Abstract
Caspases and granzyme B are proteases that share the primary specificity to cleave at the carboxyl terminal of aspartate residues in their substrates. Both, caspases and granzyme B are enzymes that are involved in fundamental cellular processes and play a central role in apoptotic cell death. Although various targets are described, many substrates still await identification and many cleavage sites of known substrates are not identified or experimentally verified. A more comprehensive knowledge of caspase and granzyme B substrates is essential to understand the biological roles of these enzymes in more detail. The relatively high variability in cleavage site recognition sequence often complicates the identification of cleavage sites. As of yet there is no software available that allows identification of caspase and/or granzyme with cleavage sites differing from the consensus sequence. Here, we present a bioinformatics tool ‘GraBCas’ that provides score-based prediction of potential cleavage sites for the caspases 1–9 and granzyme B including an estimation of the fragment size. We tested GraBCas on already known substrates and showed its usefulness for protein sequence analysis. GraBCas is available at http://wwwalt.med-rz.uniklinik-saarland.de/med_fak/humangenetik/software/index.html.
INTRODUCTION
Caspases are enzymes orchestrating the cellular pathways leading to apoptosis and inflammatory signals. Besides these functions they are supposed to be involved in other cellular processes, such as development, cell cycle, cell proliferation, cell migration and receptor internalization (1,2). Caspases are cysteine proteases with specificity for an aspartic acid residue at position P1 of the substrate. This primary specificity is shared by the serine protease granzyme B, which induces cytotoxic T lymphocyte-mediated target cell DNA fragmentation and apoptosis (3,4). Granzyme B-mediated cleavage also plays a role in induction of autoimmunity (5).
To date, at least 14 mammalian caspases can be grouped into three classes based on their substrate specificities. Group I consisting of caspases -1, -4, -5 (-14 and murine -11 and -12) cleaves the substrate sequence (W/L)EHD, group II (caspases -2, -3, -7) cleaves the DEXD motif and group III (caspase -6, -8, -9, -10) preferentially cleaves the (L/V)E(T/H)D sequence (6,7). Caspases of group I play an important role in the generation of inflammatory signals and in the immune regulation. Caspases -8, -9 and -10 are so-called initiator caspases mainly cleaving and activating procaspases, whereas caspases -3, -6 and -7 as effector caspases cleave numerous cellular proteins. The serine protease granzyme B prefers substrates with sequence IEXD, and is released by cytotoxic lymphocytes to kill virus-infected or tumor cells.
Although more than 280 caspase targets are described [for comprehensive review see (8)] many substrates still await identification and many cleavage sites of known substrates are not identified or experimentally verified. Likewise, the identification of granzyme B substrates is still at its infancy. Intracellular substrates of granzyme B include other caspases, mainly caspase 3 (9), ADPRT (ADP-ribosyltransferase 1, PARP) (10), BID (BH3 interacting domain death agonist) (11) and ICAD (DNA fragmentation factor) (12). Notably, the majority of autoantigens in systemic autoimmune diseases are efficiently cleaved by granzyme B (5).
A more comprehensive knowledge of caspase and granzyme B substrates is essential to understand the biological roles of these enzymes in more detail. The relatively high variability in cleavage site recognition sequence often complicates the identification of cleavage sites. As of yet there is no software available that allows identification of caspase and/or granzyme cleavage sites differing from the consensus sequence. The PeptidCutter program provided by the ExPasy Server (http://www.expasy.org/tools/peptidecutter) considers only the preferred peptide substrate sites. A recent tool of Lohmüller et al. (13) is restricted to caspase 3 and cathepsin B and -L substrates. Here, we present a bioinformatics tool GraBCas that provides score-based prediction of potential cleavage sites for the caspases 1–9 and granzyme B including an estimation of the fragment size. We validated our tool by scoring known substrates and demonstrated its usefulness for protein sequence analysis.
MATERIALS AND METHODS
Design of cleavage site scoring matrices
We developed position specific scoring matrices (PSSM) for the endopeptidases granzyme B and caspase 1–9 based on experimentally determined substrate specificities (6). Thornbery et al. (6) determined the substrate specificities using positional scanning synthetic combinatorial libraries. Cleavage was fluorimetrically determined with maximum value annotated with 100 and the values for the remaining amino acids given as percentage of the observed maximum rate. These experimental values provided the basis for creating our PSSM.
The values for each amino acid at position Pi are shown in Table 1. For a better readability we decided to set the maximum values to 1000 instead of 100 and adjusted the other values accordingly. For each endopeptidase the scores of the amino acids were entered in a 3 × 20 matrix. The rows of such a matrix correspond to positions P4, P3 or P2 of a possible cleavage site. Each column represents one amino acid and contains the relative frequencies of the amino acid measured in the study of Thornbery et al. (6). We are working with PSSM that can be interpreted as probability matrices. Since probabilities of value 0 should be avoided in such probability-based position scores, all entries of experimental relative frequencies with value 0 were set to 1. The amino acids cysteine and methionine were not part of the study of Thornbery et al. (6). The entries for these amino acids were also set to 1 in Table 1.
