Abstract
TargetIdentifier is a webserver that identifies full-length cDNA sequences from the expressed sequence tag (EST)-derived contig and singleton data. To accomplish this TargetIdentifier uses BLASTX alignments as a guide to locate protein coding regions and potential start and stop codons. This information is then used to determine whether the EST-derived sequences include their translation start codons. The algorithm also uses the BLASTX output to assign putative functions to the query sequences. The server is available at https://fungalgenome.concordia.ca/tools/TargetIdentifier.html.
INTRODUCTION
The generation of expressed sequence tags (ESTs) is a widely recognized gene discovery strategy. Reflecting this there were 25 556 476 EST entries deposited in GenBank as of dbEST release 020405 (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). Furthermore, The Institute for Genomic Research (TIGR) has initiated the assembly and annotation of virtual transcripts (also called tentative consensus sequences) for 73 species. This TIGR effort relies heavily upon access to the GenBank dbEST database (http://www.tigr.org/tdb/tgi/index.shtml). Two additional efforts are databases of full length cDNAs for mouse (1) and Arabidopsis (2).
EST databases are an important resource for identifying cDNAs that contain complete protein coding regions for studies of gene function. Several computational tools, compared recently by Nadershahi et al. (3), including NetStart using neural networks (4), ESTScan using a hidden Markov model (5) and ATGpr using a linear discriminant approach (6), have been developed to identify translation initiation sites and/or coding regions in cDNA-derived sequences. These programs can predict the coding regions of cDNAs for which no known orthologues are available. However, since these programs are trained using organism-specific annotated sequences, they have limited value for organisms lacking annotated sequence data. In an attempt to address this issue ATGpr_sim (7), an updated version of ATGpr, was developed. In addition to relying on annotated data for training, ATGpr_sim also uses similarity information from BLASTX (8). The ATGpr_sim server only processes one sequence per submission, hence it cannot be used to process the large number of sequences produced by EST projects.
We developed TargetIdentifier a webserver that automates the identification of full-length cDNAs within a large number of EST-derived sequences. The TargetIdentifier algorithm uses BLASTX alignments as a guide to identify full-length cDNAs and provide provisional functional assignments (9,10). Hence, TargetIdentifier does not require ‘training’ with previously annotated sequences and is useful in the analysis of sequences encoding proteins for which information of their orthologues is available. We also demonstrated that TargetIdentifier effectively identified start codons and protein coding regions in our own Aspergillus niger EST-derived data and human UniGene data from NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene).
OVERVIEW OF THE ALGORITHM AND IMPLEMENTATION
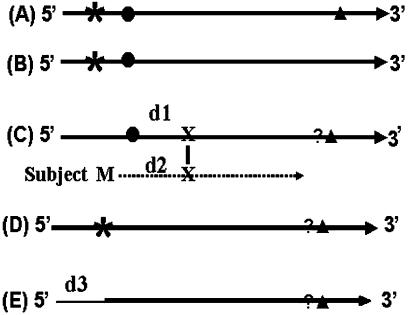
Although some polycistronic genes are found in protozoa (11), plants (12) and animals (13), almost all eukaryotic mRNAs are monocistronic. Hence a typical eukaryotic mRNA contains a 5′-untranslated terminal region (5′-UTR), a protein-coding region that begins with a translation start codon (ATG) and ends at a translation stop codon (TAA, TAG or TGA) (14) and a 3′-UTR (Figure 1A).
Figure 1.
Categories of algorithm-predicted cDNA clones. (A) A full-length sequence that includes one or more stop codons in the predicted 5′-UTR, a completely sequenced protein coding region and a 3′-UTR. (B) A sequence similar to those described in (A) except that the 3′ end of the ORF region is not sequenced. (C) A sequence having a start codon but lacking a stop codon in the 5′-UTR, whether it contains a potential translation start codon or not is determined by comparing the BLASTX alignment between its predicted protein and the subject. (D) A sequence having a stop codon in the 5′-UTR but lacking an in-frame start codon. This is an ambiguous sequence. (E) A sequence that includes a coding region but neither a stop codon nor a start codon in the sequenced portion. The length of the low quality sequence removed by Lucy (15) is taken into consideration when predicting whether or not it was a ‘possible full-length’ sequence. Asterisk: stop codon upstream of the start codon (5′ end stop codon); solid circle: predicted translation initiation codon; solid triangle: a stop codon downstream from the start codon (3′ end stop codon); question mark: indicates checking if a 3′ stop codon exists; (X): the first amino acid in the alignment of the HSP in BLASTX; (M): methionine; (d1) the length of predicted peptide from a predicted start codon to X; (d2) the length of M to X in the subject sequence of the HSP in BLASTX; (d3) length of EST sequence trimmed by Lucy, can include a portion of a vector, an adaptor and a low quality region of a cDNA sequence; thick solid line: sequences retained after processing by Lucy; thin solid line: the low quality sequence removed from the 5′ end by Lucy; dashed line: amino acid sequence of the subject in BLASTX.
