Abstract
HHpred is a fast server for remote protein homology detection and structure prediction and is the first to implement pairwise comparison of profile hidden Markov models (HMMs). It allows to search a wide choice of databases, such as the PDB, SCOP, Pfam, SMART, COGs and CDD. It accepts a single query sequence or a multiple alignment as input. Within only a few minutes it returns the search results in a user-friendly format similar to that of PSI-BLAST. Search options include local or global alignment and scoring secondary structure similarity. HHpred can produce pairwise query-template alignments, multiple alignments of the query with a set of templates selected from the search results, as well as 3D structural models that are calculated by the MODELLER software from these alignments. A detailed help facility is available. As a demonstration, we analyze the sequence of SpoVT, a transcriptional regulator from Bacillus subtilis. HHpred can be accessed at http://protevo.eb.tuebingen.mpg.de/hhpred.
INTRODUCTION
It is well known that sequence search methods such as BLAST, FASTA or PSI-BLAST (1–3) are of prime importance for biological research because functional information of a protein or gene can be inferred from homologous proteins or genes identified in a sequence search. But quite often no significant relationship to a protein of known function can be established. This is certainly the case for the most interesting group of proteins, those for which no ortholog has yet been studied.
It is less well known that in cases where conventional sequence search methods fail, the recently developed, highly sensitive methods for homology detection or structure prediction (confer, e.g. (4–11) and descriptions and links at http://bioinfo.pl/Meta/servers.html) quite often allow to make inferences from more remotely homologous relationships (12–17). If the relationship is so remote that no common function can be assumed, one can generally still derive hypotheses about possible mechanisms, active site positions and residues, or the class of substrate bound (18,19). When a homologous protein with known structure can be identified, its stucture can be used as a template to model the 3D structure for the protein of interest (5), since even remotely homologous proteins generally have quite similar 3D structure (20). The 3D model may then help to generate hypotheses to guide experiments.
The primary aim in developing HHpred was to provide biologists with a method for sequence database searching that is as easy to use as BLAST or PSI-BLAST and yet competitive in sensitivity with the most powerful servers for structure prediction that are currently available. We believe that HHpred is unique in the advantages it offers:
Speed: A search with a 300 residue sequence through the Protein Data Bank (PDB) (≈9000 HMMs) takes ∼1 min.
Databases: A wide range of regularly updated structure and protein family databases can be searched: the PDB (21), SCOP (22), Pfam (23), SMART (24), COG (25) and CDD (26).
User-friendliness: Search results are presented in an easy-to-read format that is similar to PSI-BLAST. The summary hit list includes E-values and true probabilities. Alignments contain annotation about secondary structure, consensus sequences and position-specific reliability and they can be augmented by representative sequences from the underlying multiple alignments.
Flexibility: We try to offer the user maximum control and flexibility. He can paste his own input query alignment, search in local or global alignment mode, realign alignments with other parameters and edit the query-template (multiple) alignment with which to launch the comparative modelling.
Multi-domain proteins: HHpred has been designed to work equally well for single-domain and multi-domain query sequences. It can therefore be used to predict domain boundaries.
Documentation: A comprehensive help facility is available.
Selectivity: High-scoring false positives have systematically been reduced by developing a protocol for building query and database alignments that supresses non-homologous sequences (J. Söding, to be published).
Sensitivity: HHpred is among the most sensitive servers for remote homology detection. A comparison of the new version HHpred2.1 with the servers that took part in the recent structure prediction benchmark CAFASP4 (27) can be viewed at http://protevo.eb.tuebingen.mpg.de/hhpred/hhpred_in_CAFASP4.html. In a recent study (28), in which we benchmarked HHsearch, the method for HMM–HMM comparison employed by our server, together with PSI-BLAST, HMMER, PROF_SIM and COMPASS (3,6,7,29), HHsearch was found to possesses the highest sensitivity and alignment accuracy.
