Abstract
VisANT is a web-based software framework for visualizing and analyzing many types of networks of biological interactions and associations. Networks are a useful computational tool for representing many types of biological data, such as biomolecular interactions, cellular pathways and functional modules. Given user-defined sets of interactions or groupings between genes or proteins, VisANT provides: (i) a visual interface for combining and annotating network data, (ii) supporting function and annotation data for different genomes from the Gene Ontology and KEGG databases and (iii) the statistical and analytical tools needed for extracting topological properties of the user-defined networks. Users can customize, modify, save and share network views with other users, and import basic network data representations from their own data sources, and from standard exchange formats such as PSI-MI and BioPAX. The software framework we employ also supports the development of more sophisticated visualization and analysis functions through its open API for Java-based plug-ins. VisANT is distributed freely via the web at http://visant.bu.edu and can also be downloaded for individual use.
INTRODUCTION
Networks are ideal representations for many biological processes (1), applicable to systems such as metabolism (2), gene regulation (3), signal transduction (4) and development (5). The components, or nodes, in biological networks may represent a range of biological features of interest, such as genes, proteins, macromolecular complexes and cellular pathways. These components can be connected by different types of links, just as myriad types of interactions can connect the processes within the cell. Connections can be directed or undirected; they can have physical meaning, denote general associations; they can represent shared characteristics between components. Components can also be made up of subcomponents, in which case they are compound or modular, and the connections between modular components (or modules) can exist along with interconnections between their subcomponents (6).
Choice of network representation is often dictated by the research problem at hand. Directed networks are suitable when the interactions between two components have a well-defined direction, e.g. the direction of metabolic flow from substrates to products, or the information flow from transcription factors to the genes that they regulate. Similarly, undirected networks, such as protein interaction networks, represent mutual relationships: if protein A binds to protein B, then protein B binds to protein A. This type of representation also often applies to predictions made by high-throughput proteomic or genomic analysis, or indirect links based on shared genes or protein components between pathways and complexes.
Previously, we reported a web-based application, VisANT (7,8), for simultaneous visualization and overlaying of multiple types of simple network data, enabling, for example, comparisons between experimental interactions gathered from different data sets. This early version of the tool had basic capacity for visualizing and manipulating biological network information. Here, we describe significant extensions to this concept and release VisANT 2, a more general software tool for visually integrating different types biological information based on association and connectivity data. Obtainable at http://visant.bu.edu, VisANT is free to both academic and commercial users.
The core interface of VisANT is a workbench for network analysis and visualization. Users of VisANT can upload data for any organism, such as interactions, pathways, clusters or groupings. Data can be anything that describes how genes or proteins are connected and associated, although VisANT has a controlled vocabulary for many common experimental types of data. Once uploaded, the software will display this information and allow users to (i) visually arrange, manipulate and save data in graphical form; (ii) analyze network data for topological statistics and features; and (iii) query network data for functional information from our server-side database, such as Gene Ontology (GO) function or interactions gathered from other published data sets.
In addition to simple networks, interactions in VisANT can also be defined as higher-level connections between groups of proteins, complexes, pathways or sub-networks. These ‘modular’ connections can be viewed simultaneously with connections between subcomponents, such as individual protein interactions. This complex data integration environment, therefore, significantly extends concepts of other tools, such as Cytoscape (9), Osprey (10) and the MINT viewer (11), by providing for functional annotation and scientific sharing of pathway and network information viewed at multiple scales. Interaction networks and protein complexes can be viewed, e.g. within the context of GO (12) annotations or KEGG (13) pathway assignments. We believe this capability makes VisANT an especially useful tool for integrating information from a wide variety of sources. To accompany VisANT, we have also developed a preliminary standard for exchanging files that have visual markup and annotation of network layouts, called visML. As a network specification format, visML extends concepts of similar graph languages, such as graph markup language, but contains additional features for complex and compound network components.
