Abstract
FOOTER is a newly developed algorithm that analyzes homologous mammalian promoter sequences in order to identify transcriptional DNA regulatory ‘signals’. FOOTER uses prior knowledge about the binding site preferences of the transcription factors (TFs) in the form of position-specific scoring matrices (PSSMs). The PSSM models are generated from known mammalian binding sites from the TRANSFAC database. In a test set of 72 confirmed binding sites (most of them not present in TRANSFAC) of 19 TFs, it exhibited 83% sensitivity and 72% specificity. FOOTER is accessible over the web at http://biodev.hgen.pitt.edu/Footer/.
INTRODUCTION
Identifying DNA regulatory ‘signals’ in the promoter regions of genes is still one of the challenging problems in computational biology. Part of the problem is that the transcription factor binding sites (TFBSs) are usually short DNA sequences (6–20 bp) with high degree of degeneracy. Algorithms, such as AlignACE (1), ANN-Spec (2), Consensus (3), Co-bind (4) and MEME (5) to name a few, try to address the problem of low signal-to-noise ratio by looking at sets of genes considered to be enriched in one or more TFBS motifs [for a recent comparative study of these methods, we refer the reader to the excellent review of Tompa et al. (6)]. The various oligonucleotides in the test set that could be targets of a transcription factor (TF) are scored against some ‘random’ background. The methods primarily differ on the way they calculate the background and the objective function they try to optimize. These widely used methods fall under the category generally known as de novo ‘pattern discovery’ methods. A consequence of this methodology is that usually there is not much information about the TFs that bind to the identified patterns.
Although these methods are very useful, when knowledge about the binding preferences of a TF exists, there is no reason for one to ignore it. Thus, a second category of methods has been developed, the ‘pattern identification’ methods, that use existing information about the binding preferences of certain TFs and tries to identify the exact location of the motifs in a given promoter sequence. The same problem of low signal-to-noise ratio exists here, especially when one analyzes promoter sequences from complex eukaryotes, such as human, mouse or fly. The gene regulation in these organisms is usually more complex and the promoter length can extend to many kilobases from the transcription start site (TSS). In this case, evolutionary information comes to the rescue. Homologous promoter sequences can be compared in order to identify the evolutionary conserved DNA regulatory signals. This is commonly known as phylogenetic footprinting, a term first coined by Tagle et al. (7). Two of the most widely used algorithms for analyzing mammalian sequences are rVista (8) and ConSite (9). These algorithms are using the evolutionary conservation information in a fundamentally different way: rVista scans one of the promoters for high-scoring TFBSs and then uses position conservation to eliminate false positives. ConSite scans both promoters for motifs that score higher than a position-specific scoring matrix (PSSM) score threshold and then it uses a ‘sliding window’ approach to decide about the position conservation of the putative TFBSs.
We recently developed a novel phylogenetic footprinting algorithm, named FOOTER, which combines two statistics in order to score a pair of putative regulatory sites. Our method scans both promoter sequences and for each TF, it retains the top K scoring sites (‘seed’ TFBSs) in each promoter and then it compares all against all in order to find the best matching pairs according to the two criteria. This method has been shown to perform very well in a set of 72 confirmed TFBS of 19 TFs (SN = 83%, SP = 72%) (25).
METHODS
Given two homologous promoter sequences and a number of putative motifs identified in each of them (by default FOOTER retains one top scoring motif per TF per 300 bp of promoter sequence), our method performs all pairwise comparisons of the motifs. A scoring scheme based on two statistics has been employed. The first statistic scores a pair of motifs according to their position conservation in the sequence. The second statistic scores the pair of motifs according to their agreement with the corresponding PSSM model(s). A PSSM model is the most commonly used way to represent the binding preferences of a TF (10). Typically, a set of aligned sequences is used to calculate a 4 × L weight matrix (L is the length of the pattern). In each column, the weights correspond to the log-likelihood of the preferences of the TF to each of the four bases (sometimes normalized for the background), and in some cases it has been shown that they correspond to the actual binding energies of the protein–DNA interactions (11–13).