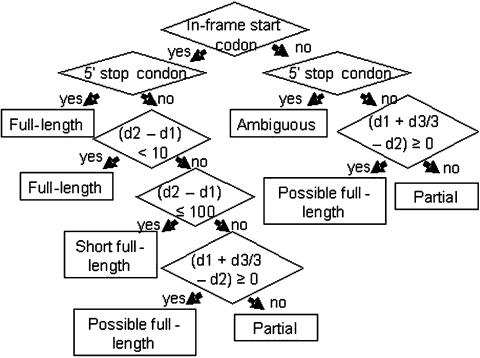
Since cDNA clones constructed using oligo-dT primers for first-strand synthesis are expected to have intact 3′ regions, clones that contain the translation initiation codon should have intact coding regions. TargetIdentifier therefore predicts whether the entire coding region is included in a cDNA clone by determining whether derived singleton and/or contig sequences include translation start codons. To accomplish this, the TargetIdentifier algorithm classifies EST-derived sequences as full-length, short full-length, possible full-length, ambiguous, partial or 3′-sequenced partial based on the decision tree presented in Figure 2 and the following definitions.
Figure 2.
A decision tree for EST-derived sequence classification. The definitions of each category of EST-derived sequences are described in detail in the text. Start codon: ATG; 5′ stop codon: stop codon (TAA, TAG, or TGA) in the 5′-UTR; d1: the predicted length of the peptide that extends from the start codon encoded methionine to the first amino acid of the query in the HSP alignment in the output of BLASTX; d2: the subject's beginning position in the HSP alignment in the output of BLASTX; d3: the estimated length of the low quality sequence removed by Lucy (15).
Full-length. A sequence is considered to include the translation start codon when it satisfies one of the following two criteria. (1) The sequence has a 5′ stop codon followed by a start codon (Figure 1A and B). (2) The sequence does not have a 5′ stop codon but has an in-frame start codon encoding a methionine that aligns to the BLASTX subject prior to the 10th amino acid (Figure 1C).
Short full-length. The sequence has an in-frame start codon encoding a methionine that aligns to a position between the 10th and the 100th amino acid of the subject sequence (Figure 1C). The program determines the location of the potential start codon relative to the start codon for the BLASTX subject sequence. An upper limit of 100 is selected, because BLAST alignments of closely related cellulases and aldehyde oxidases revealed that the length of the amino terminal region extending from the aligned core sequences rarely varies by >100 amino acids.
Possible full-length. If sequence quality at the 5′ end of an EST sequence is poor, the DNA sequence removed by the quality control program may have included the start codon. The corresponding cDNA clone is therefore categorized as ‘possible full-length’ if the low quality sequence removed is long enough to include the missing amino terminal portion of the translated query.
Ambiguous. The sequence has a 5′ stop codon but does not have a start codon (Figure 1D). This type of anomaly probably arises because of sequencing errors. This can occur in EST-derived sequences as they can often include sequence information derived from a single sequencing read.
Partial. A sequence that is not assigned to one of the above categories (Figure 1E).
3′-sequenced partial. TargetIdentifier initially processes the sequence data assuming they were obtained by sequencing from the 5′ end of the cDNA inserts. In the BLASTX report, these sequences should align with the subject sequences in a positive reading frame. Query sequences are therefore classified as ‘3′-sequenced partial’ when they align to the subject sequence in a negative reading frame (−1, −2 or −3) and are not categorized as full-length, short full-length or ambiguous.
Input
A data file containing a set of ESTs or sequences assembled from ESTs in FASTA format.
A pre-run BLASTX output for each sequence contained in the input sequence file described in 1. This can be produced by searching against a database, such as the NCBI non-redundant protein database, Swiss-Prot database or a user generated protein database. A cutoff E-value can be chosen at the time of running BLASTX. For users without access to the NCBI-blastall package for processing a batch of sequences, our server provides BLASTX searches against the UniProt/Swiss-Prot database with a limit of 1000 sequences per submission. If >1000 sequences are submitted, only the first 1000 sequences will be processed.
Two optional input files that can be included are an ace file generated by an assembler, such as Phrap (http://www.phrap.org/phrap.docs/phrap.html) and a file generated by a quality trimming program, such as Lucy (15). The ace file provides assembly information regarding the individual ESTs in a contig, and the quality file contains EST identifiers, EST length and the length of any low quality sequence removed from the 5′ end of each EST sequence in tab-delimited format.
A cutoff E-value that is set by the user to define what is a valid hit in BLASTX. If the user defined E-value is larger than the E-value used for the pre-run BLASTX output, the actual cutoff value is the value in the BLASTX output.
Options for users to choose either downloading the results or receiving the output via email.