Users of VisANT can input several basic data types, including data in standardized network and interaction data exchange formats, such as PSI-MI (14) and BioPAX (http://www.biopax.org). Development is also underway for support of the SBML (15) pathway format. Newly supported data types include:
Simple interactions: links defined as protein–protein, protein–DNA, gene–gene, etc.
Modules, groups and clusters: genes, proteins, pathways, sub-networks.
Modular interactions: complex interactions, co-localization data, shared components, pathway interactions.
Once a network data set has been imported or loaded into VisANT, the genes or proteins within it can be queried for other known and predicted interactions from published data sets, using the previously published Predictome (16) repository. Imported interactions and components define a network ‘workspace’, which can be annotated and saved for printing or sharing among others in the community. Networks can also be analyzed for topological characteristics to identify larger global properties, such as degree distribution, path length, shortest path and clustering coefficient calculations.
Output from VisANT can be saved at any point once a network has been loaded, annotated and analyzed. There are three types of output:
Graphical: JPEG-, PNG- and TIFF-formatted versions of the network.
VisML: interpreted language and exchange format for storing annotation and visual layout of complex networks.
Network statistics: statistical results and calculations of network topology.
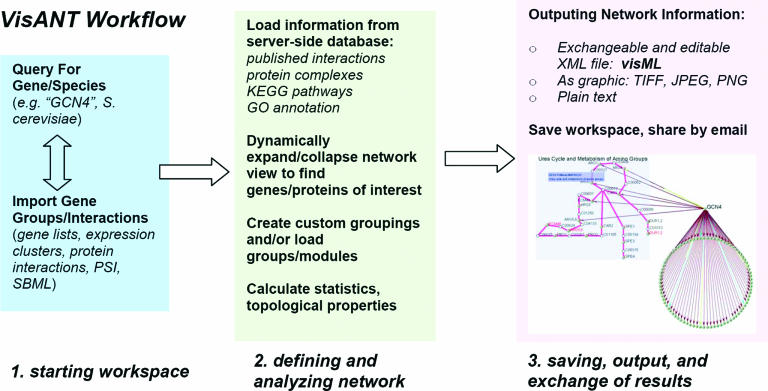
Visual annotation of networks is saved in a computer-readable format, visML, which preserves all data, markup, formatting and layout information that a user defines for a given network. With visML, work can be saved and re-loaded later, including addition or removal of any interaction data. Complex networks can also be shared and manipulated among different users, and results from different manipulations or network views can be saved to different visML files (see Figure 1).
Figure 1.
Workflow of visualizing and analyzing networks with VisANT.
USING VISANT
We give several examples of typical uses of the VisANT application. Details on how the described operations are performed are explained more thoroughly in the online Supplementary Material and on the VisANT help page, http://visant.bu.edu/help.
Simple networks: KEGG pathways and transcription factor binding
One advantage of VisANT is that it relies on a large server-side database of published information to supplement a user's own data sets. After uploading a data set of interactions between proteins, the user can see how that data set compares with other similar data sets. The server-side database also acts to control the vocabulary of gene names so that data obtained from different sources with different naming conventions can be easily interchanged. One source of information for interpreting relations is the KEGG pathway database of metabolic and signaling pathways. The workflow in Figure 1 shows how VisANT can be used to view different types of biological data. A user could start by finding published transcription factor binding interactions for the Saccharomyces cerevisiae protein Gcn4, which is known to regulate amino acid biosynthesis in yeast. Published interactions from different sources (3,17) can be viewed with the information of functions and pathways. With only a few steps, the information can be retrieved showing that the KEGG pathway for urea cycle and amine metabolism (KEGG MAP00220) has six genes, which are targeted by Gcn4, according to large-scale ChIP data (3).