Table 1.
Scoring matrices for granzyme B and caspases 1–9
| Position Pi | AA of consensus recognition motif | C | S | T | P | A | G | N | D | E | Q | H | R | K | M | I | L | V | F | Y | W | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Granzyme B | 4 | I | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1000 | 52 | 500 | 12 | 1 | 1 |
| 3 | E | 1 | 297 | 54 | 1 | 153 | 477 | 1 | 198 | 1000 | 81 | 9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | |
| 2 | P | 1 | 752 | 544 | 1000 | 576 | 16 | 624 | 144 | 288 | 576 | 544 | 1 | 1 | 1 | 16 | 96 | 304 | 144 | 16 | 16 | |
| Caspase 1 | 4 | W | 1 | 48 | 48 | 48 | 48 | 16 | 16 | 80 | 96 | 16 | 128 | 1 | 1 | 1 | 96 | 288 | 80 | 496 | 576 | 1000 |
| 3 | E | 1 | 357 | 442 | 1 | 442 | 425 | 119 | 374 | 1000 | 646 | 187 | 51 | 34 | 1 | 221 | 323 | 442 | 272 | 187 | 85 | |
| 2 | H | 1 | 144 | 396 | 144 | 180 | 18 | 72 | 36 | 72 | 108 | 1000 | 54 | 72 | 1 | 198 | 54 | 108 | 126 | 144 | 126 | |
| Caspase 2 | 4 | D | 1 | 1 | 50 | 10 | 1 | 1 | 10 | 1000 | 200 | 1 | 1 | 1 | 1 | 1 | 180 | 400 | 80 | 40 | 40 | 1 |
| 3 | E | 1 | 425 | 884 | 1 | 680 | 119 | 119 | 102 | 1000 | 680 | 153 | 408 | 221 | 1 | 119 | 187 | 646 | 255 | 187 | 153 | |
| 2 | H | 1 | 624 | 528 | 352 | 336 | 48 | 96 | 1 | 1 | 16 | 1000 | 320 | 304 | 1 | 144 | 16 | 80 | 16 | 16 | 80 | |
| Caspase 3 | 4 | D | 1 | 40 | 50 | 1 | 10 | 1 | 20 | 1000 | 40 | 1 | 10 | 1 | 1 | 1 | 10 | 1 | 20 | 10 | 1 | 1 |
| 3 | E | 1 | 306 | 357 | 1 | 357 | 85 | 153 | 255 | 1000 | 408 | 187 | 17 | 17 | 1 | 153 | 153 | 306 | 272 | 255 | 119 | |
| 2 | V | 1 | 14 | 378 | 406 | 182 | 1 | 14 | 1 | 0 | 14 | 196 | 42 | 0 | 1 | 714 | 224 | 1000 | 182 | 154 | 84 | |
| Caspase 4 | 4 | W | 1 | 80 | 208 | 144 | 96 | 48 | 80 | 288 | 256 | 96 | 48 | 1 | 1 | 1 | 304 | 848 | 224 | 384 | 352 | 1000 |
| 3 | E | 1 | 187 | 119 | 34 | 119 | 17 | 85 | 221 | 1000 | 306 | 85 | 1 | 17 | 1 | 51 | 85 | 204 | 187 | 85 | 17 | |
| 2 | H | 1 | 102 | 119 | 119 | 425 | 17 | 102 | 221 | 357 | 153 | 1000 | 51 | 1 | 1 | 595 | 17 | 119 | 85 | 102 | 51 | |
| Caspase 5 | 4 | W | 1 | 14 | 56 | 1 | 56 | 126 | 42 | 98 | 98 | 84 | 56 | 1 | 1 | 1 | 280 | 1000 | 154 | 504 | 406 | 1000 |
| 3 | E | 1 | 24 | 12 | 1 | 12 | 1 | 1 | 124 | 1000 | 12 | 12 | 1 | 1 | 1 | 12 | 12 | 12 | 12 | 12 | 1 | |
| 2 | H | 1 | 340 | 425 | 323 | 323 | 1 | 85 | 119 | 323 | 85 | 1000 | 17 | 34 | 1 | 272 | 17 | 272 | 153 | 204 | 102 | |
| Caspase 6 | 4 | V | 1 | 144 | 880 | 64 | 96 | 16 | 64 | 224 | 256 | 48 | 48 | 1 | 1 | 1 | 656 | 304 | 1000 | 80 | 48 | 48 |
| 3 | E | 1 | 48 | 48 | 16 | 80 | 16 | 16 | 176 | 1000 | 144 | 48 | 1 | 1 | 1 | 16 | 16 | 48 | 48 | 16 | 48 | |
| 2 | H | 1 | 54 | 576 | 18 | 72 | 18 | 108 | 18 | 18 | 36 | 1000 | 54 | 36 | 1 | 648 | 486 | 918 | 288 | 216 | 558 | |
| Caspase 7 | 4 | D | 1 | 117 | 78 | 1 | 39 | 26 | 26 | 1000 | 104 | 26 | 39 | 1 | 1 | 1 | 13 | 1 | 13 | 13 | 13 | 1 |
| 3 | E | 1 | 221 | 357 | 1 | 323 | 51 | 153 | 255 | 1000 | 425 | 187 | 85 | 102 | 1 | 306 | 221 | 697 | 204 | 204 | 102 | |
| 2 | V | 1 | 16 | 448 | 448 | 128 | 16 | 48 | 1 | 1 | 16 | 208 | 80 | 16 | 1 | 704 | 176 | 1000 | 160 | 128 | 48 | |
