Abstract
The identification of genes underlying human genetic disorders requires the combination of data related to cytogenetic localization, phenotypes and expression patterns, to generate a list of candidate genes. In the field of human genetics, it is normal to perform this combination analysis by hand. We report on GeneSeeker (http://www.cmbi.ru.nl/GeneSeeker/), a web server that gathers and combines data from a series of databases. All database searches are performed via the web interfaces provided with the original databases, guaranteeing that the most recent data are queried, and obviating data warehousing. GeneSeeker makes the same selection of candidate genes as the human geneticists would have performed, and thus reducing the time-consuming process to a few minutes. GeneSeeker is particularly well suited for syndromes in which the disease gene displays altered expression patterns in the affected tissue(s).
INTRODUCTION
The identification of causative genes in human genetic disorders will be accelerated by the wealth of ‘omics’ information being generated. Geneticists consult a number databases to search for these genes. Each database concentrates on a different (molecular) aspect. In addition, databases have their own user interface, different formats to present the data and sometimes even their own ontologies. Data, such as gene localization and expression patterns, may be distributed over multiple databases.
Geneticists normally collect phenotypic and/or expression data and the genes in the chromosomal region(s) of interest, and combine these to get a list of candidate genes. The rationale for this is that the gene that causes a disease is most probably expressed in the tissues affected by that disease (1–3). Using model organisms, such as the mouse, it is often possible to obtain information on genes, proteins, protein interactions and other functional attributes that can be transferred to Homo sapiens by means of synteny and protein homology relationships. The use of data from other species (such as mouse) often proves helpful in identifying the location or function of the equivalent human gene (4). GeneSeeker mimics this multi-species identification strategy (5).
MATERIALS AND METHODS
Databases used
Table 1 lists the databases that GeneSeeker queries. These are divided over database groups (DB-groups). All databases are accessed through their standard WWW interfaces except MIMMAP and OXFORD. MIMMAP is a reformatted version of the OMIM (6) gene mapping information. OXFORD is used to translate human to mouse chromosomal locations, and is described in more detail in the pre-processing section. We use SRS (Sequence Retrieval System, Lion Biosciences, Cambridge, UK) to access these two databases (7). The SRS parser was modified to allow searches for chromosomal ranges.
Table 1.
Databases accessed by the GeneSeeker
| Database | URL |
|---|---|
| DB-group 1: localization databases (human) | |
| OXFORD (15) | srs.bioasp.nl:4080 |
| MIMMAP (6) | srs.bioasp.nl:4080 |
| GDB (16) | www.gdb.org |
| DB-group 2: localization databases (mouse) | |
| MGD (15) | www.informatics.jax.org |
| Datasets used in the interface | |
| GXD thesaurus | Van Steensel et al. (10) |
| Zuerich dataset | Brewer et al. (11,12) |
| DB-group 3: expression/phenotype databases | |
| PubMed (Nature Library of Medicine, Bethesda, MD) | www.ncbi.nlm.nih.gov/pubmed |
| OMIM (6) | srs.bioasp.nl:4080 |
| UniProt (9) (Swiss-Prot, TrEMBL, etc.) | srs.bioasp.nl:4080 |
| GXD (17) | www.informatics.jax.org |
| MLC (15) | www.informatics.jax.org |
| TBASE (18) | www.informatics.jax.org (was tbase, merged January 2005) |
| ‘Link out’ database | |
| GeneCards (14) | bioinfo.weizmann.ac.il/cards/ |
Data processing
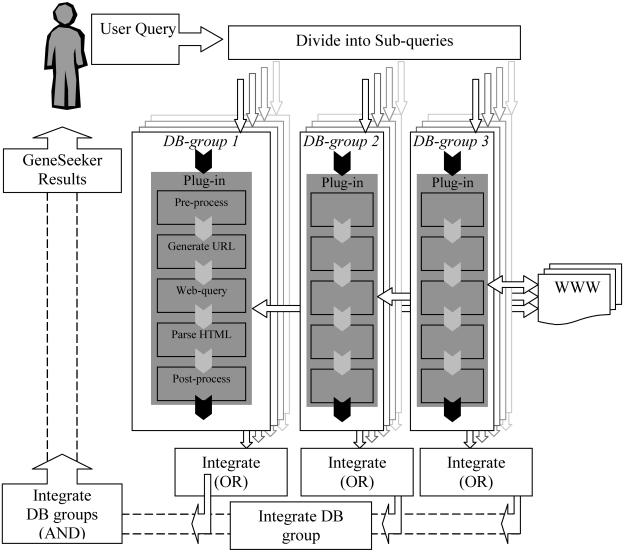
The layout of the GeneSeeker web server is shown in Figure 1. The user query consists of a chromosomal band range using standard nomenclature (e.g. 7p15–p21). This cytogenetic localization is passed through DB-group 1. Syntenic regions in the mouse are sought in DB-group 2 using an Oxford-grid. Tissues of interest or phenotypic features of a syndrome can be specified by the user as a Boolean expression that is split up and processed by DB-group 3. This modular set-up makes it easy to add extra DB-groups in the future. For every database, a plug-in was designed to perform all tasks from user-query pre-processing to query-result post-processing. These plug-ins deal with a series of technical topics, such as query reformatting, generating the correct URL, filling in the form on that database's web interface, requesting all hits rather than in chunks, parsing the database HTML output and so on.