Modular networks: GO function and protein–protein interactions
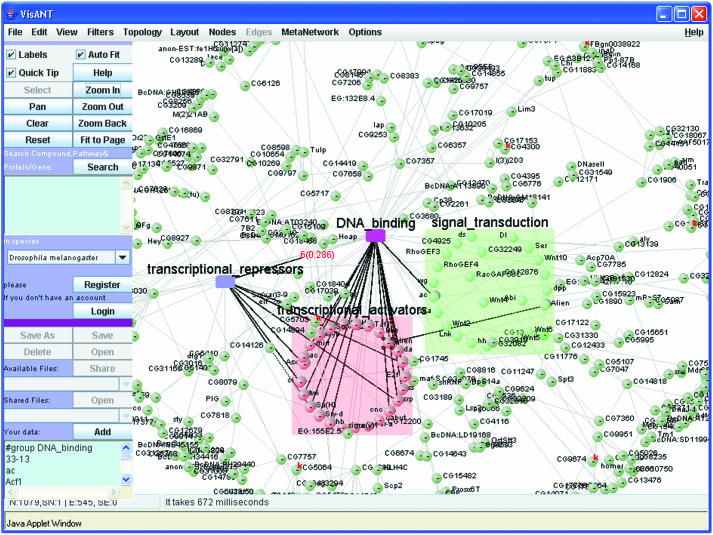
Biological networks are often modular or compound, and involve connections between groups of genes and proteins as well as between individuals. Modular networks are a useful simplification if, for example, a researcher wants to know the interactions that underlie a particular function or process in the cell. Functional organization may be revealed by how genes and proteins in different functions are connected by published interactions, or how different pathways and groups of genes or proteins have shared components. These types of modular information require more sophisticated data structures to handle the resulting networks. VisANT provides a rich visual interface to this type of problem. Figure 2 shows genes from Drosophila melanogaster assigned to GO categories of transcription activation/repression, DNA binding and signal transduction. At the same time, the view shows genes that are shared between functional categories, and the protein–protein binding evidence (18) of interactions between the functional groups.
Figure 2.
Gene Ontology function and protein–protein interaction mapping with VisANT. The colored regions contain genes and proteins annotated in the SGD yeast GO as belonging to molecular function categories of transcription activators (peach), transcription repressors (blue), DNA binding (magenta) and signal transduction (green). These functional groups can be expanded or collapsed by double-clicking with the mouse. Dark black connections between GO categories indicate shared gene or protein components of these functions, e.g. the protein alien appears both in signal transduction and in transcription activation. Gray connections are protein–protein interactions determined by high-throughput yeast two-hybrid assay by Giot et al. (18). Interactions of other types can be loaded by the user from their own sources, or from other methods in the database.
DATA INTEGRATION
VisANT performs several layers of integration with different network data sets:
Integration of nomenclature for genes/proteins. Naming conventions between different data sets can be different, and the server-side parser translates between standard nomenclatures for different common species, such as S.cerevisiae, D.melanogaster, Caenorhabditis elegans and Homo sapiens.
Integration with other annotation databases and KEGG pathways. After resolving nomenclature for genes and proteins, VisANT retrieves functional data from the GO and KEGG databases.
Integration of different types of bio-networks. VisANT supports an arbitrary number of interaction types. Users can upload different types of interactions by specifying different evidence codes that are supported in the VisANT ‘Method’ table (see Supplementary Material for full list of methods).
TOPOLOGICAL FUNCTIONS FOR NETWORK BIOLOGY
Integrated biological networks are often hybrid, meaning they contain both directed (e.g. transcription factor binding) and undirected (e.g. protein interaction) connections, and compound or modular, meaning that linked components can either be single or grouped. Unlike other software for biological network topological analysis, VisANT explicitly allows creation of mixed networks involving different types, with topological algorithms to support type.