| Caspase 8 | 4 | L | 1 | 208 | 304 | 480 | 448 | 96 | 304 | 704 | 448 | 96 | 144 | 1 | 1 | 1 | 576 | 1000 | 720 | 224 | 256 | 144 |
| 3 | E | 1 | 45 | 75 | 0 | 45 | 15 | 15 | 180 | 1000 | 150 | 45 | 1 | 1 | 1 | 45 | 15 | 105 | 45 | 45 | 15 | |
| 2 | T | 1 | 180 | 1000 | 216 | 324 | 18 | 126 | 72 | 198 | 108 | 306 | 72 | 72 | 1 | 720 | 108 | 792 | 180 | 198 | 306 | |
| Caspase 9 | 4 | L | 1 | 198 | 216 | 594 | 576 | 144 | 108 | 414 | 468 | 180 | 126 | 36 | 18 | 1 | 576 | 1000 | 684 | 252 | 216 | 144 |
| 3 | E | 1 | 85 | 136 | 51 | 85 | 17 | 17 | 272 | 1000 | 187 | 119 | 1 | 17 | 1 | 102 | 119 | 204 | 102 | 85 | 51 | |
| 2 | H | 1 | 85 | 136 | 102 | 85 | 17 | 17 | 51 | 34 | 17 | 1000 | 34 | 1 | 1 | 187 | 17 | 153 | 51 | 34 | 51 |
Amino acid preference distribution for each position Pi was extracted from Thornberry et al. (6) giving the most common amino acid a value of 1000.
Computing the scores of endopeptidase cleavage sites
For computing the score, the GraBCas program screens for tetrapeptides with Asp (D) at their last position (P1) in a given amino acid sequence. Given the tetrapeptide A4A3A2D (≈P4P3P2P1) of a potential cleavage site, its score for a given endopeptidase is computed by multiplying the corresponding matrix entries of A2 at position P2, A3 at position P3 and A4 at position P4. The product is divided by the value (10003) of the product of the consensus recognition motif for normalization and multiplied by 100, yielding a total score between 0 and 100.
Using additional filter options for granzyme B and caspase 3
To improve the power of the prediction we analyzed the amino acid distribution of known granzyme B and caspase 3 cleavage sites at positions P6–P2′ taken from the literature (see also Supplementary Material 1 and 2).
For granzyme B we found a preference for V (15×) and I (11×) at position P4, for E (9×) at position P3 and for P (11×) at position P2 in accordance with the results of Thornberry et al. (6). We detected S at position P1′ and G at position P2′, respectively in 9 out of 30 cleavage sites. The result list of the PSSM-based cleavage sites can optionally be filtered with two ‘stringency’ filters that take the occurrence of amino acids at position P2′ into account. We installed a ‘low stringency’ filter that excludes hits with the amino acids C, Q, I, M, V all of which are medium sized or large amino acids. A second ‘high stringency’ filter selects hits with a G at position P2′.
The analysis of the 59 cleavage sites of caspase 3 substrate confirmed the preferences for D at P4 (31×), E at P3 (17×) and V at P2 (16×). For P1′ we found an abundance of G (18×) and S (17×) and in lower amount A (5×) and N (4×). As for the granzyme B prediction, two additional ‘stringency’ filters for the prediction of caspase 3 cleavage sites are available. The ‘high stringency’ filter screens the predicted hits for occurrences of G, S, A or N at position P1′, and the ‘low stringency’ filter screens for absence of R, E, H, K, Q, I, L, M, F, W and Y at this position.
