Abstract
We describe new algorithms and modules for protein structure prediction available as part of the PROTINFO web server. The modules, comparative and de novo modelling, have significantly improved back-end algorithms that were rigorously evaluated at the sixth meeting on the Critical Assessment of Protein Structure Prediction methods. We were one of four server groups invited to make an oral presentation (only the best performing groups are asked to do so). These two modules allow a user to submit a protein sequence and return atomic coordinates representing the tertiary structure of that protein. The PROTINFO server is available at http://protinfo.compbio.washington.edu.
INTRODUCTION
Protein structure mediates protein function in biological processes that are essential for the survival and development of an organism. We describe a set of new modules and enhancements to our previously published PROTINFO web server (http://protinfo.compbio.washington.edu) (1) for predicting protein structure. A caveats section on the module page is constantly updated to reflect performance and accuracy issues.
PROTEIN TERTIARY STRUCTURE PREDICTION
There are two primary categories of methods for 3D (tertiary structure) modelling: comparative modelling (CM) and de novo prediction (AB). In CM (which includes distant homology and fold recognition), the methodologies rely on the presence of one or more evolutionarily related template protein structures that are used to construct models. In the AB category, there is no strong dependence on database information, and prediction methods are based on general principles that govern protein structure and energetics. The categories vary in difficulty, and consequently methods in each of these categories produce models with different levels of accuracy relative to the experimental structures.
The 3D modelling methods are based on our published research (1–9) and use some software developed as part of the RAMP suite of programs. The source code for the RAMP software, along with more detailed documentation, is accessible from our software distribution server (http://software.compbio.washington.edu/ramp/).
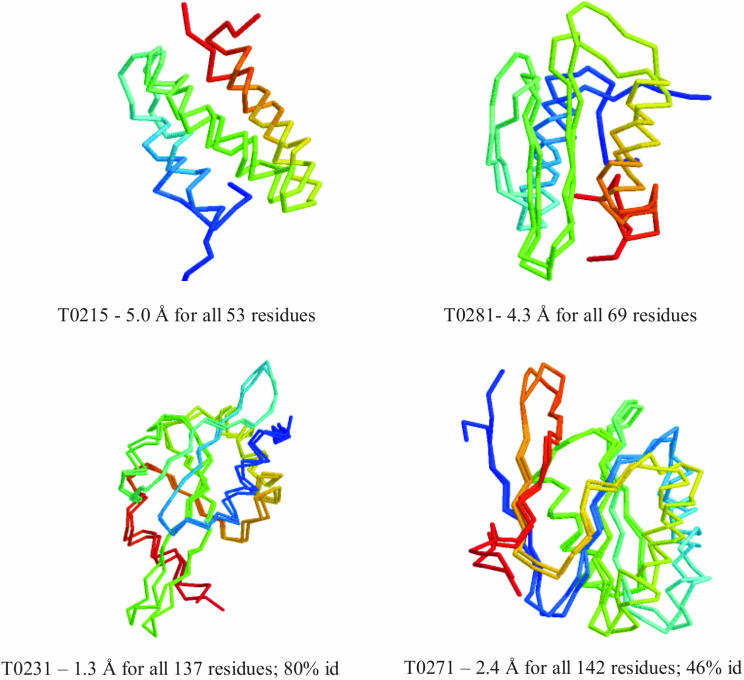
Protein structure prediction methods are rigorously evaluated by the Critical Assessment of Structure Prediction methods (CASP, and CAFASP for ‘fully automated’) experiments held every two years (10) (http://predictioncenter.llnl.gov). We have taken part in all six CASP experiments, including the most recent one (CASP6) that finished in December 2004 (5,7,11,12). The results provide a benchmark as to what level of model accuracy we can expect from our methodologies. Our server modules were one of four server groups invited to make an oral presentation at CASP6 (Figure 1), with the organizers judging the performance of our servers as being particularly good for targets with no obvious detectable homology. Detailed analysis of our CASP6 predictions in relationship to those made by others is at http://predictioncenter.llnl.gov/casp6/. Figure 1 illustrates the general accuracy of our methods using a few examples. The changes in both our CM and AB prediction protocols since CASP5 are described in detail below.
Figure 1.
Examples of selected CASP6 AB (top) and CM (bottom) predictions made by the PROTINFO server. All models are model 1. The superposition of the model and the corresponding experimental structure is shown, along with the Cα RMSD relative to the experimental structure. The percentage identity of the alignment between the target and the most similar template sequence is given for the bottom two CM targets.