Node degree and distribution
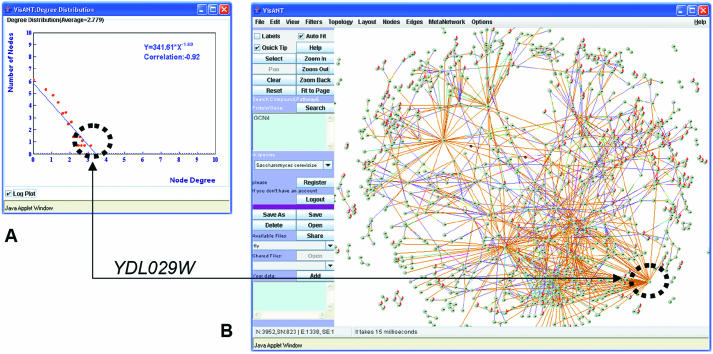
The degree of a network component, k, is the number of connections it has with other components. The distribution of degrees among components is useful for characterizing the topology and scale of a network, and often has meaningful biological interpretation. In protein interaction and genetic interaction networks, for example, the degree of a hub is often its importance and essentiality for cell function. For directed networks, such as transcription factor binding networks, the degree is separated into ‘in’-degree and ‘out’-degree, depending on the directions of interaction between two given components. Degree is also a feature that distinguishes hubs (highly connected nodes) from leaves or orphans (weakly or non-connected nodes) in the network. In VisANT, users can see a scatter plot and log-linear regression fit of the degree distribution, p(k), of the network. The degree exponent (γ) of the log-linear regression, where p(k)k−γ, is a measure of the network's ‘scale-free’ property (19,20). The VisANT degree plot is dynamically linked to the network view (selection in one window maps onto corresponding points in the other, see Figure 3).
Figure 3.
Dynamic linking for finding topological features. The network of synthetic lethal genetic interactions (28) in yeast contains 823 genes and 3952 interactions. A plot of the degree distribution of genes in this network allows users to quickly identify which genes have the highest (or lowest) connectivity. In this example, the gene shown is YDL029W, which has synthetic lethal interactions with 58 neighbors.
Clustering coefficients and distribution
The clustering coefficient (C = 2n/[k(k − 1)]), where n is the number of links between k neighbors, measures the tendency of a network to have highly connected clusters. Fully connected sets of nodes have C = 1, because all nodes are highly connected. In large-scale mass spectrometric networks in yeast, this property can be used to identify groups of proteins involved in the assembly of the ribosome (21). The exponential degree of the log-linear fit C(k) = k−γ can be used to characterize the hierarchical structure of a network (22,23). VisANT provides scatter plots and log-linear regression fit of the clustering coefficients in a network, allowing users to identify densely connected clusters of nodes (the direction in hybrid networks is ignored in calculating C). This plot, like the degree plot, is dynamically linked to the network view.
Shortest path lengths and distribution
In studying the function of pathways, the property of interest is often how a given gene or protein is related to (or responds to) an up- or downstream signal. Given a large data set of interactions, it may be useful in some contexts to find the most direct path between two genes, proteins, complexes or pathways; for example, the overall lengths of such pathways may be related to the immediacy or breadth of signal response (24). The average shortest path also indicates the well-known ‘small-world’ property of many real-life networks (25). Networks in VisANT are analyzed by breadth-first-searching for both the shortest path between two given components as well as the distribution of shortest paths between all components.
Detection of network motifs
Certain patterns and motifs have been shown to occur with more frequency in biological networks than would be expected by chance alone (26). This leads to the hypothesis that such motifs, e.g. feed-forward loops, have functional characteristics that correspond to their structure (27). Identifying topological features in networks is an important part of understanding the relationship between structure and function of these motifs. VisANT supports searching of basic motif types, such as feedback and feed-forward loops, and development is in progress for the detection of other arbitrary motifs, and for assessing the statistical significance of these patterns in large networks. Motif detection is performed in the manner described previously (3).