The two statistics FOOTER employs consist of the P-values of the observed data, under the null hypothesis that the two sites are unrelated. The position-related score is calculated using the following formula:
| 1 |
where DXY is the random variable denoting the distance between two putative sites, d is the observed distance of the particular putative sites (measured from the 3′ closest conserved region boundary), N is the effective promoter length (i.e. the promoter length minus L − 1, where L is the length of the pattern). Equation 1 calculates the tail probability that two high-scoring ‘signals’ will be found by chance at a distance d or less in the promoter with effective length N.
The PSSM-related score is calculated using the following formula:
| 2 |
where M1 and M2 are the PSSM models for the two species; S and T are random variables following the models' score distributions; and s and t are the observed PSSM scores. The PFS score is calculated using Gaussian approximation of mean and standard deviation estimated through random samplings from the PSSM model distributions. The results of the samplings are stored in each model. Similarly to PFD, Equation 2 calculates the corresponding tail probability under the null hypothesis that the two high-scoring ‘hits’ are due to chance alone.
We have developed a novel scoring scheme that combines the above two statistics in a single similarity measure. The combined score, PF, consists of a weighted log-likelihood transformation:
| 3 |
The weights wD and wS are positive numbers that sum to one (current default values: wD = 0.85; wS = 0.15). Summation of the logarithms is valid, since the individual PF scores are tail probabilities based on the null hypothesis that the human and the mouse patterns are not true binding sites, and hence the individual tail probabilities should be independent. Note that higher PF values correspond to higher probability that the human and mouse patterns are true sites. Since PF is the negative weighted average of the logarithms of P-values, exponentiation of −PF will return the weighted average P-value (WAP), which we use as a threshold cutoff in the server.
Algorithm
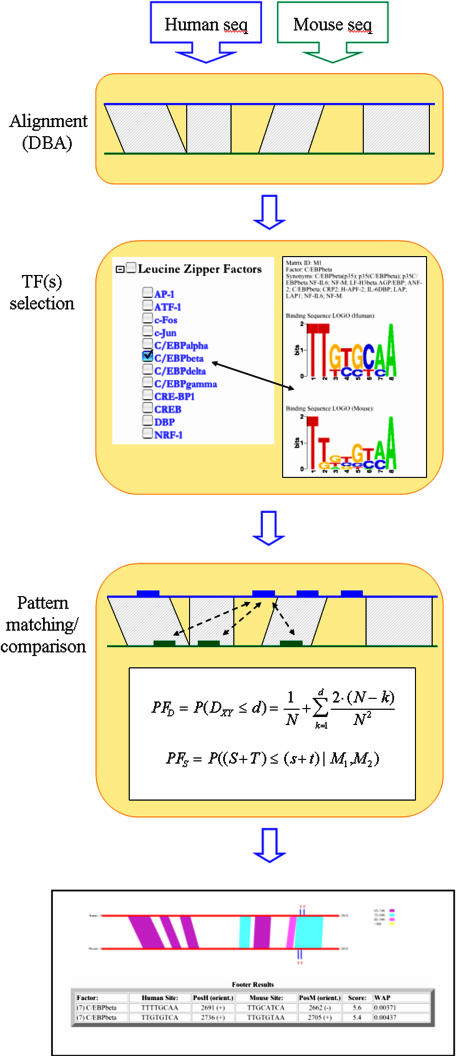
Once the promoter sequences have been specified and the PSSM models have been selected, FOOTER runs program DBA (14) to calculate the alignment between them (Figure 1). Then, a series of conserved and non-conserved regions are defined. Subsequently, the promoter sequences are scanned with each of the selected species- or mammalian-specific PSSM models and the top K ‘seed’ sites (user-defined parameter) are retained in each. These sites are analyzed pairwise, scored according to Equation 3 and matched using a greedy algorithm. The pairs that score above a user-specified WAP threshold are reported in the output. We should note here that since FOOTER compares pairwise all ‘seed’ sites in the two promoters (irrespectively of the DNA conservation in their surrounding region), it eliminates the need for a sliding window to identify ‘conserved’ sites.
Figure 1.
Flowchart of the execution of web-tool FOOTER. Protein or DNA sequences can be entered in the input. In the first case, a series of BLAST (15) searches will be performed to identify the homologous promoter sequences. The DNA sequences will be aligned and putative motifs will be compared pairwise as we describe in the text.