METHODS AND INPUT PARAMETERS
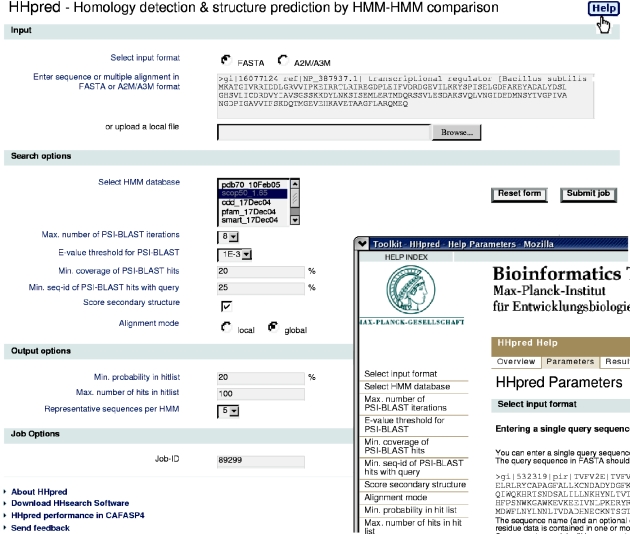
In the first step, an alignment of homologs is built for the query sequence by multiple iterations of PSI-BLAST searches against the non-redundant database from NCBI. The maximum number of PSI-BLAST iterations and the E-value threshold can be specified on the start page (Figure 1). Instead of a single sequence, the user may also enter a multiple alignment to jumpstart PSI-BLAST, or he can choose to skip the PSI-BLAST iterations altogether by choosing zero for the maximum number of PSI-BLAST iterations.
Figure 1.
Start page for the HHpred server at http://protevo.eb.tuebingen.mpg.de/hhpred with part of a help window overlaid.
The user can further specify a minimum coverage of the query by the PSI-BLAST matches. With a value of 50%, at least half of the query residues must be aligned (‘covered’) with residues from the matched sequence in order for it to enter into the profile. Similarly, a minimum sequence identity of the PSI-BLAST match to the query sequence can be demanded. Our benchmarks (data not published) have shown that a value between 20 and 25% improves selectivity without compromising sensitivity. The final alignment from PSI-BLAST is annotated with the predicted secondary structure and confidence values from PSIPRED (30).
In the next step, a profile HMM is generated from the multiple alignment that includes the information about predicted secondary structure. A profile HMM is a concise statistical description of the underlying alignment. For each column in the multiple alignment that has a residue in the query sequence, an HMM column is created that contains the probabilities of each of the 20 amino acids, plus 4 probabilities that describe how often amino acids are inserted and deleted at this position (insert open/extend, delete open/extend). These insert/delete probabilites are translated into position-specific gap penalties when an HMM is aligned to a sequence or to another HMM.
The query HMM is then compared with each HMM in the selected database. The database HMMs have been precalculated and also contain secondary structure information, either predicted by PSIPRED, or assigned from 3D structure by DSSP (31). The database search is performed with the HHsearch software for HMM–HMM comparison (28). Compared to methods that rely on pairwise comparison of simple sequence profiles, HHsearch gains sensitivity by using position-specific gap penalties. If the default setting ‘Score secondary structure’ is active, a score for the secondary structure similarity is added to the total score. This increases the sensitivity for homologous proteins considerably (28). As a possible drawback, it may lead to marginally significant scores for structurally analogous, but non-homologous proteins.
The user can choose between local and global alignment mode. In global mode alignments extend in both directions up to the end of either the query or the database HMM. No penalties are charged for end gaps. In local mode, the highest-scoring local alignment is determined, which can start and end anywhere with respect to the compared HMMs. It is recommended to use the local alignment mode as a default setting since it has been shown in our benchmarks to be on average more sensitive in detecting remote relationships as well as being more robust in the estimation of statistical significance values. A global search might be appropriate when one expects the database entries to be (at least marginally) similar over their full length with the query sequence. In most cases it will be advisable to run a search in both modes to gain confidence in one's results.