GraBCas software tool
The GraBCas program was written in Java™ and is available as an application or as an applet. Both are available at http://wwwalt.med-rz.uniklinik-saarland.de/med_fak/humangenetik/software/index.html. If your browser does not support Java™ you need to install the Java Runtime Environment (JRE) 1.4.x, which can be downloaded at http://java.sun.com.
The graphical user interface is easy to use. There are several register cards for each endopeptidase and one register card presenting the input form, where the amino acid sequence can be pasted and a cutoff for the PSSM scores can be chosen. After pressing the OK-button in the input form, the program calculates the scores of potential cleavage sites for all endopeptidases and presents them in the corresponding register card sorted with the highest scoring sites on top. The user can open an additional window for viewing the positions of the predicted cleavage sites within the amino acid sequence. The window also shows the fragment length and size in kDa (0.11 kDa per amino acid) of the predicted fragments.
As described above, for caspase 3 and granzyme B additional filter options are available in their register cards. The two filter types for these enzymes, a ‘high-stringency’ and a ‘low-stringency’ filter, are based on the extended substrate specificity. For granzyme B the amino acids at position P2′ were taken into account in addition to the positions P4–P1. For caspase 3, amino acids at position P1′ are evaluated.
Sensitivity–specificity plots
For determining the specificity and sensitivity of the GraBCas predictions we used the known cleavage sites of granzyme B (4–6,9–12) summarized in Table 2 and the known non-substrates of granzyme B (5) presented in Table 4. Due to the lack of information on known non-substrates for caspase 3 the sensitivity–specificity plot could only be calculated for granzyme B (Figure 1).
Table 2.
Analysis of cleavage sites of known granzyme B substrates with GraBCas
| Granzyme B substrate | Acc_number | Known cleavage site | Score by GraBCas | P6–P2′ of cleavage site |
|---|---|---|---|---|
| AARS: alanyl-tRNA synthetase | NP_001596 | VADP (632) | 7,65 | SLVAPDRL |
| ADPRT: ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase) | NP_001609 | VDPD (536) | 9,9 | AAVDPDSG |
| BID: BH3 interacting domain death agonist | NP_001187 | IEAD (75) | 57,6 | GRIEADSE |
| CASP3: caspase 3, apoptosis-related cysteine protease | NP_004337 | IETD (175) | 54,4 | CGIETDSG |
| CASP7: caspase 7, apoptosis-related cysteine protease | NP_001218 | IQAD (198) | 4,6656 | DGIQADSG |
| CENPB: centromere protein B, 80 kDa | NP_001801 | VDSD (457) | 7,4448 | GDVDSDEE |
| CHD4: chromodomain helicase DNA binding protein 4 | NP_001264 | VDPD (1312) | 9,9 | ESVDPDYW |
| DFFA: DNA fragmentation factor, 45 kDa, alpha polypeptide | NP_004392 | DETD (117) | 0,0544 | MEVTGDAG |
| DFFA: DNA fragmentation factor, 45 kDa, alpha polypeptide | NP_004392 | VTGD (6) | 0,0432 | DVDETDSG |
| FBL: fibrillarin | NP_001427 | VGPD (184) | 23,85 | DIVGPDGL |
| FLNA: filamin A, alpha (actin binding protein 280) | NP_001447 | ? | 11,4048 | TEIDQDKY |
| G22P1: thyroid autoantigen 70 kDa (Ku antigen) | NP_001460 | ISSD (79) | 22,3344 | KIISSDRD |
| GRIA3: glutamate receptor, ionotrophic, AMPA 3 | NP_000819 | ISND (416) | 18,5328 | QQISNDSA |
| HARS: histidyl-tRNA synthetase | NP_002100 | LGPD (48) | 2,4804 | AQLGPDES |
| IARS: isoleucine-tRNA synthetase | NP_002152 | VTPD (983) | 2,7 | LDVTPDQS |
| L4 100K [Human adenovirus C] | AAQ19301 | IEQD (48) | 57,6 | VIIEQDPG |
| MKI67: antigen identified by monoclonal antibody Ki-67 | NP_002408 | VCTD (1481) | 0,0272 | TPVCTDKP |
| NUMA1: nuclear mitotic apparatus protein 1 | NP_006176 | VATD (1705) | 4,1616 | FQVATDAL |
| PMS1: PMS1 postmeiotic segregation increased 1 | NP_000525 | ISAD (496) | 17,1072 | SEISADEW |
| PMS2: PMS2 postmeiotic segregation increased 2 | NP_000526 | VEKD (493) | 0,05 | AEVEKDSG |
| PMSCL2: polymyositis/scleroderma autoantigen 2, 100 kDa | NP_002676 | VEQD (252) | 28,8 | QQVEQDMF |
| POLR1A: polymerase (RNA) I polypeptide A, 194 kDa | NP_056240 | ICPD (448) | 0,1 | SVICPDMY |
| POLR2A: polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa | NP_000928 | ITPD (370) | 5,4 | TVITPDPN |
| PRKDC: protein kinase, DNA-activated, catalytic polypeptide | NP_008835 | VGPD (2698) | 23,85 | KSVGPDFG |
| SNRP70: small nuclear ribonucleoprotein 70 kDa polypeptide (RNP antigen) | NP_003080 | LGND (409) | 1,5477696 | EGLGNDSR |
| SRP72: signal recognition particle 72 kDa | NP_008878 | VTPD (573) | 2,7 | PKVTPDPE |
| SSB: Sjogren syndrome antigen B (autoantigen La) | NP_003133 | LEED (220) | 1,4976 | QKLEEDAE |
| TOP1: topoisomerase (DNA) I | NP_003277 | IEAD (15) | 57,6 | SQIEADFR |
| UBE4B: ubiquitination factor E4B (UFD2 homolog, yeast) | NP_006039 | VDVD (123) | 3,0096 | SQVDVDSG |
| UBTF: upstream binding transcription factor, RNA polymerase I | NP_055048 | VRPD (220) | 0,05 | LKVRPDAT |
The bold printed amino acids in the extended cleavage site indicate hits with a G residue at position P2′ detected by the high stringency filter. Numbers in brackets indicate cleavage site position in the amino acid sequence.
