Abstract
The FFAS03 server provides a web interface to the third generation of the profile–profile alignment and fold-recognition algorithm of fold and function assignment system (FFAS) [L. Rychlewski, L. Jaroszewski, W. Li and A. Godzik (2000), Protein Sci., 9, 232–241]. Profile–profile algorithms use information present in sequences of homologous proteins to amplify the patterns defining the family. As a result, they enable detection of remote homologies beyond the reach of other methods. FFAS, initially developed in 2000, is consistently one of the best ranked fold prediction methods in the CAFASP and LiveBench competitions. It is also used by several fold-recognition consensus methods and meta-servers. The FFAS03 server accepts a user supplied protein sequence and automatically generates a profile, which is then compared with several sets of sequence profiles of proteins from PDB, COG, PFAM and SCOP. The profile databases used by the server are automatically updated with the latest structural and sequence information. The server provides access to the alignment analysis, multiple alignment, and comparative modeling tools. Access to the server is open for both academic and commercial researchers. The FFAS03 server is available at http://ffas.burnham.org.
INTRODUCTION
The most effective methods of protein structure and function predictions are based on establishing a homology between the protein of interest and an already characterized protein. The standard sequence–sequence comparison methods, however, rapidly lose sensitivity in the ‘twilight zone’ of 30% or less sequence identity (1). The sensitivity of homology recognition can be improved by using information present in the families of protein sequences connected with detectable homology. In this approach, one compares a protein sequence with a protein family represented by a sequence profile [e.g. in PSI-BLAST (2)]. A next step in this strategy is to compare two sequence profiles.
The fold and function assignment system (FFAS) is a profile–profile comparison algorithm developed in 2000 by our group (3). Profile–profile scoring was used earlier to align short blocks (4), and FFAS extended this approach to allow for gaps and align entire proteins. Profile–profile alignment algorithms surpass sequence-sequence and profile-sequence alignment algorithms in terms of sensitivity (3) and alignment accuracy (5). FFAS is regularly assessed in CASP (6) and CAFASP (7) competitions and continually benchmarked in LiveBench (8) experiment. In the last LiveBench, it was ranked as the most sensitive of all sequence-based methods in the category of difficult fold prediction (see http://bioinfo.pl/Meta/results.pl?B=LiveBench&V=9). Development of FFAS was followed by many similar methods that differ in the way two profiles are compared with each other (9–13).
FFAS ALGORITHM
Each profile–profile alignment method includes four steps: (i) preparation of the multiple sequence alignment, (ii) calculation of a profile, (iii) alignment of profile with sequence profiles from the database such as PDB and (iv) estimation of the statistical significance of the alignment score.
In FFAS method, the multiple sequence alignment is prepared using PSI-BLAST (2). Five iterations of PSI-BLAST are performed against the NR85S database of protein sequences (NR85S database is described in Table 1).
Table 1.
The databases used by the FFAS03 server
| Database | Source of data | Preparation |
|---|---|---|
| NR85S (sequences) | NCBI, SEED | Protein sequences from the NCBI NR database and predicted open reading frames from unfinished bacterial genomes (kindly provided by Ross Overbeek) are clustered at 85% of sequence identity with the CD-HIT program (15). Regions of low complexity are masked with SEG (16). |
| PDB (profiles) | Protein Data Bank | FFAS profiles of all unique proteins (clustered at 99% identity level) from the PDB (17), including prereleased entries. |
| PFAM (profiles) | PFAM website | FFAS profiles of all PFAM (18) domains longer than 25 residues. |
| COG (profiles) | NCBI | FFAS profiles of all domains from COG database longer than 25 residues (19). |
| SCOP (profiles) | SCOP–ASTRAL website | FFAS profiles of SCOP domain sequences with <40% sequence identity to each other. SCOP protein sequences clustered at 40% of sequence identity have been downloaded from the Astral website (20). |
| JCSG (profiles) | JCSG website | FFAS profiles of all sequences of active targets of the Joint Center for Structural Genomics (21). |
In the second step, all sequences found by PSI-BLAST with E-value <−0.005 are used for profile calculation. Weights are assigned to sequences based on their similarity to other sequences in the multiple sequence alignment (3).