EXAMPLE ANALYSIS
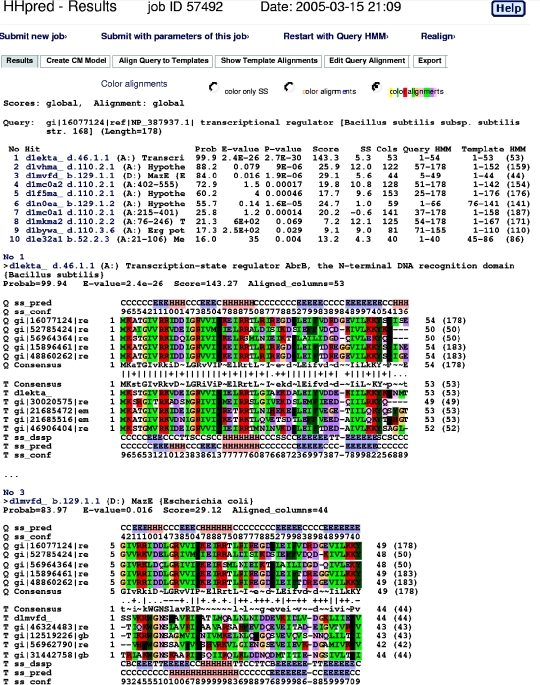
As an example we analyze the sequence of Stage V sporulation protein T (SpoVT) from Bacillus subtilis that is known to regulate forespore-specific σG-dependent transcription (32) (annotated as ‘transcriptional regulator’ in GenBank). Input parameters are set as shown in Figure 1. The results consist of two parts (Figure 2): a summary list with matching database sequences (‘templates’) and a list of query–template alignments below.
Figure 2.
Search results for SpoVT from Bacillus subtilis. The summary hit list at the top shows that SpoVT consists of two domains: the N-terminal domain is very similar to AbrB (rank 1) and clearly homologous to MazE (rank 3) and the C-terminal domain is similar to GAF and PAS domains (rank 2, 4–9). In the summary hit list, column ‘Prob’ gives the probability that the hit is homologous to the query. This is the principle measure of statistical significance. In the alignments below, the sequences marked ‘Q’ (‘T’) refer to the query (template) alignment. Sequences ‘ss_pred’ and ‘ss_conf’ denote the PSI-PRED secondary structure prediction and confidence values, ‘ss_dssp’ is the secondary strcuture assigned by DSSP. Upper an lower case amino acids in the consensus sequences indicate high (≳60%) and moderate (≳40%) conservation, respectively. Symbols indicating the quality of the column–column match: ‘|’ very good, ‘+’ good, ‘·’ neutral, ‘−’ bad and ‘=’ very bad.
The first column of the summary hit list has indices that link to the corresponding alignment further down. Next are the first 30 characters from the description of the HMM. The ‘Prob’ column lists the probability in percent that the database match is a true positive, i.e. that it is homologous to the query sequence at least in some core part. This is the most relevant statistical measure of significance and can be interpreted quite literally. The true-positive probability is a conservative measure in the sense that it corrects for occasional high-scoring false positives. (The major cause for high-scoring false positives are corrupted alignments that contain non-homologous sequences which slipped in during the automized alignment-building with PSI-BLAST.) [See (28) for details.] The E-values in HHpred are defined in the same way as in BLAST or PSI-BLAST. (The E-value for a sequence match is the expected number of false positives per database search with a score at least as good as the score of this sequence match.) But it is important to note that, in contrast to the true-positive probability, HHpred E-values do not take into account the secondary structure similarity. Hits can therefore be significant by the true-positive probability criterion even when the E-value is ∼1. The P-value is equal to the E-value divided by the number of HMMs in the searched database. The ‘Score’ column gives the total score that includes the score from the secondary structure comparison which is listed in the next column (‘SS’). ‘Cols’ contains the total number of matched columns in the query–template alignment and the remaining columns describe the range of aligned residues in the query and template.
From the summary list in Figure 2 it is evident that the SpoVT protein consists of two domains, one from residue 1 to ∼51 and the other from residue 52 to 178. The N-terminal domain has two significant hits in SCOP at rank 1 and 3. The first hit is the DNA-binding domain of transition-state regulator AbrB (33), a known close homolog of SpoVT. AbrB is a protein that is broadly represented in bacterial species and is involved in switching from exponential growth to stationary phase by integrating a great number of environmental factors. The second hit is to MazE, the antidote of the antidote-toxin addiction module MazEF (34). How can both AbrB and MazE be homologous to the query if they are not even classified into the same class, let alone fold or superfamily, by the SCOP database? Can the match with MazE be a false positive despite the rather significant 84% probability?
