Abstract
The identification of the genes that participate at the biological interface of two species remains critical to our understanding of the mechanisms of disease resistance, disease susceptibility and symbiosis. The sequencing of complementary DNA (cDNA) libraries prepared from the biological interface between two organisms provides an inexpensive way to identify the novel genes that may be expressed as a cause or consequence of compatible or incompatible interactions. Sequence classification and annotation of species origin typically use an orthology-based approach and require access to large portions of either genome, or a close relative. Novel species- or clade-specific sequences may have no counterpart within existing databases and remain ambiguous features. Here we present a web-service, Éclair, which utilizes support vector machines for the classification of the origin of expressed sequence tags stemming from mixed host cDNA libraries. In addition to providing an interface for the classification of sequences, users are presented with the opportunity to train a model to suit their preferred species pair. Éclair is freely available at http://eclair.btk.fi.
INTRODUCTION
The identification of the genes, their corresponding proteins and the concomitant protein networks mediating successfully between two species and resulting in either disease or symbiosis remains critical to contemporary research (1–4). While genome-wide expression-profiling approaches have been applied to successfully identify the genes that are differentially regulated within either organism (5–7), not all species are endowed with the luxury of whole genome arrays or even completely sequenced genome scaffolds. In the many ‘more exotic’ interacting species pairs where neither genome is sequenced, there remains a need to sample the pools of genes expressed at the biological interface. Expressed sequence tag (EST) sequencing has come to the forefront as a robust but relatively inexpensive method for sampling the protein encoding genes that are expressed within a tissue, reviewed in (8). Dissection techniques may be used to prepare tissue homogeneous for each of the test organisms, e.g. from within a plant–nematode interaction. Complementary DNA (cDNA) that is homogeneous for species origin may be prepared from such tissue. This scenario becomes much more complicated when we wish to consider finer biological interfaces such as those within plant bacterial interactions where the bacteria may exist as an intracellular parasite, or where a fungal genome may co-exist with a host plant as an endophyte. In such cases it is easiest to prepare and sequence cDNA libraries that contain mixed content from both genomes. Dozens of such cDNA libraries already appear within the large publicly available EST sequence databases for plant pathogen pairs including soybean and Phytophthora sojae (9), Biomphalaria glabrata and Schistosoma mansoni (10), Medicago truncatula and Glomus versiforme, Populus tremula x P.tremuloides and Amanita muscaria, Gerbera hybrid and Botrytis, Oryza sativa and Magnaporthe grisea.
The sceptre of such mixed libraries has already been raised, and bioinformatics solutions that can assign sequences to one of the defined parental species with varying degrees of success have been described (11–14). These solutions, however, remain firmly within the realm of the bioinformatics laboratory. They require large BLAST databases, or require robust training and test datasets. This demands the pre-processing of sequence to strip redundancy from the collection and to identify the probable protein coding sequence (CDS) that can be used to build classifiers based on e.g. the underlying codon and amino acid usages.
Here we present an integrated bioinformatics solution, Éclair, that can be automatically trained and tested for the classification of the species origin for ESTs sequenced from mixed cDNA libraries. In addition to providing a framework upon which a classification model may be produced and used, the Éclair web server also provides pre-computed models for a series of the more common host:pathogen and host:host pairs that have been encountered within our research.
ECLAT
The Eclat solution (14) to the problem of differentiating species origin for ESTs sequenced from mixed libraries uses a support vector machine (SVM) method for classification. The Eclat SVM is trained to discriminate between species on the basis of codon frequencies—this requires robust training and test sequence data from both species. These data should stem from either the genomes that have to be classified or their close taxonomic relatives, and should be homogeneous for species origin. Eclat provides internal methodology to predict the CDS, but can also use CDS predictions generated by other methods. The SVM model is then used by the classifier methods to assign a CDS to one of the parental species. Eclat has been empirically shown to offer superior classification rates when compared with other approaches (14).