Figure 1.
Overview of GeneSeeker. The query, which consists of a cytogenetic localization, a phenotypic description and expression data, is divided over the three DB-groups that use the database-specific plug-ins to deal with all topics ranging from user-query pre-processing to post-processing of the query output. Results from each DB-group are merged with a Boolean OR. The results of the three DB-groups are combined as specified in the user query.
The name of a gene can vary from database to database. The gene for the multi-drug resistance-associated protein 1, for example, is stored as ABCC1, MRP or MRP1, depending on the database used. These gene nomenclature problems have to be solved because GeneSeeker depends on the gene names in the combination steps. For each DB-group the results are integrated with a Boolean OR. The resulting gene lists of the three DB-groups are combined according to the Boolean logic specified in the user query.
Implementation issues
Parallelization
The database plug-ins run in parallel to minimize the waiting time. A queuing system prevents excessive loads on remote servers. The plug-ins return the results of the queries to GeneSeeker as a list containing the gene names and corresponding database hyperlinks.
Mouse–human synteny
An Oxford grid (8) is used to find the homologous genes and gene regions in the mouse genome for all human chromosome locations entered by the user. A human chromosomal band range is translated into the corresponding mouse chromosome locations. Two mouse locations are combined if the genetic distance is shorter than a user-specified value (defaults to 10 cM). We regenerate this Oxford grid weekly to ensure that the latest synteny information is used in each query.
Gene nomenclature
Inconsistent gene nomenclature is resolved using gene synonym information from UniProt database (9). We use the MGD human homologues information to interconvert mouse and human gene names. We maintain local copies of these conversion tables because nearly all queries require that gene nomenclature problems be solved.
User interface
The GeneSeeker interface consists of the query form shown in Figure 2 and an options form that usually requires no user input. A genetic localization and the phenotypic/expression terms should be entered for a meaningful search. Databases that generate more noise than signal can be removed from the query. The user can also suppress the display of housekeeping genes or a specified list of genes. The options form contains a thesaurus (10) that can help the user to select the correct expression terms: for example, when the user is interested in a genetic trait that results in abnormalities in the brain, selection of the ‘brain’ category returns the hints ‘brain or hindbrain or forebrain…’. Hints for the genetic localization data can be found in a table containing frequently aberrant chromosomal bands in specific disorders taken from literature (11,12). The user can be notified on request about the completion of GeneSeeker searches by email. All parameters are linked to help screens. The results are presented in four tables (Figure 3).
Figure 2.
An example of a GeneSeeker query. Analyses of Trismus-Pseudocamptodactyly syndrome (TPC; MIM 158 300) has been linked to 17p12–p13.1 (13). TPC is characterized by defects in muscle tissue mainly in limb and/or mouth. The options form is data not shown.
Figure 3.
The output of GeneSeeker for the Trismus-Pseudocamptodactyly syndrome query (see Figure 2). It has been shown that mutations in the MYH8 gene can cause TPC (13). Top left table: genes that agree perfectly with the user query. Top right table: genes found in mouse syntenic regions that cannot be mapped automatically on the human genome, but match the expression pattern. Bottom left table: genes found in mouse syntenic regions that match the expression pattern, but map on the human genome outside the candidate cytogenetic region. Bottom right table: human genes in the candidate cytogenetic region that do not match the phenotype/expression pattern. All genes are hyperlinked to the underlying database, and, when possible, to GeneCards (14).
RESULTS AND DISCUSSION
The GeneSeeker offers a user-friendly quick scan of several databases that are commonly used by geneticists to identify candidate genes for specific Mendelian diseases. As such, GeneSeeker uses those databases that are most appropriate for the questions asked. Several aspects are likely to change in the near future as genomics and genetics develop. For example, our usage of an Oxford grid can be improved or replaced as soon as consensus is reached about the localization of genes on the mouse and human genomes among the various databases. Expression pattern information (e.g. microarray data) is growing rapidly, and is expected to become useful for GeneSeeker in the near future. At the moment, publicly available expression information is still sparse, scattered and not yet standardized.
