Abstract
Off-target effects are one of the most serious problems in RNA interference (RNAi). Here, we present dsCheck (http://dsCheck.RNAi.jp/), web-based online software for estimating off-target effects caused by the long double-stranded RNA (dsRNA) used in RNAi studies. In the biochemical process of RNAi, the long dsRNA is cleaved by Dicer into short-interfering RNA (siRNA) cocktails. The software simulates this process and investigates individual 19 nt substrings of the long dsRNA. Subsequently, the software promptly enumerates a list of potential off-target gene candidates based on the order of off-target effects using its novel algorithm, which significantly improves both the efficiency and the sensitivity of the homology search. The website not only provides a rigorous off-target search to verify previously designed dsRNA sequences but also presents ‘off-target minimized’ dsRNA design, which is essential for reliable experiments in RNAi-based functional genomics.
INTRODUCTION
RNA interference (RNAi) is now widely used to knockdown gene expression in a sequence-specific manner, making it a powerful tool for studying gene function (1–3). The process of RNAi is mediated by double-stranded RNA (dsRNA) that contains a sequence homologous to the target mRNA. Long dsRNA introduced into the cell is cleaved by the enzyme Dicer into short-interfering RNA (siRNA) followed by incorporation into the RNA-induced silencing complex (RISC), which is responsible for target mRNA degradation (4).
One of the most serious problems in RNAi is ‘off-target’ silencing effects (5). Off-target silencing effects are caused by siRNA (introduced directly into cells, or produced in vivo from long dsRNA) that has sequence similarities with unrelated genes. In Caenorhabditis elegans, Drosophila or plants, RNAi experiments are usually performed using long dsRNAs. In these cases, there is a high risk of cross-suppression or co-suppression between closely related genes that share a highly conserved region.
To minimize the possibility of off-target effects, it is necessary to perform an off-target search to design dsRNA or siRNA that has limited sequence similarities with unrelated genes. Recently, fast and sensitive off-target search software for siRNA design has been reported (6,7), but commonly used siRNA design servers are not useful in performing off-target searches for long dsRNAs. DEQOR server uses BLAST to perform off-target searches for endoribonuclease-prepared siRNAs (8), although BLAST frequently fails to identify off-targets (6). Therefore, we have developed a new web-based online software system, dsCheck, to provide fast and accurate off-target searches for long dsRNA sequences. The software ‘dices’ the input sequence into an siRNA cocktail and performs an exhaustive scan for each siRNA to find off-target gene candidates, simulating the biochemical process of dsRNA-mediated RNAi in vivo. dsCheck also provides efficient design of ‘off-target minimized’ dsRNA by avoiding regions that share a considerable number of diced siRNAs with a specific off-target gene, and monitoring the total number of off-target hits. The software should be especially useful for checking whether previously designed dsRNAs have off-target gene candidates, as well as for designing target-specific dsRNA when off-target effects are suspected.
METHODS
Off-target search strategies for long dsRNA
The key idea of the program follows the biochemical process of dsRNA-mediated RNAi shown in Figure 1A. The input dsRNA sequence is diced into 19 nt substrings of an siRNA cocktail, and an exhaustive off-target search is performed for all individual siRNAs using the siDirect engine, which makes it possible to enumerate the complete set of off-targets in a reasonable amount of time (7). In dsCheck, the in silico dicing size is set to 19, as a complete match at the 19 nt double-stranded region of an siRNA is sufficient for the target mRNA degradation. For example, an input 500 bp dsRNA sequence is processed into 482 substrings each 19 nt in length, which are subjected to the off-target search individually. In the next step, all the hits with a complete match (i.e. 19/19 matches), one mismatch (18/19 matches) or two mismatches (17/19 matches) are counted individually for every off-target gene candidate and sorted in descending lexicographic order for the output.
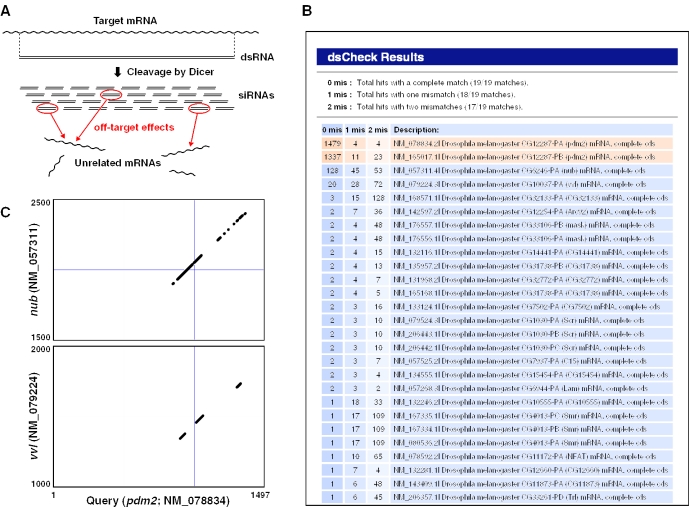
Figure 1.