Table 4.
Analysis of cleavage sites of known non-substrates of granzyme B with GraBCas
| Granzyme B non-substrate | Acc_number | Best hit | Score by GraBCas |
|---|---|---|---|
| TRIM21: 52 kD Ro/SSA autoantigen | NP_003132 | LDPD (294) | 1,0296 |
| SSA2: 60 kD Ro/SSA autoantigen | NP_004591 | VTTD (427) | 1,4688 |
| XRCC5: ATP-dependent DNA helicase II Ku80 | NP_066964 | FGTD (62) | 0,3113856 |
| VCL: vinculin isoform VCL | NP_003364 | LQSD (98) | 0,3167424 |
| VCL: vinculin isoform meta-VCL | NP_054706 | LQSD (98) | 0,3167424 |
| TUBB2: tubulin, beta 2 | NP_001060 | VISD (26) | 0,0376 |
| CRP: C-reactive protein, pentraxin-related | NP_000558 | LSPD (187) | 1,5444 |
| SERPINA1: serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | NP_000286 | LAED (26) | 0,2291328 |
| SERPINA1: serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | NP_001002235 | LAED (26) | 0,2291328 |
| SERPINA1: serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | NP_001002236 | LAED (26) | 0,2291328 |
| GSTA1: glutathione S-transferase A1 | NP_665683 | VEID (61) | 0,8 |
| PYGB: brain glycogen phosphorylase | NP_002853 | IEED (129) | 28,8 |
| TF: transferring | NP_001054 | VTLD (82) | 0,2592 |
| LTF: lactotransferrin | NP_002334 | VTLD (79) | 0,2592 |
| LYZ: lysozyme precursor | NP_000230 | RSTD (71) | 0,0161568 |
| ORM1: orosomucoid 1 precursor | NP_000598 | LAFD (133) | 0,1145664 |
| F2: coagulation factor II precursor (Thrombin B-chain) | NP_000497 | LDED (306) | 0,2965248 |
Numbers in brackets indicate cleavage site position in the amino acid sequence.
Figure 1.
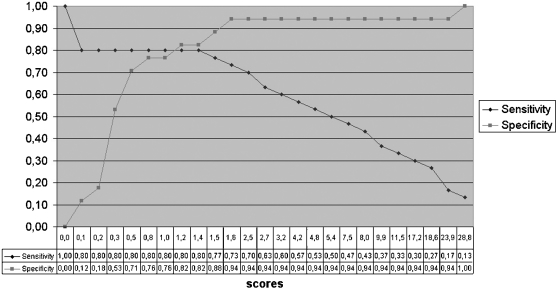
Sensitivity–specificity plot for granzyme B. x-axis: scores by the GraBCas program; y-axis: percentage of specificity or sensitivity.
The x-axis of the plots represents the cutoff values (with respect to the PSSM scores), while the y-axis represents the percentage of the specificity or sensitivity of the predictions made by GraBCas, respectively. The specificity is computed as follows:
The true negatives are the known non-substrates where the maximal PSSM score of all tetrapeptides ending with a D is smaller than the chosen cutoff value. A specificity of 1 means that all known non-substrates were below the cutoff, i.e. all known non-substrates were correctly classified as negatives.
The sensitivity is defined as:
where true positives are the known cleavage sites with a score larger than the chosen cutoff value. A sensitivity of 1 means that all cleavage sites of our test set (Table 2) have a score higher than the chosen cutoff and that they have been correctly classified as positives.
