Abstract
The biological data explosion of the ‘omics’ era requires fast access to many data types in rapidly growing data banks. The MRS server allows for very rapid queries in a large number of flat-file data banks, such as EMBL, UniProt, OMIM, dbEST, PDB, KEGG, etc. This server combines a fast and reliable backend with a very user-friendly implementation of all the commonly used information retrieval facilities. The MRS server is freely accessible at http://mrs.cmbi.ru.nl/. Moreover, the MRS software is freely available at http://mrs.cmbi.ru.nl/download/ for those interested in making their own data banks available via a web-based server.
INTRODUCTION
Massively parallel high-throughput experiments are generating increasing data volumes at an ever more rapid pace. Institutes, such as the EBI (http://www.ebi.ac.uk/) or the NCBI (http://www.ncbi.nih.gov/), provide large series of tools to search in many different data banks. Obviously, these data banks contain only the publicly available data. If a user wants to search in-house data, the in-house software is required, and either the query results or the data banks must be merged.
The popular SRS software (1) is hosted for public access by 44 institutes that, together, provide access to 1205 data banks (http://downloads.lionbio.co.uk/publicsrs.html). Typically, each of these sites has one or more database managers, who run mirror scripts to maintain up to date copies of (a subset of) the possible data banks. After each update, the data banks are indexed (typically overnight) and made available to the users. SRS has separate data and index entries. Therefore, the data bank must be off-line during the indexing. Alternatively, two copies of the data can be maintained, one for on-line usage and another for off-line indexing. The latter overwrites the former as soon as the indexing has been completed. Data banks, such as EMBL (2) or GenBank (3), have reached such a size that only large institutes like those 44 that host SRS can afford to keep these data on-line.
We have designed the MRS software for simple, rapid in-house access to biological data banks. MRS is a Perl (http://www.perl.org/) plug-in that allows for rapid access to data from Perl scripts. MRS-files contain both the raw data and the indices so that it is guaranteed that the data and the search indices always remain synchronous. The disk space required by MRS to store the raw data and the indices is typically less than half the space required to store the uncompressed raw data. Multiple MRS-files for one kind of data can be merged. Therefore, one can download the MRS-files for public data from the MRS WWW pages and merge them with MRS-files that were generated in-house from private data.
MRS has been optimized for speed and ease of use. For example, a search for ‘lysozyme’ in 12 MRS-files, including EMBL, PDB (4), UniProt (5), etc., typically takes 0.02 s on a single processor PC, whereas combined searches like ‘chloride AND channel’ typically take 0.15 s. Similar searches using the EBI search engines typically take several seconds.
An MRS server is available at http://mrs.cmbi.ru.nl/. Scientists from academia and industry can freely use this server to search presently in 14 data banks. All materials needed to build one's own MRS server are available at http://mrs.cmbi.ru.nl/download/. The present distribution includes pre-indexed MRS-files for UniProt and the protein structure related data banks. More data banks will be included soon, and a series of example Perl scripts that index data banks is available.
METHODS
The MRS system can be decomposed into the following components:
data bank update,
data structure,
indexing,
searching,
result presentation.
Data bank updates are carried out with a series of Makefiles and Perl scripts. MRS users will not need these files because it will be faster and easier for them to obtain the pre-indexed MRS-files. The Makefiles and Perl scripts are nevertheless available from the MRS download website for bioinformaticians interested in extending their in-house MRS server.