IMPLEMENTATION
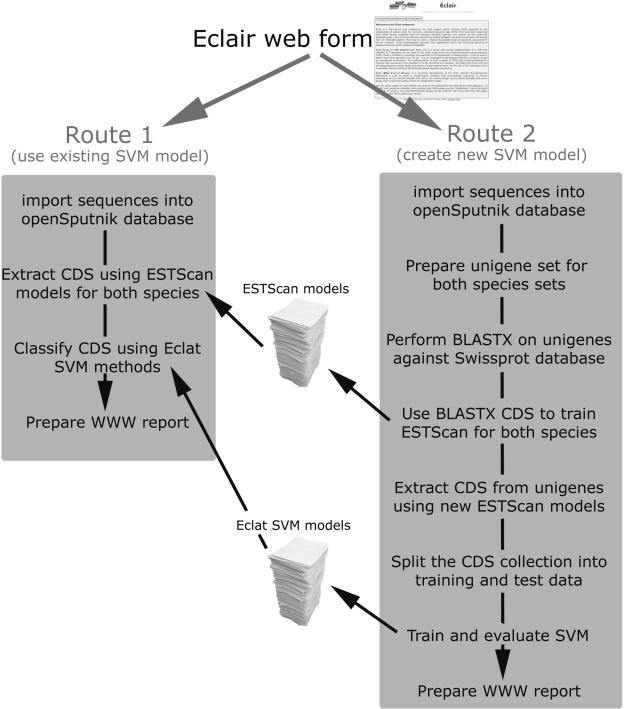
Éclair is a web-service that builds upon the functionality provided by Eclat to allow a user to estimate the probable origin of ESTs from within a mixed sequence collection. Éclair utilizes the core analytical pipeline from the openSputnik software (15). The logic flow for the Éclair web application is summarized in Figure 1. The core openSputnik software and the Éclair adapters are implemented using the JAVA programming language. The web display and interfaces are written in Python and are implemented as a Zope product. The upload of sequences to the Éclair service will create a series of case scripts that are run within a distributed Linux environment using the Sun GridEngine software for job scheduling. There are two approaches for the use of Éclair.
Figure 1.
A schematic showing the workflow as applied by the Éclair web-server to classify EST sequences for species origin. There are two routes by which Éclair may be used. In Route 1, a user applies an already existing model to classify sequences. In the second scenario, Route 2, the application is trained. A user uploads homogeneous sequence collections and these are used to prepare the required models for ESTScan and the Eclat SVM. Both methods produce extensive WWW reporting to indicate sequence origins and to indicate the sensitivity and selectivity of the underlying models.
In the first instance (Route 1), a user has a collection of EST sequences that have been sequenced from the biological interface of two organisms. Each of the species has already been trained within the Éclair system so no new model needs to be produced. The user uploads the sequences to the Éclair server along with information on the species these sequences should be classified to. An openSputnik project is created and the sequences are imported. From each EST sequence, a CDS is predicted through the ESTScan application (16) using each of the species-specific ESTScan models. The resultant CDSs are classified using the Eclat SVM and the results are placed back in the openSputnik database. A result page is produced that identifies the sequences predicted to stem from each of the genomes, along with basic statistics as to how successful the SVM model was at the time of preparation.
In the second instance (Route 2), a user has a sequence collection associated with a species pair that is novel to Éclair. To produce and test an Eclat SVM, training data must be supplied; the user, therefore, uploads homogeneous data stemming from each of the species and sequence data from the mixed library. openSputnik projects are created for each of the datasets and sequences are inserted into the underlying database. To remove any codon bias within the more abundant transcripts, the training data are clustered and assembled using the sequence assembly pipeline within the openSputnik application. The CDS is identified from the unigene sequences by performing a BLASTX (17) against the Swissprot database, filtering the results arbitrarily at 1 × 10−8 and selecting, where applicable, the best result. These CDSs are used to train an ESTScan model which in turn is used with ESTScan to predict the CDS from the remaining unigenes. Repeated random sampling of the CDS sets are used to split the dataset into training and test sets. Training data is used to produce an Eclat SVM, while the test data is used to evaluate the efficacy of the resulting model. The results from the repeated random samples are retained and are displayed with the classification results to indicate any probable error.
APPLICATION OF ÉCLAIR
We have tested the Éclair web service using EST data from the host pairs shown in Table 1. The number of sequences used for testing and training is shown along with some statistics that demonstrate the efficiency with which Éclair has classified sequences during the repeated training and testing cycles. Using the species pair with most underlying EST sequences (Lycopersicon esculentum and Phytophthora infestans) we have additionally tested the effect of the number of sequences used to train the model with the sensitivity of the final model (data not shown). This reveals that at least 1000 unigene training sequences for each species should be the minimum number applied to obtain an optimal SVM model, but only as few as 100 training sequences for each species are required to establish a model that has >80% sensitivity and selectivity. The efficiency of any model is of course largely dependent upon the underlying differences in both codon usage and amino acid usage.
Table 1.