The value of the comparison score between positions n and m from the two profiles is calculated as a dot product of the nth column from the first profile and the mth column from the second profile. After assigning values to all positions, the matrix is normalized. The optimal alignment is calculated by a dynamic programming algorithm (14). In the last step, the raw alignment score is translated into the final FFAS score by comparing it with the distribution of raw scores obtained for pairs of unrelated proteins. A detailed description of the first version of FFAS method is given in (3). The current version is based on the same approach with modifications in profile comparison and scoring system and will be a subject of a separate publication.
FFAS03 SERVER
Overview
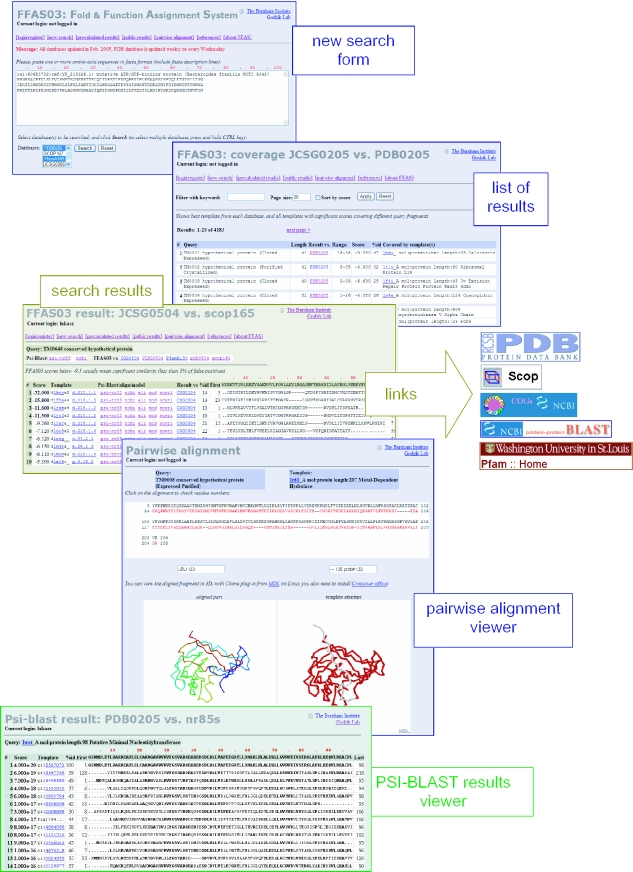
FFAS03 server allows searching of five databases of protein profiles (see Table 1): PDB (17), PFAM (18), COG (19), SCOP (22) and active targets of Joint Center for Structural Genomics (21). The user can select one or more of these databases using a selection window in the new search form (see Figure 1).
Figure 1.
The overview of FFAS pages.
By default, the results are shown on a (public results) page, but private password-protected user accounts can also be used. All results contain links to the appropriate pages from the website of the corresponding database and links to additional analysis and modeling tools (see Figure 1).
Submitting a job to FFAS03
The FFAS03 server accepts sequences between 25 and 2000 residues, but best results are obtained for proteins with lengths between 50 and 500 residues and containing no more than two domains. Sequences longer that 500 residues and/or expected to contain multiple protein domains should be split into shorter fragments (see below). Sensitivity of FFAS method decreases for sequences shorter than 50 residues.
To initiate a search, a user pastes the protein sequence(s) into the input field of the new search form (available through [new search] link located in the upper part of the FFAS03 page), selects profile database(s) to be searched (multiple databases can be selected by holding a CTRL key) and clicks a search button. Information on the status of the search is displayed and updated automatically every 20 s. When the search is completed, the browser displays the contents of the account where the search results are stored. The profile–profile alignment of two arbitrary amino acid sequences can be calculated by following a [pairwise alignment] link located in the upper part of the FFAS03 page.