In its present form, GeneSeeker is best suited for syndromes in which one can assume aberrant or absent gene expression in the affected tissues. GeneSeeker allows the user to query heterogeneous databases and obtain good candidate genes for the disease of interest based on positional, expression and model data (5). With the present hardware set-up GeneSeeker can perform ∼1000 searches per day.
Acknowledgments
We thank David Thomas for helpful corrections to the manuscript. This work was supported by NWO/Unilever, the Irene Kinderziekenhuis Foundation and the EU FP6 Programme (LHSG-CT-2003-503265). Funding to pay the Open Access publication charges for this article was provided by Radboud University Nijmegen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackshaw S., Fraioli R.E., Furukawa T., Cepko C.L. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 2.den Hollander A.I., van Driel M.A., de Kok Y.J., van de Pol D.J., Hoyng C.B., Brunner H.G., Deutman A.F., Cremers F.P. Isolation and mapping of novel candidate genes for retinal disorders using suppression subtractive hybridization. Genomics. 1999;58:240–249. doi: 10.1006/geno.1999.5823. [DOI] [PubMed] [Google Scholar]
- 3.Dryja T.P. Gene-based approach to human gene–phenotype correlations. Proc. Natl Acad. Sci. USA. 1997;94:12117–12121. doi: 10.1073/pnas.94.22.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang A.P., Nishimura D., Searby C., Elbedour K., Carmi R., Ferguson A.L., Secrist J., Braun T., Casavant T., Stone E.M., et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet–Biedl syndrome (BBS3) Am. J. Hum. Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Driel M.A., Cuelenaere K., Kemmeren P.P., Leunissen J.A., Brunner H.G. A new web-based data mining tool for the identification of candidate genes for human genetic disorders. Eur. J. Hum. Genet. 2003;11:57–63. doi: 10.1038/sj.ejhg.5200918. [DOI] [PubMed] [Google Scholar]
- 6.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etzold T., Ulyanov A., Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- 8.Edwards J.H. The Oxford Grid. Ann. Hum. Genet. 1991;55:17–31. doi: 10.1111/j.1469-1809.1991.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 9.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Steensel M.A., Celli J., van Bokhoven J.H., Brunner H.G. Probing the gene expression database for candidate genes. Eur. J. Hum. Genet. 1999;7:910–919. doi: 10.1038/sj.ejhg.5200405. [DOI] [PubMed] [Google Scholar]
- 11.Brewer C., Holloway S., Zawalnyski P., Schinzel A., FitzPatrick D. A chromosomal deletion map of human malformations. Am. J. Hum. Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewer C., Holloway S., Zawalnyski P., Schinzel A., FitzPatrick D. A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality—and tolerance of segmental aneuploidy—in humans. Am. J. Hum. Genet. 1999;64:1702–1708. doi: 10.1086/302410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veugelers M., Bressan M., McDermott D.A., Weremowicz S., Morton C.C., Mabry C.C., Lefaivre J.F., Zunamon A., Destree A., Chaudron J.M., et al. Mutation of perinatal myosin heavy chain associated with a Carney complex variant. N. Engl. J. Med. 2004;351:460–469. doi: 10.1056/NEJMoa040584. [DOI] [PubMed] [Google Scholar]
- 14.Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E., et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blake J.A., Richardson J.E., Bult C.J., Kadin J.A., Eppig J.T. MGD: the Mouse Genome Database. Nucleic Acids Res. 2003;31:193–195. doi: 10.1093/nar/gkg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letovsky S.I., Cottingham R.W., Porter C.J., Li P.W. GDB: the Human Genome Database. Nucleic Acids Res. 1998;26:94–99. doi: 10.1093/nar/26.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringwald M., Eppig J.T., Begley D.A., Corradi J.P., McCright I.J., Hayamizu T.F., Hill D.P., Kadin J.A., Richardson J.E. The Mouse Gene Expression Database (GXD) Nucleic Acids Res. 2001;29:98–101. doi: 10.1093/nar/29.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woychik R.P., Wassom J.S., Kingsbury D., Jacobson D.A. TBASE: a computerized database for transgenic animals and targeted mutations. Nature. 1993;363:375–376. doi: 10.1038/363375a0. [DOI] [PubMed] [Google Scholar]