A list of the host: pathogen pairs that are available through the Éclair web-server and basic statistics that illustrate the effectiveness of the underlying Eclat SVM models
| Pathogen genome | Host genome | Test data | Host | PP | HP | HP | HH | Evaluation of the SVM model | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | Pathogen | Host | |||||||||
| Sensitivity | Selectivity | Sensitivity | Selectivity | ||||||||
| Blumeria graminis | Hordeum vulgare | 292.9 | 1254.4 | 206.4 | 79.2 | 72.7 | 1061.2 | 0.72 (0.07) | 0.74 (0.03) | 0.94 (0.01) | 0.93 (0.02) |
| Globodera pallida | Solanum tuberosum | 727.7 | 1247.0 | 605.7 | 73.4 | 64.7 | 1096.2 | 0.89 (0.01) | 0.90 (0.01) | 0.94 (0.01) | 0.94 (0.01) |
| Haemonchus contortus | Ovis aries | 838.6 | 861.4 | 771.2 | 67.0 | 55.5 | 796.5 | 0.92 (0.02) | 0.93 (0.03) | 0.93 (0.03) | 0.92 (0.02) |
| Heterodera glycines | Glycine max | 1246.4 | 1239.5 | 1166.3 | 64.3 | 182.9 | 890.4 | 0.95 (0.03) | 0.86 (0.08) | 0.83 (0.16) | 0.93 (0.03) |
| M.grisea | O.sativa | 742 | 1241.3 | 640.5 | 116.4 | 98.9 | 974.8 | 0.85 (0.02) | 0.87 (0.01) | 0.91 (0.01) | 0.89 (0.01) |
| Manduca sexta | Nicotiana tabacum | 164.2 | 1237.3 | 133.8 | 35.0 | 11.5 | 898.7 | 0.79 (0.03) | 0.92 (0.02) | 0.99 (0.00) | 0.96 (0.01) |
| Meloidogyne incognita | Gossypium arboreum | 1247 | 1245.6 | 1073 | 100.8 | 138.2 | 969.4 | 0.91 (0.02) | 0.89 (0.01) | 0.88 (0.01) | 0.91 (0.02) |
| Neurospora crassa | Arabidopsis thaliana | 542.4 | 1251.8 | 503.9 | 34.7 | 44.8 | 1075.1 | 0.94 (0.03) | 0.92 (0.01) | 0.96 (0.01) | 0.97 (0.01) |
| Phytophthora infestans | L.esculentum | 1241.1 | 1240.1 | 1117.6 | 123.5 | 52.8 | 1074.1 | 0.90 (0.01) | 0.95 (0.01) | 0.95 (0.01) | 0.90 (0.01) |
| P.sojae | Glycine max | 1248.7 | 1242.1 | 1227 | 32.7 | 40.6 | 1025.0 | 0.97 (0.01) | 0.97 (0.01) | 0.96 (0.01) | 0.97 (0.01) |
Following the unigene assembly and CDS prediction steps in Éclair the dataset was randomly sampled 10 times to produce representative data sets for Eclat training. The sampled data was split so that 75% of the sequences were used for model training; with the remaining 25% of sequences used for testing. The average number of sequences used for testing are shown for the pathogen and host datasets. Following creation of the Eclat SVM model, the retained test sequences were classified, the average numbers of sequences classified are shown in columns PP, PH, HP and HH where PP represents a pathogen sequence classified as a pathogen sequence, PH represents a pathogen sequence classified as a host sequence and so on. The results of the simulations are summarized as sensitivity and selectivity for both the pathogen and host sequence models, the standard deviations from the 10 replicates are shown in brackets.
FUTURE DIRECTIONS
The Éclair pipeline is to be fully integrated with the openSputnik database to provide an additional annotative resource for EST collections sequenced from mixed cDNA libraries. This will provide detailed classification for the large numbers of already existing sequences of ‘unclear’ origin. The openSputnik association will also provide information on and context with complete and draft genome assemblies. EST sequences that can be anchored at high confidence to annotated genes will be automatically excluded from the processing by the Éclair pipeline thereby increasing the quality of classification.
The Éclair web-server will be further developed by the inclusion of additional Eclat SVM models for new host::pathogen pairs as they are encountered within our research, when they are requested by the community or when models are created by users. This will hopefully shift the current bias for plant genomes towards a more comprehensive platform for host interactions.