FFAS03 results
FFAS03 search results are displayed in a block ‘master–slave’ alignment format (see Figure 1). Lower FFAS scores indicate higher confidence of the prediction. According to our benchmarks, predictions with scores lower than −9.5 (shown in bold font on the results page) contain <3% of false positives.
In addition to the alignments, the results page contains corresponding scores, sequence identities and starting and ending residue numbers. It also contains links to PDB SCOP, COG or PFAM entries corresponding to all aligned sequences. The PSI-BLAST multiple sequence alignments used to calculate FFAS profiles can be displayed. Users can also automatically start PSI-BLAST search at the NCBI website with any sequence or sequence region included in the alignment by clicking [ncbi] link. The individual pairwise alignments between sequences can be analyzed by clicking the [ali] link. For templates from PDB and SCOP databases, one can build a homology model of the query protein by clicking the [scwrl] link. This link points to the SCRWL (23) web server and provides it with the alignment between the query and the template. The page also contains links to the precalculated FFAS03 results related to the sequences shown on the page. These links allow extensive exploration of the sequence similarity neighborhood of the query and enable intermediate searches (see below).
Suggested strategies for remote homology predictions with FFAS03
When the direct FFAS03 search does not give compelling results, one can still obtain interesting prediction by applying additional strategies and reliability criteria. The same strategies can be used to further verify reliable FFAS results.
Splitting a query sequence into domains
All sequences expected to contain multiple domains should be split into fragments corresponding to putative domains. Specialized algorithms for protein domain prediction, such as DomPred (24) and GlobPlot (25), are available on the web. Moreover, the FFAS03 server itself can be used to assign putative domains to the sequence. Quite often FFAS03 detects similarity between some fragment of the query sequence and a protein or protein domain from one of the databases. Then, naturally, any fragment of the query, which is not included in the corresponding alignment and is longer than 50 residues, can be treated as a putative new domain. Such fragments can be subject of separate FFAS03 searches. PSI-BLAST generated multiple alignment available on the server can also provide hints about domain boundaries.
The quality of the profiles
FFAS predictions obtained with profiles based on a large number of homologs are more reliable than predictions made with profiles based on only few homologs (3). Therefore, it is informative to examine the multiple sequence alignment used to calculate FFAS profiles (available via the [psi-nr85] link). The most reliable profiles are based on numerous homologs with the same conserved regions present in most of the aligned sequences. On the contrary, profiles calculated from the PSI-BLAST alignments containing a large number of low-complexity or coiled-coil proteins and lacking well-conserved sequence motifs often yield false-positive predictions.
Intermediate sequence search
PSI-BLAST search used to calculate the FFAS profile strongly depends on the sequence used to initiate it. Results obtained for homologs of the original query can provide additional information. Consequently, prediction results can be improved by submitting additional FFAS03 jobs started from distant homologs of the original query. Results of this intermediate sequence strategy are, nevertheless, significantly less reliable than results of direct searches, since the probability of false-positive prediction accumulates.
The consistency of the predictions
By checking structural and functional consistency of predictions, one can gain additional insights about their reliability. For example, results of FFAS03 searches against SCOP database contain structural classification codes. If the same region of the query is aligned with two domains classified as different folds, then one or both of these predictions are incorrect (unless fold classification is incorrect). Other simple criteria of reliability include functional analogies between the query and the template (if both are functionally annotated) and the conservation of the active site.
Server updates
Since the most important purpose of the FFAS03 server is to provide up-to-date fold assignments for submitted protein sequences, the database of protein profiles corresponding to sequences from the PDB database is automatically updated once a week. User results and precalculated results are, however, not automatically updated. In order to update such results, one has to resubmit the queries to the server.
All the databases used by the FFAS03 server undergo a full update every 3 months. Full update includes downloading and clustering of the current NR and SEED databases, calculation of all protein profiles for sequences from PDB, PFAM, COG, SCOP and JCSG databases and updating precalculated results.