Output
The TargetIdentifier output is tab-delimited and can be opened as a spreadsheet with Microsoft Excel. The output file includes: a summary of the results obtained for the whole set of EST or EST-derived sequences and a detailed report for each sequence predicted to fall within the various categories. The detailed report includes the following fields: (i) the name of the subject protein in the high score pair (HSP) of the BLASTX alignment; (ii) a query identifier; (iii) the HSP E-value; (iv) a prediction of whether the EST or EST-derived query sequence is full-length, short full-length, possible-full length, ambiguous, partial or 3′-sequenced partial; (v) start codon position; (vi) the strand and the sequence status of the query sequences regarding whether or not the protein coding region has been completely sequenced and (vii) HSP heading information taken from the BLASTX output that includes the subject definition line, length, score, E-value, identities, positives and reading frame. To sort genes by gene name, the algorithm removes the terms ‘probable’, ‘putative’, ‘possible’ and ‘similar to’ from the subject definition.
ACCURACY EVALUATION
To evaluate TargetIdentifier, we used the human UniGene set and our own EST-derived A.niger unigene set of contigs and singletons. The human UniGene set (Build #160, Homo sapiens, February 16, 2003) was searched using BLASTX against the full-length human protein sequences (total 8956) downloaded from the Swiss-Prot database. TargetIdentifier predicted that there were 7210 full-length, 66 short full-length, 376 (5′) partial, 400 3′-sequenced partial and 81 ambiguous sequences in the human UniGene set. We used a random number generator (http://www.random.org) to select a total of 270 human UniGene sequences and compared the TargetIdentifier output with manually obtained results. This comparison showed that TargetIdentifier correctly sorted 93% of the sequences into the full-length, short full-length, possible full-length, ambiguous and partial categories. We also assessed the TargetIdentifier predictions using our EST-derived A.niger assembly set. To assemble this dataset the EST sequence chromatograms were traced by Phred (16), vector and low quality regions were removed by Lucy (15) and the ESTs were assembled by Phrap (http://www.phrap.org/). The accuracy of TargetIdentifier was assessed using 98 EST assemblies that encode predicted protein sequences sharing >90% identity with an A.niger protein entry at GenBank. This revealed that of the 55 sequences classified as full-length by TargetIdentifier, 54 were correctly predicted (98%). The human Unigene sequences, the 98 A.niger EST-assemblies and the TargetIdentifier prediction data are available at https://fungalgenomics.concordia.ca/methods/tools/EST_annotation/index.php.
SUMMARY
TargetIdentifier is a webserver that uses BLASTX alignments to identify full-length cDNAs from an EST-derived dataset. We have evaluated the prediction accuracy with the human UniGene set and our own set of assembled A.niger ESTs, and found that it is >90% accurate. TargetIdentifier can therefore be used to search EST-derived datasets for sequences encoding specific functionalities and predict whether or not a cDNAclone harboring the complete coding region has been identified.
Acknowledgments
We thank Yueqin Chen for his work on adding the Lucy program to the sequence quality control pipeline, Jian Sun for assisting with the EST assembly and Wei Ding for assisting with the server interface development. This work was supported by Genome Quebec and Genome Canada. Funding to pay the Open Access publication charges for this article was provided by Genome Quebec and Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bono H., Kasukawa T., Furuno M., Hayashizaki Y., Okazaki Y. FANTOM DB: database for functional annotation of RIKEN mouse cDNA clones. Nucleic Acids Res. 2002;30:116–118. doi: 10.1093/nar/30.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seki M., Narusaka M., Kamiya S., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 3.Nadershani A., Fahrenkrug S.C., Ellis L.B.M. Comparison of computational methods for identifying translation initiation sites in EST data. BMC Bioinformatics. 2004;5:14. doi: 10.1186/1471-2105-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen A.G., Nielsen H. Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997:226–233. [PubMed] [Google Scholar]
- 5.Iseli C., Jongeneel C.V., Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:138–148. [PubMed] [Google Scholar]
- 6.Salamov A., Nishikawa T., Swindells M.B. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–390. doi: 10.1093/bioinformatics/14.5.384. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa T., Ota T., Isogai T. Prediction whether a human cDNA sequence contains initiation codon by combining statistical information and similarity with protein sequences. Bioinformatics. 2000;16:960–967. doi: 10.1093/bioinformatics/16.11.960. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Gish W., States D.J. Identification of protein coding regions by database similarity search. Nature Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 10.Liang F., Holt I., Pertea G., Karamycheva S., Salzberg S., Quackenbush J. An optimized protocol for analysis of EST sequences. Nucleic Acids Res. 2000;28:3657–3665. doi: 10.1093/nar/28.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell D.A., Thomas S., Sturm N.R. Transcription in kinetoplastid protozoa: why be normal. Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Leader D.J., Clark G.P., Watters J., Beven A.F., Shaw P.J., Brown J.W. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNA. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. reviews 0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou H., Holmes M.H. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B., Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]