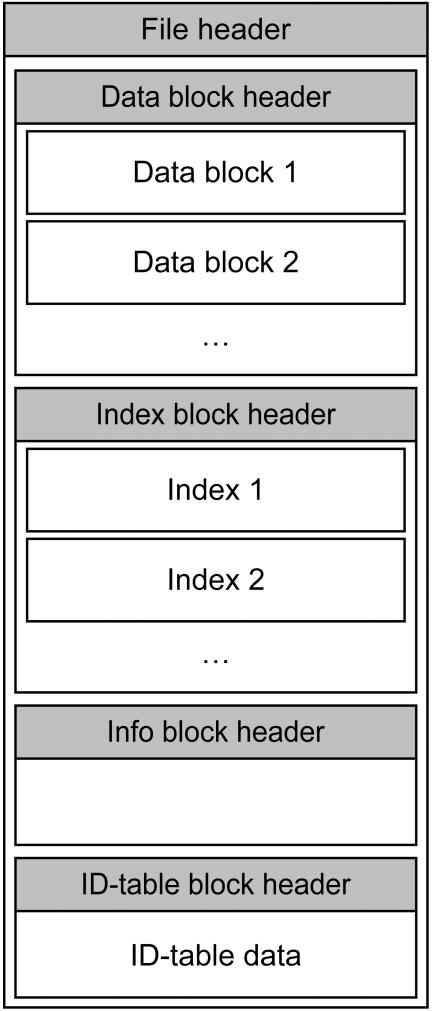
Figure 1 shows the MRS data structure. An MRS-file consists of a header, and four blocks, each consisting of a block header and data. The main header only contains pointers to the four blocks and a series of counters, such as the number of indexed entries, etc. The four blocks are called ‘Data’, ‘Index’, ‘Info’ and ‘ID-table’. Each block contains its own block header which holds some administrative information regarding the MRS version used, pointers to the records in that block, a table with pointers for rapid access to the data in that block, etc.
Figure 1.
MRS-file data structure.
The Data block consists of multiple data parts. This subdivision was made to allow for parallel indexing and for easy merging of public and private data. Compression information is stored for each separate data part to allow for this merging. Each data part holds the complete raw data bank entries compressed with zlib (http://www.gzip.org/zlib/), bzip2 (http://sources.redhat.com/bzip2/) or Huffman (6). The MRS manager decides which compression to use.
For each data field, the Index block contains one index that consists of a B-Tree (7) optionally augmented with an inverted index (8) that allows for very rapid access to files that contain a certain index. B-trees are especially useful for the type of query as performed by MRS (8). The Info block is at present still empty. It is reserved for future applications, such as storing copyright information.
The ID-table holds the mapping from MRS-entries to the IDs of entries in the indexed data bank. The reverse mapping, from the IDs of the data bank entries to the internal MRS entry numbers, is stored in the Index block.
Indexing is performed with a Perl script that calls a series of data bank-specific plug-ins written in Perl. Each multiple instances of the plug-in can index a section of the data, which, after completion, is written into a MRS-file. (After completing all sections, the resulting MRS-files are merged into a single new MRS-file.) A plug-in holds a parser object with information about the data fields to be indexed. One index is constructed for each data field. For example, indexing UniProt results in 14 indices. Indexing a new data bank typically requires a 100-line Perl script. Several indexing scripts are available as examples at the MRS download site.
Searching is carried out by a Perl script that uses the indices to find records that contain the keywords requested by the user. A so-called ‘full text search’ is executed as a union of the searches over all indices. Searches always result in an iterator object that holds for each data bank hit all information needed to obtain the raw data for the requested entry. A series of intelligent filters is used to remove double occurrences in case a data bank is queried at the same time as updates for that data bank (e.g. EMBL and EMBLnew).
A CGI script, written in Perl, that uses one plug-in for each data bank, performs the presentation of the query results to the user. This script uses a configuration file that holds for each data bank all information about its visual display. The default visualization modus is the raw ASCII data as is found in the data bank. Users can write fancy display modi for individual data banks and add these to the configuration file. The visualization scripts can, on-the-fly, add hyperlinks to data banks indexed in the same MRS environment. Hyperlinks to remote data banks require more intricate programming.