User accounts
FFAS03 server allows a user to create a password-protected account by clicking [login/register] link located in the upper part of the FFAS03 page. Existing user accounts are accessible from the same form. After login all results of the searches performed by the user are automatically stored in her/his account.
APPLICATIONS
Selected examples of publications inspired by FFAS predictions
FFAS method predicted twilight-zone similarity between NB-ARC domain present in APAF-1 and CED4 proteins and the family of AAA+ family of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes (27). The model explained some known experimental data about CED4-mediated caspase activation and, at the same time, suggested experiments that could test existing hypotheses. The FFAS prediction was recently confirmed by the experimental determination of the APAF-1 structure (26).
FFAS03 server was used to build 3D models of tubulin cofactors, including several previously unannotated domains of cofactors B–E (28). It identified the new HEAT and Armadillo domains in cofactor D and an unusual spectrin-like domain in cofactor C and a new subfamily of ubiquitin-like domains in tubulin cofactors B and E. Some of these observations were recently confirmed by experiment (29).
The models based on FFAS03 alignments were used in the molecular replacement phasing method to solve over 50 crystallographic structures from the JCSG consortium. The accuracy of the alignment used to build search models for molecular replacement is often critical to the convergence of the method. Molecular replacement search models based on the FFAS alignments allowed determination of several protein structures impossible to solve with less accurate models (30).
Consensus methods and meta-servers
FFAS03 predictions are being used by other prediction methods, such as Robetta (31) and 3D-Jury (32). They are also available through homology prediction meta-servers, such as Bioinfobank Metaserver at http://bioinfo.pl/meta. Meta-servers and consensus methods use the email service of FFAS03 available at http://ffas.ljcrf.edu/ffas/mailffas03.html. Because of the limited throughput of the FFAS03 server, the authors of meta-servers and consensus methods are asked to get approval from the authors before connecting their methods to FFAS03 email service.
FUTURE PLANS
Besides maintenance and regular updates of the FFAS03 server, we are planning to improve existing elements of the server and to develop several new features.
First, the pool of sequence profiles available for searches will be increased. The protein databases available for FFAS03 searches need to be more adequately represented by sequence profiles. Currently, each COG or PFAM domain is represented only by one sequence profile. For many diverse protein families, one profile is probably not sufficient. Such families can be better represented by clustering their sequences at some level of similarity and calculating separate sequence profiles representing each cluster.
Second, sequence profiles representing new databases, such as PFAMB and PRODOM, can be added to the server. Since FFAS03 searches are CPU demanding, such extension will require significant hardware upgrade.
Third, since the number of precalculated and private search results stored on the server is growing, we are planning to implement an advanced query system, which would allow filters, such as “Display FFAS results containing the word ‘apoptotic’ in the description of any FFAS hit” or ‘Show only the results containing bacterial proteins’.
Finally, the most interesting challenge is the improvement of the FFAS algorithm itself. It is related to one of the most intriguing and, still not solved problems of computational biology that is deciphering the relationship between proteins sequence and its structure. Ironically the success of profile–profile methods only emphasized the problem—the methods based solely on sequence information still give results comparable with the results of the methods utilizing structural information (and, most such methods also rely on profile–profile comparison). It seems that, so far, structural information is most beneficial in consensus methods, such as 3D-Jury, where it is used to evaluate the consistence of the alignments obtained with different methods.
We are planning new computational experiments concentrated on the most intriguing cases of close structural similarity which could not be predicted before both structures were determined. Structural genomics initiative aimed at solving structures of proteins without detectable sequence similarity to known structures repeatedly provides such examples.
The authors would like to encourage input from the server users concerning the possible improvements of the server.