Input data types
FOOTER accepts two types of input data (Figure 1): either a single protein sequence (human or mouse/rat) or two DNA sequences (presumed human and mouse promoters). The input sequence files should be in FASTA format. If the specified input sequence is a human (mouse) protein, FOOTER will employ a BLASTP search (15) in the human/mouse proteome to identify its homologous mouse (human) protein, then it will use these protein sequences to perform TBLASTN searches against Unigene database (16) to identify the longest mRNA sequences. Finally, these mRNA sequences will be used in BLASTN searches against the corresponding genomes in order to identify the locations of the TSSs and thus automatically retrieve the corresponding promoter sequences.
FOOTER parameters
The input parameters for FOOTER are the wD and wS weights (see Equation 3) that should sum to one (if not, the program will automatically adjust them proportionally); the WAP threshold (default value is 0.005, which corresponds to a FOOTER combined score of 7.6); and the number of seed sites that will be initially retained and analyzed (default: an average of one site per 300 bp). In the case that a protein sequence is specified in input (see above), the user should also specify the promoter length (upstream and downstream sequence from the TSS) to be analyzed.
Testing
FOOTER has been tested in a set of 72 confirmed TFBS of 19 TFs. FOOTER was able to predict correctly 60 of these binding sites (SN = 83%) while it made an additional (unverified) 24 predictions. A table with the results mentioned above is provided at http://biodev.hgen.pitt.edu/Footer/webNAR05/Table.pdf. A more detailed description and extensive evaluation of the algorithm has been submitted for publication (25).
IMPLEMENTATION
FOOTER is written in Perl (CGI). For the graphical representation of the aligned promoters (PNG file), the PG package of Perl is used. The program uses program DBA (14) for the promoter alignment, which is currently its main performance bottleneck. DBA alignment time depends on the size of the promoter region, though it usually takes 45–75 s for a 3 kb promoter region. Once the two promoters have been aligned, FOOTER requires only a couple of seconds to identify the optimal matching patterns for a 3 kb region (on a Dell PowerEdge 2650 machine with 2.8 GHz dual-processor Xeon machine with HT technology and 2 GB of RAM). The time increases linearly with the number of TFs that it considers and exponentially with the number of seed patterns. With the default parameters, the complete search, including automatic identification of the promoter regions and alignment using DBA, does not usually require more than 3 min for a promoter length of 3 kb. FOOTER is available at http://biodev.hgen.pitt.edu/Footer/.
RESULTS
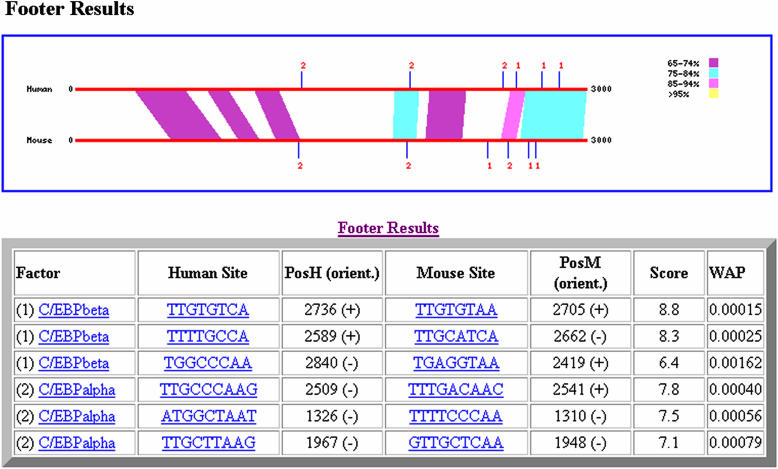
The final result of the program is a list of predicted sites in a table format (Figure 2). The results include the name of the TF, the sequence and position of the predicted TFBS in both the human and mouse promoters, the FOOTER calculated score (Equation 3) and the WAP value. This table can be copied into a spreadsheet program and analyzed further. In addition, FOOTER produces a PNG image with the alignment of the two sequences, color-coded by percent identity. The PNG image also displays all predicted sites. Finally, the results page provides links to the individual promoter sequences, the DBA alignment output and a summary of the program run, including all predicted sites (not just those above the cutoff).