DESCRIPTION OF THE WEB INTERFACE
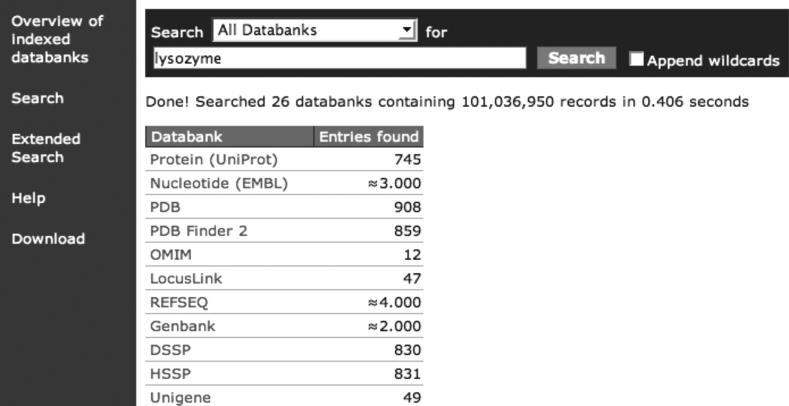
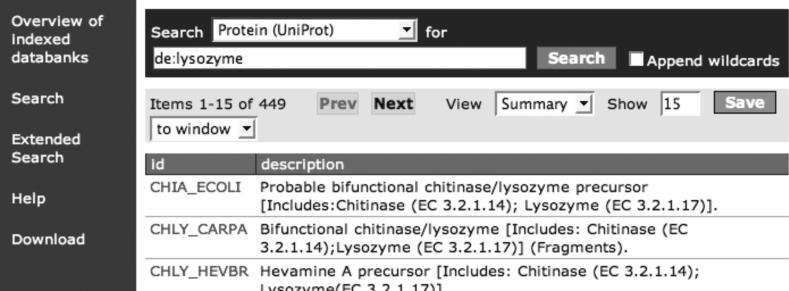
Figure 2 shows the results of a multi data bank search for ‘lysozyme’. Figure 3 shows the result for the narrower search ‘lysozyme in the DE field of a UniProt entry’. The field names used for indexing are listed under ‘Overview of indexed data banks’. The output visualization script selects the text in the ‘description per hit’. Even though the WWW form is highly intuitive and self-explanatory, an extensive user manual is available at the MRS site. This manual also explains the extended query form and the use of AND, OR and NOT that are not described here.
Figure 2.
Query form and results for a ‘full text search’ on all data banks.
Figure 3.
Query for and results for a ‘single field search’ on one data bank.
The MRS server has been incorporated in the BioSapiens (http://www.biosapiens.info/) DAS server. A SOAP interface is available for remote usage bypassing the WWW interface.
DISCUSSION
Many query and retrieval systems exist in the WWW. Some, like SRS, provide access to a massive number of extensively hyperlinked data. On the other hand, the NAR special volumes on servers and databases have, over the years, listed a long series of systems that allows for complex queries on single, often small, databases. MRS has a unique position because it allows for very fast and robust simple queries on any number of data banks. Additionally, installing and maintaining an MRS server is simple and requires less disk space than storing the raw data.
Future extensions of MRS include queries on numerical values, distributed queries over the grid, improved data visualization, the use of ontologies and thesauri and, of course, more indexed data banks.
The full availability of the MRS system is likely to help us achieve these goals quickly.
Acknowledgments
This work has been supported by NWO (FlexWork: 050.50.204) and the EC FP6 project BioSapiens (LHSG-CT-2003-503265). Funding to pay the Open Access publication charges for this article was provided by Radboud University, Nijmegen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Etzold T., Ulyanov A., Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–28. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- 2.Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro M., Cochrane G., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffman D.A. A method for the construction of minimum-redundancy codes. Proc. IRE. 1952;40:1098–1101. [Google Scholar]
- 7.Knuth D.E. The Art of Computer Programming, Vol. 3: Sorting and Searching. 2nd edn. Boston, MA: Addison Wesley Longman Publishing Co., Inc.; 1998. [Google Scholar]
- 8.Witten I.H., Moffat A., Bell T.C. Managing Gigabytes: Compressing and Indexing Documents and Images. 2nd edn. San Francisco, CA: Morgan Kaufmann Publishing; 1999. Chapter 2. [Google Scholar]