Acknowledgments
The authors would like to thank their colleagues at Joint Center for Structural Genomics and the Burnham Institute, and in particular Drs Robert Schwarzenbacher and Yuzhen Ye, for support and help in maintaining and development of the server and the method. Funding to pay the Open Access publication charges for this article was provided by The Burnham Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holm L., Ouzounis C., Sander C., Tuparev G., Vriend G. A database of protein structure families with common folding motifs. Protein Sci. 1992;1:1691–1698. doi: 10.1002/pro.5560011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden T.L., Schaeffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rychlewski L., Jaroszewski L., Li W., Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaroszewski L., Rychlewski L., Godzik A. Improving the quality of twilight-zone alignments. Protein Sci. 2000;9:1487–1496. doi: 10.1110/ps.9.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moult J., Fidelis K., Zemla A., Hubbard T. Critical assessment of methods of protein structure prediction (CASP)-round V. Proteins. 2003;53(Suppl. 6):334–339. doi: 10.1002/prot.10556. [DOI] [PubMed] [Google Scholar]
- 7.Fischer D., Barret C., Bryson K., Elofsson A., Godzik A., Jones D., Karplus K.J., Kelley L.A., MacCallum R.M., Pawowski K., et al. CAFASP-1: critical assessment of fully automated structure prediction methods. Proteins. 1999;(Suppl. 3):209–217. doi: 10.1002/(sici)1097-0134(1999)37:3+<209::aid-prot27>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 8.Bujnicki J.M., Elofsson A., Fischer D., Rychlewski L. LiveBench-1: continuous benchmarking of protein structure prediction servers. Protein Sci. 2001;10:352–361. doi: 10.1110/ps.40501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yona G., Levitt M. Within the twilight zone: a sensitive profile–profile comparison tool based on information theory. J. Mol. Biol. 2002;315:1257–1275. doi: 10.1006/jmbi.2001.5293. [DOI] [PubMed] [Google Scholar]
- 10.Sadreyev R.I., Baker D., Grishin N.V. Profile–profile comparisons by COMPASS predict intricate homologies between protein families. Protein Sci. 2003;12:2262–2272. doi: 10.1110/ps.03197403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlson T., Wallner B., Elofsson A. Profile–profile methods provide improved fold-recognition: a study of different profile–profile alignment methods. Proteins. 2004;57:188–197. doi: 10.1002/prot.20184. [DOI] [PubMed] [Google Scholar]
- 12.Marti-Renom M.A., Madhusudhan M.S., Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G., Dunbrack R.L., Jr Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Li W., Jaroszewski L., Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 16.Wootton J.C., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 17.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandonia J.M., Hon G., Walker N.S., Lo Conte L., Koehl P., Levitt M., Brenner S.E. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T., et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl Acad. Sci. USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Canutescu A.A., Shelenkov A.A., Dunbrack R.L., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsden R.L., McGuffin L.J., Jones D.T. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 2002;11:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linding R., Russell R.B., Neduva V., Gibson T.J. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl S.J., Li W., Chao Y., Schwarzenbacher R., Shi Y. Structure and mechanism of the apoptotic protease activating factor 1 (Apaf-1) Nature. 2005 doi: 10.1038/nature03465. in press. [DOI] [PubMed] [Google Scholar]
- 27.Jaroszewski L., Rychlewski L., Reed J.C., Godzik A. ATP-activated oligomerization as a mechanism for apoptosis regulation: fold and mechanism prediction for CED-4. Proteins. 2000;39:197–203. doi: 10.1002/(sici)1097-0134(20000515)39:3<197::aid-prot10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 28.Grynberg M., Jaroszewski L., Godzik A. Domain analysis of the tubulin cofactor system: a model for tubulin folding and dimerization. BMC Bioinformatics. 2003;4:46. doi: 10.1186/1471-2105-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lytle B.L., Peterson F.C., Qiu S.H., Luo M., Zhao Q., Markley J.L., Volkman B.F. Solution structure of a ubiquitin-like domain from tubulin-binding cofactor B. J. Biol. Chem. 2004;279:46787–46793. doi: 10.1074/jbc.M409422200. [DOI] [PubMed] [Google Scholar]
- 30.Schwarzenbacher R., Godzik A., Grzechnik S.K., Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1229–1236. doi: 10.1107/S0907444904010145. [DOI] [PubMed] [Google Scholar]
- 31.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginalski K., Elofsson A., Fischer D., Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]