(A) Biochemical process of dsRNA-mediated RNAi. Off-target effects are caused by ‘diced’ siRNAs that have sequence similarities with unrelated genes. (B) The output for the 1497 bp query sequence of the Drosophila pdm2 gene (NM_078834, coding region). Significant hits against two off-target genes, nub (NM_057311) and vvl (NM_079224) were detected. (C) pdm, nub and vvl share a highly conserved POU domain. Each dot represents a position with 17/19 or more matches.
Figure 1B shows a typical output for a 1497 bp query sequence of the Drosophila POU domain protein, pdm2 (NM_078834, coding region). The result shows significant hits against pdm2 (two splicing variants: NM_078834 and NM_165017), and two unrelated genes, nub (NM_057311) and vvl (NM_079224). These proteins share the highly conserved POU domain shown in Figure 1C, indicating a high risk of cross-suppression by dsRNA targeting this region.
Designing off-target minimized dsRNA sequences
To design off-target minimized dsRNA sequences, one approach would be to suppose that the off-target effects are caused by a considerable number of collaborative hits by diced siRNAs on the same gene, and to select a region that minimizes the maximum number of collaborative off-target hits, which are defined as complete or partial matches of multiple 19 nt substrings against the same off-target gene. According to this criterion, dsCheck starts by selecting a region that minimizes the maximum number of ‘complete match’ collaborative off-target hits. If multiple regions are optimal, it also examines the maximum number of ‘partial match’ collaborative off-target hits to select the best one. If the complete match, collaborative hits on a sequence exceed 80% of the total number of diced 19 nt substrings, dsCheck regards the sequence as the intended target gene.
Some dsRNA sequences include 19 nt substrings that may react with a large number of off-target genes, which differs from the collaborative silencing effects acting on a single off-target gene. An additional criterion is necessary to evaluate the silencing effect of one siRNA sequence on many off-targets, although the effect may not be as serious as the collaborative silencing effect, as the concentration of single siRNA is low in diced siRNA cocktails. One reasonable measure would be the total number of off-target hits for each 19 nt substring of designed dsRNA. To attract attention to this risk, dsCheck displays a warning if the total number of off-target hits exceeds a specified threshold.
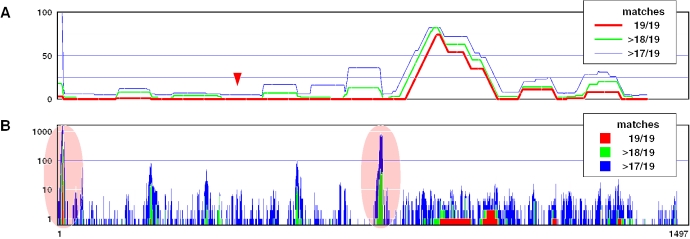
Figure 2 illustrates how dsCheck designs target-specific dsRNA for the Drosophila pdm2 gene (NM_078834, coding region). Given that the length of dsRNA is 100 bp, dsCheck returns the positions 424–523 for the target-specific region that successfully avoids the collaborative silencing effects on the major off-target genes nub (NM_057311) and vvl (NM_079224).
Figure 2.
Designing ‘off-target minimized’ dsRNA for the Drosophila pdm2 gene (NM_078834, coding region). (A) The maximum number of ‘collaborative off-target hits’ by 82 adjacent 19 nt substrings in 100 bp dsRNAs. The arrowhead indicates the recommended region for designing an off-target minimized dsRNA of 100 bp in length. (B) Total number of off-target hits. The 19 nt substrings in the shaded area may react with a large number of off-target genes.
Efficacy of each diced siRNA
In mammalian RNAi, the efficacy of each siRNA varies widely depending on its sequence; hence, several groups have reported guidelines for the selection of siRNAs (9–12). However, in Drosophila cells, it is reported that most, if not all, siRNA sequences may act as effective silencers (9). Incorporation of siRNA efficacy prediction may run the risk of underestimating off-target effects in non-mammalian RNAi. Therefore, all siRNA sequences are treated equally in dsCheck.
Database maintenance
Currently, off-target searches can be performed against the Drosophila, C.elegans, Arabidopsis and Oryza sativa mRNA sequences stored in the NCBI RefSeq database (13). Since off-target searches demand a substantial number of mRNA sequences that are likely to cover the entire set of transcripts, we plan to incorporate additional species when ample cDNA collections are available.
Acknowledgments
This work was supported in part by the Special Coordination Fund for Promoting Science and Technology to K.S., the Leading Project for Biosimulation to S.M. and Grants-in-Aid for Scientific Research to K.U.-T., K.S. and S.M. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: short RNAs that silence gene expression. Nature Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 3.Mello C.C., Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 4.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 5.Jackson A.L., Linsley P.S. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Naito Y., Yamada T., Ui-Tei K., Morishita S., Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–W129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada T., Morishita S. Accelerated off-target search algorithm for siRNA. Bioinformatics. 2005;21:1316–1324. doi: 10.1093/bioinformatics/bti155. [DOI] [PubMed] [Google Scholar]
- 8.Henschel A., Buchholz F., Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–W120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Ueda R., Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 11.Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 12.Chalk A.M., Wahlestedt C., Sonnhammer E.L. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 13.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]