RESULTS AND DISCUSSION
We analyzed the cleavage sites of known substrates of granzyme B and caspase 3 to compare the experimentally identified peptide specificity with the cleavage site predicted by the program GraBCas.
In total, we collected 29 substrates with 30 cleavage sites for granzyme B (Table 2) and 47 substrates with 59 cleavage sites for caspase 3 (Table 3) and computed the GraBCas scores of the cleavage sites. For granzyme B we collected additionally 17 sequences which are non-substrates of this endopeptidase (Table 4), computed the scores of all putative cleavage sites in these sequences and extracted the best hit by GraBCas for each of these non-substrates.
Table 3.
Analysis of cleavage sites of known caspase 3 substrates with GraBCas
| Caspase 3 substrate | Acc_number | Known cleavage site | Score by GraBCas | P6–P2′ of cleavage site |
|---|---|---|---|---|
| ADD1: adducing 1 (alpha) | NP_001110 | DDSD (633) | 0,357 | TGDDSDAA |
| APAF1: apoptotic protease activating factor | NP_001151 | SVTD (271) | 0,462672 | DKSVTDSV |
| ARHGDIB: Rho GDP dissociation inhibitor (GDI) beta | NP_001166 | DELD (19) | 22,4 | DDDELDSK |
| ATP2B4: ATPase, Ca++ transporting, plasma membrane 4 | NP_001675 | DEID (1080) | 71,4 | GLDEIDHA |
| BAD: BCL2-antagonist of cell death | NP_004313 | EQED (14) | 0,001632 | PSEQEDSS |
| BAX: BCL2-associated X protein | NP_004315 | FIQD (33) | 0,002142 | QGFIQDRA |
| BCL2: B-cell CLL/lymphoma 2 | NP_000624 | DAGD (34) | 0,0357 | EWDAGDVG |
| BCL2L1: BCL2-like 1 | NP_001182 | HLAD (61) | 0,027846 | SWHLADSP |
| BCL2L1: BCL2-like 2 | NP_001182 | SSLD (76) | 0,274176 | HSSSLDAR |
| BIRC2: baculoviral IAP repeat-containing 2 | NP_001157 | ENAD (372) | 0,111384 | GEENADPP |
| BLM: Bloom syndrome | NP_000048 | TEVD (415) | 5 | LLTEVDFN |
| BRCA1: breast cancer 1, early onset | NP_009225 | DLLD (1154) | 3,4272 | PDDLLDDG |
| CAMK4: calcium/calmodulin-dependent protein kinase IV | NP_001735 | YWID (31) | 0,0084966 | PDYWIDGS |
| CAMK4: calcium/calmodulin-dependent protein kinase IV | NP_001735 | PAPD (176) | 0,0144942 | ATPAPDAP |
| CDC2L1: cell division cycle 2-like 1 (PITSLRE proteins) | NP_001778 | YVPD (391) | 0,0124236 | GDYVPDSP |
| CDC6: CDC6 cell division cycle 6 homolog (Saccharomyces cerevisiae) | NP_001245 | SEVD (442) | 4 | VISEVDGN |
| CDC6: CDC6 cell division cycle 6 homolog (S.cerevisiae) | NP_001245 | LVRD (99) | 0,0055692 | RRLVFDNQ |
| CDKN1A: cyclin-dependent kinase inhibitor 1A (p21, Cip1) | NP_000380 | DHVD (112) | 18,7 | EEDHVDLS |
| CSEN: calsenilin, presenilin binding protein, EF hand transcription factor | NP_038462 | DSSD (64) | 0,4284 | GSDSSDSE |
| CTNNB1: catenin (cadherin-associated protein), beta 1, 88 kDa | NP_001895 | DLMD (764) | 0,0153 | AQDLMDGL |
| CTNNB1: catenin (cadherin-associated protein), beta 1, 88 kDa | NP_001895 | YPVD (751) | 10 | ADYPVDGL |
| CTNNB1: catenin (cadherin-associated protein), beta 1, 88 kDa | NP_001895 | ADID (83) | 0,18207 | QVADIDGQ |
| CTNNB1: catenin (cadherin-associated protein), beta 1, 88 kDa | NP_001895 | TQFD (115) | 0,37128 | PSTQFDAA |
| CTNNB1: catenin (cadherin-associated protein), beta 1, 88 kDa | NP_001895 | SYLD (32) | 0,22848 | QQSYLDSG |
| DFFA: DNA fragmentation factor, 45 kDa, alpha polypeptide | NP_004392 | DAVD (224) | 35,7 | EVDAVDTG |
| DFFA: DNA fragmentation factor, 45 kDa, alpha polypeptide | NP_004392 | DETD (117) | 37,8 | DVDETDSG |
| DRPLA: dentatorubral-pallidoluysian atrophy (atrophin-1) | NP_001931 | DSLD (109) | 6,8544 | DLDSLDGR |