De novo prediction
Our protocol for the automated prediction of protein structure from sequence alone is very different from the one used in the previous version of PROTINFO. Structures are generated using a simulated annealing search phase that minimizes a target scoring function. Moves are derived from a synthetic function that produces continuous ϕ/ψ angular distributions similar to the empirically observed distribution for that secondary structure type. In contrast to fragment-based methods [which we used at CASP5 (1)], this is accomplished without copying any angles or coordinates. The angle-distribution-based move generation method is both new and unique to our group. We have also added two new phases to our simulations in addition to the main search phase. In cases where strands are present, a pre-condensation phase encourages strand pairing, increasing the likelihood of proper strand formation by several orders of magnitude. Because our simulations include side chains, and we have a continuous main chain representation, we can explore subtle differences in conformations. To exploit this, we have added a post-minimization phase that uses Brent's method and small angular moves to search the local energy minimum and further reduce the target function. Finally, the target function itself has been optimized resulting in a 10–15-fold increase in speed without loss of accuracy (0.99 correlation to original function), allowing exploration of many more conformations in the same time.
The selection procedure has also been improved. In addition to the original scoring functions, several new functions are introduced and used for decoy filtering. ‘Alp’, ‘Phipsi’ and ‘Sol’ are based on the probability of a residue adopting a particular virtual torsion angle, ϕ/ψ state and degree of exposure to water, respectively. ‘Coord’ is based on the probability of a residue being within a prescribed cut-off distance with respect to other hydrophobic, hydrophilic and neutral residues, ‘Conseq’ determines the probability of a pair of residue having a particular distance between them, taking into account the degree of conservation of the residues and whether their distance in sequence is above or below six-residue cut-off. ‘Curv’ determines the probability of having a given triplet of residues being within a certain distance from each other. Finally, ‘Rad’ is based on the probability of a given residue being at a certain distance from the centre of the protein.
There are altogether 15 scoring functions [8 that existed in the previous version of the server (1) and the 7 described above]. Each function is individually normalized by subtracting its own mean of scores for all the decoys and dividing by the standard deviation. These normalized functions are then combined to form a set of 19 hierarchical filters, used in filtering the decoys and in forming consensus among the remaining decoys. Finally, a new iterative density protocol, where the centre of the cluster is recalculated as outliers are discarded, is used to choose the final five conformers (9).
Comparative modelling
If a user supplies only a sequence, the server does a search using a variety of sequence-only methods and then uses the ‘hits’ returned as seeds for a multiple sequence alignment. A user may also specify a template structure and its alignment to the target sequence. Initial models are then built for each alignment to a template and the resulting models are scored using an all-atom function (3,9,13). Loops and side chains are built on the best scoring models using a frozen approximation (4). A sophisticated graph-theory search to mix and match between various main chain and side chain conformations is used to generate the final model when appropriate (3). The primary difference in this module, as compared with what we published in (1), is that the mixing and matching using a clique finding algorithm (14) has been implemented and made fully automated. This server's main strengths are in building non-conserved regions of main chains (typically loops) and side chains. For best template detection and alignments, we suggest that they first be obtained from the Bioinfo meta-server (http://bioinfo.pl/meta/) (15) and submitted using the optional input fields.
OTHER MODULES
Two other modules available as part of the PROTINFO server, already described elsewhere, are PsiCSI (1,8) for the secondary structure prediction and PIRSpred (16–19) for the prediction of HIV drug effectiveness.
INPUT AND OUTPUT FORMATS AND BEHAVIOUR
Input formats and behaviour
Sequences must be specified in a single line using the one-letter amino acid notation. Splitting up longer sequences into domains if knowledge of the domain boundaries available is prudent. This is because the complexities of most calculations are generally exponentially proportional to the lengths of the sequences, and most prediction methods are calibrated to work on domains. The programs currently perform a limited amount of automatic domain parsing, which will be enhanced in the future.
Very short (<30 residues) and very long sequences are not likely to generate reliable predictions. Any PDB files submitted optionally must generally start with residue 1 and the residues must be numbered consecutively without any chain breaks. There is some support for cleaning up the PDB files submitted.
Output formats and behaviour
Following the convention used in the experiments on the CASP, up to five models for each tertiary prediction module (CM, AB) will be returned (in the CASP format). Under certain conditions (e.g. when no clear relationship to a template is discerned), both methods may be executed by the PROTINFO server regardless of the method requested. Detailed output is available for both as part of the file that is emailed back to the recipient.
CALCULATION TIMES AND CURRENT USAGE FOR ALL MODULES
The PIRSpred and PsiCSI modules run within seconds and return results immediately. The publicly available tertiary structure prediction modules are typically executed on a cluster with 64 dedicated CPUs. Our goal is to ensure that the prediction time for each sequence is <24 h (CM predictions will most probably take only a few hours), but this depends on the number of sequences submitted and their lengths. There is a feature to monitor the progress of submissions.
Currently, the tertiary structure prediction modules sometimes receive 30–40 sequences per day, which requires far more capacity than the computational resources allocated to handling them. Thus, a response might not be sent for several days in a worst-case scenario. Nonetheless, given the detailed model building capabilities of the server, we feel it is a useful resource for the study of protein structure. We expect to dedicate more computational resources in the near future.
FUTURE WORK
Enhancements planned for the near future include a module to predict the tertiary structure of proteins given noisy or limited NMR data along with our de novo methods; a module to assess binding energies/affinities of substrate–protein interactions of any protein; and a module for protein–protein docking calculations.
