)RI-mediated mast cell activation. Immunity 12, 525–35 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Geldman A., Pallen C. J., Protein tyrosine phosphatases in mast cell signaling. Methods Mol Biol 1220, 269–86 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Fernandes R. A., et al. , A cell topography-based mechanism for ligand discrimination by the T cell receptor. Proceedings of the National Academy of Sciences 116, 14002–14010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger S. A., Mak T. W., Paige C. J., Leukocyte common antigen (CD45) is required for immunoglobulin E-mediated degranulation of mast cells. J Exp Med 180, 471–6 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chisholm K. M., et al. , Mast cells in systemic mastocytosis have distinctly brighter CD45 expression by flow cytometry. Am J Clin Pathol 143, 527–34 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Pyenta P. S., Holowka D., Baird B., Cross-correlation analysis of inner-leaflet-anchored green fluorescent protein co-redistributed with IgE receptors and outer leaflet lipid raft components. Biophys J 80, 2120–32 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holowka D., Thanapuasuwan K., Baird B., Short chain ceramides disrupt immunoreceptor signaling by inhibiting segregation of Lo from Ld Plasma membrane components. Biol Open 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacheco A., et al. , C2-Phytoceramide Perturbs Lipid Rafts and Cell Integrity in Saccharomyces cerevisiae in a Sterol-Dependent Manner. PLoS One 8, e74240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gidwani A., Brown H. A., Holowka D., Baird B., Disruption of lipid order by short-chain ceramides correlates with inhibition of phospholipase D and downstream signaling by FcϵRI. J Cell Sci 116, 3177–3187 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Theoharides T. C., Kempuraj D., Potential Role of Moesin in Regulating Mast Cell Secretion. Int J Mol Sci 24, 12081 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balagopalan L., Raychaudhuri K., Samelson L. E., Microclusters as T Cell Signaling Hubs: Structure, Kinetics, and Regulation. Front Cell Dev Biol 8, 608530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yonemura S., Tsukita S., Tsukita S., Direct Involvement of Ezrin/Radixin/Moesin (ERM)-binding Membrane Proteins in the Organization of Microvilli in Collaboration with Activated ERM Proteins. J Cell Biol 145, 1497–1509 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanono A., Garbett D., Reczek D., Chambers D. N., Bretscher A., EPI64 regulates microvillar subdomains and structure. J Cell Biol 175, 803–813 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veatch S. L., Chiang E. N., Sengupta P., Holowka D. A., Baird B. A., Quantitative nanoscale analysis of IgE-FcεRI clustering and coupling to early signaling proteins. J Phys Chem B 116, 6923–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balagopalan L., Kortum R. L., Coussens N. P., Barr V. A., Samelson L. E., The Linker for Activation of T Cells (LAT) Signaling Hub: From Signaling Complexes to Microclusters. Journal of Biological Chemistry 290, 26422–26429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman M. A., Shoag J., Wu J., Koretzky G. A., Disruption of SLP-76 Interaction with Gads Inhibits Dynamic Clustering of SLP-76 and FcεRI Signaling in Mast Cells. Mol Cell Biol 26, 1826–1838 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holowka D., Baird B., Roles for lipid heterogeneity in immunoreceptor signaling. Biochim Biophys Acta 1861, 830–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kovářová M., et al. , Structure-Function Analysis of Lyn Kinase Association with Lipid Rafts and Initiation of Early Signaling Events after Fcɛ Receptor I Aggregation. Mol Cell Biol 21, 8318–8328 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernstein B. W., Bamburg J. R., ADF/Cofilin: a functional node in cell biology. Trends Cell Biol 20, 187–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M. S., Huse M., Phollow the phosphoinositol: Actin dynamics at the B cell immune synapse. Journal of Cell Biology 221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Head B. P., Patel H. H., Insel P. A., Interaction of membrane/lipid rafts with the cytoskeleton: Impact on signaling and function. Biochimica et Biophysica Acta (BBA) - Biomembranes 1838, 532–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Viola A., Gupta N., Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nat Rev Immunol 7, 889–896 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Hrdinka M., Horejsi V., PAG - a multipurpose transmembrane adaptor protein. Oncogene 33, 4881–4892 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Hálová I., et al. , Cross-talk between Tetraspanin CD9 and Transmembrane Adaptor Protein Non-T Cell Activation Linker (NTAL) in Mast Cell Activation and Chemotaxis. Journal of Biological Chemistry 288, 9801–9814 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliver J. M., Seagrave J. C., Pfeiffer J. R., Feibig M. L., Deanin G. G., Surface functions during mitosis in rat basophilic leukemia cells. J Cell Biol 101, 2156–66 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J., Veatch S. L., Baird B., Holowka D., Molecular mechanisms of spontaneous and directed mast cell motility. J Leukoc Biol 92, 1029–1041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code used in the analysis is available with the previous publication (13). Data and documentation are available upon reasonable request from the corresponding authors.