To elucidate this, we can look at the SpoVT–MazE alignment below. Five representative (i.e. maximally diverse) sequences from each of the two underlying alignments are shown for each HMM. (Their amino acids can be colored by biochemical properties by pressing one of the radio buttons entitled ‘color alignments’ above the summary hit list.) First we note that the predicted secondary structure of SpoVT (sequence ‘Q ss_pred’) agrees very well with the actual secondary structure of MazE determined by the program DSSP (sequence ‘T ss_dssp’). Second, the hydrophobicity pattern in the aligned HMMs looks quite similar, which is especially evident with the coloring. Third, the HMM–HMM alignment contains a single gap in MazE at a position where also some sequences in SpoVT exhibit a gap. All in all, the alignment looks very much like what one would expect for a distant homologous relationship.
The conflict posed by the manifest homology between MazE and AbrB and their grossly different structural topology prompted us to undertake a thorough bioinformatic investigation of the AbrB-like superfamily and to redetermine the AbrB structure by NMR (M. Coles and S. Djuranovic et al., manuscript submitted, PDB ID: 1YFB). Indeed, we found that the published structure of AbrB (PDB ID: 1EKT) is incorrect and that the correct structure for AbrB places it in the same superfamily as MazE.
Hits 2 and 4–9 in the summary list are all proteins from the same SCOP fold d.110. Clicking on the SCOP family IDs opens a window with the corresponding entry in SCOP. Irrespective of the specific significance values, the fact that so many quite divergent members from the same two superfamilies d.110.2 (GAF-domain) and d.110.3 (PAS-domain) appear among the best hits strongly indicates that these are not high-scoring chance hits but true homologs. Whether the C-terminal domain looks more like a GAF or a PAS domain, we can now generate an approximate structural model that could help us to guide experiments to investigate what regulatory substrate this domain may actually bind (32).
By clicking ‘Create CM Model’ one can select the templates to be used for comparative modelling. HHpred then returns a multiple alignment in PIR format with the query sequence and the selected templates. This aligment may be edited by the user and then fed to the MODELLER software (35), accessible via the MPI toolkit for users of HHpred.
A very useful feature is the possibility to view and manually improve the query alignment that was used to generate the query HMM; via the tab ‘Edit Query Alignment’ the user can modify the query alignment that appears in a text field and start a new search with the modified alignment.
By pressing ‘Realign’ at the top, the user may also realign the identified templates in the summary hit list with different parameters without the need to rerun the database search. One can change the alignment mode from global to local, set the number of representative sequences or use filters to narrow down the set of sequences allowed into the query and template alignments. If the user wants to search another database with the same query HMM, she can select ‘Restart with Query HMM’.
CONCLUSION
Whenever biologists cannot get satisfactory results from BLAST, PSI-BLAST or other database searches due to insignificant matches with proteins of known structure or function, they should consider using one of the recently developed sensitive structure prediction and homology detection servers (4–11) that are listed, for instance, on the LiveBench/CAFASP site at http://bioinfo.pl/Meta/servers.html. Among these servers, HHpred offers a high degree of flexibility and user-friendliness combined with excellent sensitivity. In contrast to methods based on profile–profile comparison, HHpred exploits the information that is contained in insert and delete probabilities by including them in a statistical framework. But the speed of HHpred is perhaps the most important advantage, considering that the best-ranked servers in CAFASP4 generally take hours or even days to return a prediction. The speed enables the user to tweak the performance and gain confidence in the results by modifying input alignments, search parameters or selected databases on a trial and error basis.