Network randomization
An add-on to VisANT provides functions to randomize a given network. The randomization functions were developed using the plug-in API of VisANT, described later in the paper. The functions provide randomization criteria for both directed and undirected networks. Randomization of undirected networks preserves the same number of nodes and edges, but edges are randomly distributed, and the resulting network is approximated by a Poisson distribution in large sparsely graphs. Directed network randomization preserves the same number of nodes and same in-degree and out-degree for each node, but the directed edges are randomly distributed among the nodes. Directed randomized networks can serve as a reference for the statistical analysis of topological features (e.g. degree distribution and clustering coefficient) of the network under study. For example, combined with motif detection function, the randomization function can help evaluate the statistical significance of the detected motifs. These functions will be extended in the future to aid in calculation of complete statistical significance of more complex network features.
THE VisANT PROJECT
Applications of networks to new biological problems will require a larger set of functions than VisANT currently provides. We have, therefore, engineered VisANT to have an open plug-in architecture for extending the software to meet the future requirements of network biology. Plug-ins can be developed that access all of the software layers of VisANT: the network data model, server-side database connectivity and network visualization. Additional information on plug-ins can be obtained at http://visant.bu.edu.
We have implemented VisANT to run as an applet, which will be convenient for most users. Researchers who are interested in downloading the application to run on local machines can do so from the main web site. Data integration features, such as downloading data sets and gene name lookup in this case, still rely on the VisANT server-side engine, but for security purposes when running VisANT locally, users can also view and manipulate visML files strictly on their own machines. Currently, visML is the native file format of VisANT, but we are currently expanding the visML specification to allow exchange of annotated and marked-up networks.
The VisANT project is designed to accommodate rich visual annotation of functional and modular networks in different species. In addition to software features in the VisANT application, we anticipate further integration with other species and newly characterized genomes as this becomes available.
AVAILABILITY
Full user manual and tutorials are available on the VisANT website, http://visant.bu.edu. The full application is available as locally installable executable.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by NIH grant 1P2 0GM66401-01 and NIGMS grant A08-POGM66401A.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 2.Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 3.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 4.Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 5.Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 6.Segal E., Shapira M., Regev A., Pe'er D., Botstein D., Koller D., Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z., Mellor J., Wu J., DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z., Mellor J., DeLisi C. Current Protocols in Bioinformatics. NJ: John Wiley & Sons; 2004. Analyzing networks with VisANT. [DOI] [PubMed] [Google Scholar]
- 9.Ideker T., Ozier O., Schwikowski B., Siegel A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl. 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 10.Breitkreutz B.J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 12.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M., Goto S., Kawashima S., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 15.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 16.Mellor J.C., Yanai I., Clodfelter K.H., Mintseris J., DeLisi C. Predictome: a database of putative functional links between proteins. Nucleic Acids Res. 2002;30:306–309. doi: 10.1093/nar/30.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 19.Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabasi A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 20.Albert R., Jeong H., Barabasi A.L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 21.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 22.Ravasz E., Barabasi A.L. Hierarchical organization in complex networks. Phys. Rev. E. Stat. Nonlin. Soft. Matter Phys. 2003;67:026112. doi: 10.1103/PhysRevE.67.026112. [DOI] [PubMed] [Google Scholar]
- 23.Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabasi A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 24.Babu M.M., Luscombe N.M., Aravind L., Gerstein M., Teichmann S.A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Del Sol A., O'Meara P. Small-world network approach to identify key residues in protein–protein interaction. Proteins. 2005;58:672–682. doi: 10.1002/prot.20348. [DOI] [PubMed] [Google Scholar]
- 26.Itzkovitz S., Milo R., Kashtan N., Ziv G., Alon U. Subgraphs in random networks. Phys. Rev. E. Stat. Nonlin. Soft. Matter Phys. 2003;68:026127. doi: 10.1103/PhysRevE.68.026127. [DOI] [PubMed] [Google Scholar]
- 27.Shen-Orr S.S., Milo R., Mangan S., Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 28.Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.