Figure 2.
Example of FOOTER output. The predicted sites are presented in table format and in the PNG formatted figure. The figure displays the alignment of the two promoter sequences, colour-coded by conservation percentage.
Current limitations/future improvements
At the present stage, FOOTER has two limitations. One is the availability of PSSM models. We currently use models we constructed using TRANFAC (17) binding sites. In this way, we have calculated mammalian PSSM models for 127 TFs. With the development of high-throughput techniques for binding site identification, such as ChIP (18) and SELEX (19), construction of mammalian-specific matrix for many TFs will not be a problem. Recently, well-curated sets of binding sites have started to become publicly available (20). Nevertheless, we plan to add a feature to FOOTER that will allow for a user-defined PSSM model to be uploaded and used to scan the promoter sequences. We also plan to hyperlink the PNG image so that by moving the cursor over it, the user will receive information on various features of the predictions.
We have noticed that DBA (14) sometimes becomes slow in aligning long DNA sequences. For this purpose, we are currently exploring other algorithms (21,22) and strategies in order to further speed up FOOTER performance. For example, another way to speed the performance is to use pre-calculated alignments or databases of aligned promoter regions [e.g. (23,24)].
Acknowledgments
P.V.B. was partly supported by NSF grant MCB0316255. Funding to pay the Open Access publication charges for this article was provided by intramural funds of the Department of Computational Biology and the University of Pittsburg Cancer Institute, School of Medicine, University of Pittsburgh.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hughes J.D., Estep P.W., Tavazoie S., Church G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 2.Workman C.T., Stormo G.D. ANN-Spec: a method for discovering transcription factor binding sites with improved specificity. Pac. Symp. Biocomput. 2000:467–478. doi: 10.1142/9789814447331_0044. [DOI] [PubMed] [Google Scholar]
- 3.Hertz G.Z., Hartzell G.W., III, Stormo G.D. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- 4.GuhaThakurta D., Stormo G.D. Identifying target sites for cooperatively binding factors. Bioinformatics. 2001;17:608–621. doi: 10.1093/bioinformatics/17.7.608. [DOI] [PubMed] [Google Scholar]
- 5.Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 6.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 7.Tagle D.A., Koop B.F., Goodman M., Slightom J.L., Hess D.L., Jones R.T. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 8.Loots G.G., Ovcharenko I., Pachter L., Dubchak I., Rubin E.M. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenhard B., Sandelin A., Mendoza L., Engstrom P., Jareborg N., Wasserman W.W. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 11.Benos P.V., Lapedes A.S., Stormo G.D. Probabilistic code for DNA recognition by proteins of the EGR family. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- 12.Benos P.V., Bulyk M.L., Stormo G.D. Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benos P.V., Lapedes A.S., Fields D.S., Stormo G.D. SAMIE: statistical algorithm for modeling interaction energies. Pac. Symp. Biocomput. 2001;6:115–126. doi: 10.1142/9789814447362_0013. [DOI] [PubMed] [Google Scholar]
- 14.Jareborg N., Birney E., Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingender E. TRANSFAC, TRANSPATH and CYTOMER as starting points for an ontology of regulatory networks. In Silico Biol. 2004;4:55–61. [PubMed] [Google Scholar]
- 18.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 19.Choo Y., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl Acad. Sci. USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nix D.A., Eisen M.B. GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinformatics. 2005;6:9. doi: 10.1186/1471-2105-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palaniswamy S.K., Jin V.X., Sun H., Davuluri R.V. OMGProm: a database of orthologous mammalian gene promoters. Bioinformatics. 2005;21:835–836. doi: 10.1093/bioinformatics/bti119. [DOI] [PubMed] [Google Scholar]
- 24.Zhao F., Xuan Z., Liu L., Zhang M.Q. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–D107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran D.L., Feingold E., Dominick J., Wright M., Harnaha J., Trucco M., Giannoukakis N., Benos P.V. FOOTER: a quantitative comparative genomics method for efficient recognition of cis-regulatory elements. Genome Res. 2005 doi: 10.1101/gr.2952005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]