| EIF2S1: eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa | NP_004085 | AEVD (301) | 1 | ENAEVDGD |
| EIF2S1: eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa | NP_004085 | DGDD (304) | 0,0085 | EVDGDDDA |
| FNTA: farnesyltransferase, CAAX box, alpha | NP_002018 | VSLD (59) | 0,137088 | GFVSLDSP |
| GCLC: glutamate-cysteine ligase, catalytic subunit | NP_001489 | AVVD (499) | 0,306 | GNAVVDGC |
| GSN: gelsolin (amyloidosis, Finnish type) | NP_000168 | DQTD (403) | 15,4224 | DPDQTDGL |
| HD: huntingtin (Huntington disease) | NP_002102 | DSVD (513) | 30,6 | WEAQRDSH |
| HNRPU: heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | NP_004492 | SALD (100) | 0,319872 | GISALDGD |
| IL16: interleukin 16 (lymphocyte chemoattractant factor) | NP_004504 | SSTD (510) | 0,462672 | LNSSTDSA |
| IL18: interleukin 18 (interferon-gamma-inducing factor) | NP_001553 | LESD (36) | 0,0014 | ENLESDYF |
| KRT18: keratin 18 | NP_000215 | VEVD (238) | 2 | LTVEVDAP |
| MAPT: microtubule-associated protein tau | NP_005901 | DMVD (421) | 0,1 | SIDMVDSP |
| MDM2: Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) | NP_002383 | DVPD (361) | 12,4236 | GFDVPDCK |
| NFKBIA: nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NP_065390 | DRHD (32) | 0,3332 | LDDRHDSG |
| NUMA1: nuclear mitotic apparatus protein 1 | NP_006176 | DSLD (1712) | 6,8544 | SIDSLDLS |
| PAK2: p21 (CDKN1A)-activated kinase 2 | NP_002568 | SHVD (212) | 0,748 | GDSHVDGA |
| POLE: polymerase (DNA directed), epsilon | NP_006222 | DQLD (189) | 9,1392 | IADQLDNI |
| POLE: polymerase (DNA directed), epsilon | NP_006222 | DMED (1185) | 10 | APDMEDFG |
| PPP2R1A: protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | NP_055040 | DEQD (218) | 1,4 | ASDEQDSV |
| PRKCD: protein kinase C, delta | NP_006245 | DMQD (329) | 0,0014 | GEDMQDNS |
| PRKCM: protein kinase C, mu | NP_002733 | CQND (378) | 5,712 | AECQNDSG |
| PRKCQ: protein kinase C, theta | NP_006248 | DEVD (354) | 100 | PLDEVDKM |
| PRKCZ: protein kinase C, zeta | NP_002735 | DGVD (239) | 0,0085 | VIDGMDGI |
| PRKCZ: protein kinase C, zeta | NP_002735 | EETD (210) | 1,512 | PSEETDGI |
| PRKDC: protein kinase, DNA-activated, catalytic polypeptide | NP_008835 | DEVD (2713) | 100 | PGDEVDNK |
| PSEN2: presenilin 2 (Alzheimer disease 4) | NP_000438 | DSYD (329) | 4,7124 | EEDSYDSF |
| RB1: retinoblastoma 1 (including osteosarcoma) | NP_000312 | DEAD (886) | 18,2 | GSDEADGS |
| RFC1: replication factor C (activator 1) 1, 145 kDa | NP_002904 | DEVD (722) | 100 | IMDEVDGM |
| ROCK1: Rho-associated, coiled-coil containing protein kinase 1 | NP_005397 | DETD (1113) | 37,8 | SADETDGN |
| SNRP70: small nuclear ribonucleoprotein 70 kDa polypeptide (RNP antigen) | NP_003080 | DGPD (341) | 3,451 | GPDGPDGP |
| SPTBN1: spectrin, beta, non-erythrocytic 1 | NP_003119 | DEVD (1457) | 100 | STDEVDSK |
| SPTBN1: spectrin, beta, non-erythrocytic 2 | NP_003119 | ETVD (2146) | 1,428 | MAETVDTS |
| VIM: vimentin | NP_003371 | DSVD (85) | 30,6 | KGDEVDGV |
The bold printed amino acids in the extended clevage site indicate hits detected by the high stringency filter. Numbers in brackets indicate clevage site position in the amino acid sequence.
The sensitivity–specificity plot for granzyme B is shown in Figure 1. When using a cutoff value of 1.2 in the GraBCas program, we obtain a sensitivity of ∼80% and a specificity of ∼82%. The cutoff value can be adjusted if a higher specificity or sensitivity is needed for the cleavage site prediction.