Acknowledgments
We thank past and present members of the Levitt, Moult and Samudrala groups, and John Mittler, for their intellectual guidance and support that helped in the development of these servers. This work was supported in part by a Searle Scholar Award, NSF Grant DBI-0217241, NIH Grant GM068152 and the University of Washington's Advanced Technology Initiative in Infectious Diseases. Funding to pay the Open Access publication charges for this article was provided by a Searle Scholar Award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hung L.-H., Samudrala R. PROTINFO: secondary and tertiary protein structure prediction. Nucleic Acids Res. 2003;31:3736–3737. doi: 10.1093/nar/gkg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samudrala R., Xia Y., Levitt M., Huang E. A combined approach for ab initio construction of low resolution protein tertiary structures from sequence. In: Altman R., Dunker A., Hunter L., Klein T., Lauderdale K., editors. Proceedings of the Pacific Symposium on Biocomputing. Honolulu, Hawaii: World Scientific Press; 1999. pp. 505–516. [DOI] [PubMed] [Google Scholar]
- 3.Samudrala R., Moult J. A graph-theoretic algorithm for comparative modelling of protein structure. J. Mol. Biol. 1998;279:287–302. doi: 10.1006/jmbi.1998.1689. [DOI] [PubMed] [Google Scholar]
- 4.Samudrala R., Moult J. Determinants of side chain conformational preferences in protein structures. Protein Eng. 1998;11:991–997. doi: 10.1093/protein/11.11.991. [DOI] [PubMed] [Google Scholar]
- 5.Samudrala R., Xia Y., Huang E., Levitt M. Ab initio protein structure prediction using a combined hierarchical approach. Proteins. 1999;S3:194–198. doi: 10.1002/(sici)1097-0134(1999)37:3+<194::aid-prot24>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Samudrala R., Huang E., Koehl P., Levitt M. Side chain construction on near-native main chains for ab initio protein structure prediction. Protein Eng. 2000;7:453–457. doi: 10.1093/protein/13.7.453. [DOI] [PubMed] [Google Scholar]
- 7.Samudrala R., Levitt M. A comprehensive analysis of 40 blind protein structure predictions. BMC Struct. Biol. 2002;2:3–18. doi: 10.1186/1472-6807-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung L.-H., Samudrala R. Accurate and automated assignment of secondary structure with PsiCSI. Protein Sci. 2003;12:288–295. doi: 10.1110/ps.0222303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Fain B., Levitt M., Samudrala R. Improved protein structure selection using decoy-dependent discriminatory functions. BMC Struct. Biol. 2004;4:296. doi: 10.1186/1472-6807-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moult J., Hubbard T., Fidelis K., Pedersen J. Critical Assessment of Methods of Protein Structure Prediction (CASP): Round III. Proteins. 1999;3(Suppl):2–6. [PubMed] [Google Scholar]
- 11.Samudrala R., Pedersen J., Zhou H., Luo R., Fidelis K., Moult J. Confronting the problem of interconnected structural changes in the comparative modelling of proteins. Proteins. 1995;23:327–336. doi: 10.1002/prot.340230307. [DOI] [PubMed] [Google Scholar]
- 12.Samudrala R., Moult J. Handling context-sensitivity in protein structures using graph theory: bona fide prediction. Proteins. 1997;29(Suppl.):43–49. doi: 10.1002/(sici)1097-0134(1997)1+<43::aid-prot7>3.3.co;2-z. [DOI] [PubMed] [Google Scholar]
- 13.Samudrala R., Moult J. An all-atom distance dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- 14.Bron C., Kerbosch J. Algorithm 457: finding all cliques of an undirected graph. Commun. ACM. 1973;16:575–577. [Google Scholar]
- 15.Ginalski K., Elofsson A., Fischer D., Rychlewski L. 3D-Jury: a simple approach to improve protein structure prediction. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 16.Wang K., Jenwitheesuk E., Samudrala R., Mittler J. Simple linear model provides highly accurate genotypic predictions of hiv-1 drug resistance. Antivir. Ther. 2004;9:343–352. [PubMed] [Google Scholar]
- 17.Jenwitheesuk E., Samudrala R. Prediction of HIV-1 protease inhibitor resistance using a protein-inhibitor flexible docking approach. Antivir. Ther. 2005;10:157–166. [PubMed] [Google Scholar]
- 18.Jenwitheesuk R., Wang K., Mittler J., Samudrala R. Improved accuracy of hiv-1 genotypic susceptibility interpretation using a consensus approach. AIDS. 2004;18:1858–1859. doi: 10.1097/00002030-200409030-00020. [DOI] [PubMed] [Google Scholar]
- 19.Jenwitheesuk E., Wang K., Mittler J., Samudrala R. Pirspred: a web server for reliable hiv-1 protein-inhibitor resistance/susceptibility prediction. Trends Microbiol. 2005;13:150–151. doi: 10.1016/j.tim.2005.02.003. [DOI] [PubMed] [Google Scholar]