AVAILABILITY
The Éclair system is a freely available web resource and may be used anonymously by all scientific users. An email address can be supplied and the system will alert the user to visit a URL to retrieve the results of the completed analysis; this URL is also supplied at submission time. The only caveats that are imposed are with the creation of new Eclat SVM models. The process of clustering, assembly and training is computationally expensive and before a job is executed it is subject to checks by an annotator. We would welcome the opportunity to further develop Éclair and Eclat within the context of collaborative projects—please contact the authors for details.
Acknowledgments
This work was supported by Academy of Finland grant 107333 to S.R. and BFAM 031U212C (BMBF) and TE 380/1-1 (DFG) grants to I.V.T. Funding to pay the Open Access publication charges for this article was provided by Academy of Finland grant 107333.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thomas S.R., Elkinton J.S. Pathogenicity and virulence. J. Invertebr. Pathol. 2004;85:146–151. doi: 10.1016/j.jip.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura H., Reich S., Ito A., Saitoh H., Kamoun S., Winter P., Kahl G., Reuter M., Kruger D.H., Terauchi R. Gene expression analysis of plant host–pathogen interactions by SuperSAGE. Proc. Natl Acad. Sci. USA. 2003;100:15718–15723. doi: 10.1073/pnas.2536670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokes T. Transcriptional responses to plant pathogen interactions. Trends Plant Sci. 2001;6:50–51. doi: 10.1016/s1360-1385(00)01841-0. [DOI] [PubMed] [Google Scholar]
- 4.Staskawicz B.J. Genetics of plant–pathogen interactions specifying plant disease resistance. Plant Physiol. 2001;125:73–76. doi: 10.1104/pp.125.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan J., Dunning F.M., Bent A.F. Probing plant–pathogen interactions and downstream defense signaling using DNA microarrays. Funct. Integr. Genomics. 2002;2:259–273. doi: 10.1007/s10142-002-0080-4. [DOI] [PubMed] [Google Scholar]
- 6.Marathe R., Guan Z., Anandalakshmi R., Zhao H., Dinesh-Kumar S.P. Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 2004;55:501–520. doi: 10.1007/s11103-004-0439-0. [DOI] [PubMed] [Google Scholar]
- 7.Moran P.J., Cheng Y., Cassell J.L., Thompson G.A. Gene expression profiling of Arabidopsis thaliana in compatible plant–aphid interactions. Arch. Insect Biochem. Physiol. 2002;51:182–203. doi: 10.1002/arch.10064. [DOI] [PubMed] [Google Scholar]
- 8.Rudd S. Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci. 2003;8:321–329. doi: 10.1016/S1360-1385(03)00131-6. [DOI] [PubMed] [Google Scholar]
- 9.Qutob D., Hraber P.T., Sobral B.W., Gijzen M. Comparative analysis of expressed sequences in Phytophthora sojae. Plant Physiol. 2000;123:243–254. doi: 10.1104/pp.123.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowak T.S., Woodards A.C., Jung Y., Adema C.M., Loker E.S. Identification of transcripts generated during the response of resistant Biomphalaria glabrata to Schistosoma mansoni infection using suppression subtractive hybridization. J. Parasitol. 2004;90:1034–1040. doi: 10.1645/GE-193R1. [DOI] [PubMed] [Google Scholar]
- 11.Hraber P.T., Weller J.W. On the species of origin: diagnosing the source of symbiotic transcripts. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0037. RESEARCH0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiang T., Goodwin P.H. Distinguishing plant and fungal sequences in ESTs from infected plant tissues. J. Microbiol. Methods. 2003;54:339–351. doi: 10.1016/s0167-7012(03)00067-8. [DOI] [PubMed] [Google Scholar]
- 13.Maor R., Kosman E., Golobinski R., Goodwin P., Sharon A. PF-IND: probability algorithm and software for separation of plant and fungal sequences. Curr. Genet. 2003;43:296–302. doi: 10.1007/s00294-003-0394-3. [DOI] [PubMed] [Google Scholar]
- 14.Friedel C.C., Jahn K.H., Sommer S., Rudd S., Mewes H.W., Tetko I.V. Support vector machines for separation of mixed plant-pathogen EST collections based on codon usage. Bioinformatics. 2005;21:1383–1388. doi: 10.1093/bioinformatics/bti200. [DOI] [PubMed] [Google Scholar]
- 15.Rudd S. openSputnik—a database to ESTablish comparative plant genomics using unsaturated sequence collections. Nucleic Acids Res. 2005;33:D622–D627. doi: 10.1093/nar/gki040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iseli C., Jongeneel C.V., Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:138–148. [PubMed] [Google Scholar]
- 17.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]