A closer look at the sensitivity–specificity plot shows that the best score (28.8 for IEED in glycogen phosphorylase) of the alleged non-substrates is extremely high. The top value of the best hit IEED is due to the fact that this tetrapeptide has three identical positions with the granzyme B consensus recognition motif IEPD. Furthermore, the amino acid E on P2 has a middle-sized value and the tetrapeptides LEED, IEAD and IETD are known substrates of granzyme B. We assume that glycogen phosphorylase is probably a substrate of granzyme B. This warrants further experimental analysis.
We also studied the occurrences of amino acids at position P1′ and P2′ of the known cleavage sites of granzyme B and caspase 3. Additional filtering options have been added to GraBCas that are based on these statistics. For granzyme B, we detected G at position P2′ in 9/30 cleavage sites. This confirms the results of Harris et al. (14), who found for recombinant rat granzyme B a specificity for G at P2′. We did not, however, confirm the proposed total absence of charged amino acids at P1′, in that we found E three times, R two times and K and D one time, each.
For caspase 3, we found in total 44/59 (75%) cleavage sites with G, S, A or N at position P1′. These results are in good accordance with the results of Stennicke et al. (15). Absent amino acids included the charged residues R, E, H, K and the large residues Q, I, L, M, F, W and Y.
With GraBCas we provide a position specific scoring scheme for the prediction of cleavage sites for granzyme B and caspases 1–9. GraBCas offers an easy to use, concise user interface in register card format. The design of GraBCas specifically acknowledged the high variability of cleavage site recognition sequences. We validated our tool by scoring known substrates and demonstrated its usefulness for protein sequence analysis. GraBCas may contribute to a more comprehensive knowledge of caspase and granzyme B substrates and a better understanding of the biological roles of these enzymes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This study was supported by a grant of the Center of Bioinformatics/Saarbrücken supported by the Deutsche Forschungsgemeinschaft and by a grant from the Deutsche Krebshilfe (10-1966-Me4). Funding to pay the Open Access publication charges for this article was provided by the University of Saarland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Los M., Stroh C., Janicke R.U., Engels I.H., Schulze-Osthoff K. Caspases: more than just killers? Trends Immunol. 2001;22:31–34. doi: 10.1016/s1471-4906(00)01814-7. [DOI] [PubMed] [Google Scholar]
- 2.Algeciras-Schimnich A., Barnhart B.C., Peter M.E. Apoptosis-independent functions of killer caspases. Curr. Opin. Cell Biol. 2002;14:721–726. doi: 10.1016/s0955-0674(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 3.Heusel J.W., Wesselschmidt R.L., Shresta S., Russell J.H., Ley T.J. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 4.Sharif-Askari E., Alam A., Rheaume E., Beresford P.J., Scotto C., Sharma K., Lee D., DeWolf W.E., Nuttall M.E., Lieberman J., Sekaly R.P. Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation. EMBO J. 2001;20:3101–3113. doi: 10.1093/emboj/20.12.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casciola-Rosen L., Andrade F., Ulanet D., Wong W.B., Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J. Exp. Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thornberry N.A., Rano T.A., Peterson E.P., Rasper D.M., Timkey T., Garcia-Calvo M., Houtzager V.M., Nordstrom P.A., Roy S., Vaillancourt J.P., Chapman K.T., Nicholson D.W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Calvo M., Peterson E.P., Rasper D.M., Vaillancourt J.P., Zamboni R., Nicholson D.W., Thornberry N.A. Purification and catalytic properties of human caspase family members. Cell Death Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 8.Fischer U., Janicke R.U., Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darmon A.J., Nicholson D.W., Bleackley R.C. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature. 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 10.Andrade F., Roy S., Nicholson D., Thornberry N., Rosen A., Casciola-Rosen L. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- 11.Sutton V.R., Davis J.E., Cancilla M., Johnstone R.W., Ruefli A.A., Sedelies K., Browne K.A., Trapani J.A. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 2000;192:1403–1414. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas D.A., Du C., Xu M., Wang X., Ley T.J. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 13.Lohmuller T., Wenzler D., Hagemann S., Kiess W., Peters C., Dandekar T., Reinheckel T. Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol. Chem. 2003;384:899–909. doi: 10.1515/BC.2003.101. [DOI] [PubMed] [Google Scholar]
- 14.Harris J.L., Peterson E.P., Hudig D., Thornberry N.A., Craik C.S. Definition and redesign of the extended substrate specificity of granzyme B. J. Biol. Chem. 1998;273:27364–27373. doi: 10.1074/jbc.273.42.27364. [DOI] [PubMed] [Google Scholar]
- 15.Stennicke H.R., Renatus M., Meldal M., Salvesen G.S. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.