Acknowledgments
We would like to thank Michael Remmert for his valuable help in setting up the web interface. We thank Sergej Djuranovic for first pointing out the HHpred prediction for the SpoVT C-terminal domain. J.S. is indebted to Alex Diemand for assistance in preparing the screenshots. Many thanks to all users who helped to improve our software with their questions and feedback. Funding to pay the Open Access publication charges for this article was provided by the Max-Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Pearson W.R. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rychlewski L., Zhang B., Godzik A. Fold and function predictions for Mycoplasma genitalium proteins. Fold Des. 1998;3:229–238. doi: 10.1016/S1359-0278(98)00034-0. [DOI] [PubMed] [Google Scholar]
- 6.Yona G., Levitt M. Within the twilight zone: a sensitive profile–profile comparison tool based on information theory. J. Mol. Biol. 2002;315:1257–1275. doi: 10.1006/jmbi.2001.5293. [DOI] [PubMed] [Google Scholar]
- 7.Sadreyev R.I., Grishin N.V. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 8.vonÖhsen N., Sommer I., Zimmer R. Profile–profile alignment: a powerful tool for protein structure prediction. Pac. Symp. Biocomput. 2003:252–263. [PubMed] [Google Scholar]
- 9.Panchenko A.R. Finding weak similarities between proteins by sequence profile comparison. Nucleic Acids Res. 2003;31:683–689. doi: 10.1093/nar/gkg154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer D. 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51:434–441. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- 11.Ginalski K., Elofsson A., Fischer D., Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 12.Venclovas C., Thelen M.P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng M., Ginalski K., Rychlewski L., Grishin N. Protein domain of unknown function DUF1023 is an alpha/beta hydrolase. Proteins. 2005;59:1–6. doi: 10.1002/prot.20388. [DOI] [PubMed] [Google Scholar]
- 14.Ginalski K., Rychlewski L., Baker D., Grishin N.V. Protein structure prediction for the male-specific region of the human Y chromosome. Proc. Natl Acad. Sci. USA. 2004;101:2305–2310. doi: 10.1073/pnas.0306306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rand T.A., Ginalski K., Grishin N.V., Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl Acad. Sci. USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlak S.D., Radlinska M., Chmiel A.A., Bujnicki J.M., Skowronek K.J. Inference of relationships in the ‘twilight zone’ of homology using a combination of bioinformatics and site-directed mutagenesis: a case study of restriction endonucleases Bsp6I and PvuII. Nucleic Acids Res. 2005;33:661–671. doi: 10.1093/nar/gki213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara D., Skolnick J. Microbial genomes have over 72% structure assignment by the threading algorithm PROSPECTOR_Q. Proteins. 2004;55:464–473. doi: 10.1002/prot.20044. [DOI] [PubMed] [Google Scholar]
- 18.Todd A.E., Orengo C.A., Thornton J.M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 19.Pawlowski K., Jaroszewski L., Rychlewski L., Godzik A. Sensitive sequence comparison as protein function predictor. Pac. Symp. Biocomput. 2000:42–53. doi: 10.1142/9789814447331_0005. [DOI] [PubMed] [Google Scholar]
- 20.Kinch L., Grishin N. Evolution of protein structures and functions. Curr. Opin. Struct. Biol. 2002;12:400–408. doi: 10.1016/s0959-440x(02)00338-x. [DOI] [PubMed] [Google Scholar]
- 21.Bourne P.E., Addess K.J., Bluhm W.F., Chen L., Deshpande N., Feng Z., Fleri W., Green R., Merino-Ott J.C., Townsend-Merino W., et al. The distribution and query systems of the RCSB protein data bank. Nucleic Acid Res. 2004;32:D223–D225. doi: 10.1093/nar/gkh096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Sonnhammer E.L., Eddy S.R., Birney E., Bateman A., Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting C.P., Schultz J., Milpetz F., Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;24:229–232. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41–41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A., Panchenko A., Shoemaker B., Thiessen P., Geer L., Bryant S. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer D., Rychlewski L., Dunbrack R.L.J., Ortiz A.R., Elofsson A. CAFASP3: the third critical assessment of fully automated structure prediction methods. Proteins. 2003;53:503–516. doi: 10.1002/prot.10538. [DOI] [PubMed] [Google Scholar]
- 28.Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 29.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 30.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Dong T.C., Cutting S.M., Lewis R.J. DNA-binding studies on the Bacillus subtilis transcriptional regulator and AbrB homologue, SpoVT. FEMS Microbiol. Lett. 2004;233:247–256. doi: 10.1016/j.femsle.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly M., Devine K.M. Expression of AbrB, a transition state regulator from Bacillus subtilis, is growth phase dependent in a manner resembling that of Fis, the nucleoid binding protein from Escherichia coli. J. Bacteriol. 1997;179:522–529. doi: 10.1128/jb.179.2.522-529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada K., Hanaoka F., Burley S.K. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 35.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]