Abstract
enoLOGOS is a web-based tool that generates sequence logos from various input sources. Sequence logos have become a popular way to graphically represent DNA and amino acid sequence patterns from a set of aligned sequences. Each position of the alignment is represented by a column of stacked symbols with its total height reflecting the information content in this position. Currently, the available web servers are able to create logo images from a set of aligned sequences, but none of them generates weighted sequence logos directly from energy measurements or other sources. With the advent of high-throughput technologies for estimating the contact energy of different DNA sequences, tools that can create logos directly from binding affinity data are useful to researchers. enoLOGOS generates sequence logos from a variety of input data, including energy measurements, probability matrices, alignment matrices, count matrices and aligned sequences. Furthermore, enoLOGOS can represent the mutual information of different positions of the consensus sequence, a unique feature of this tool. Another web interface for our software, C2H2-enoLOGOS, generates logos for the DNA-binding preferences of the C2H2 zinc-finger transcription factor family members. enoLOGOS and C2H2-enoLOGOS are accessible over the web at http://biodev.hgen.pitt.edu/enologos/.
INTRODUCTION
The sequence logo, introduced initially by Schneider and Stephens (1), has become a popular method for the visual representation of DNA and amino acid sequence patterns. Traditionally, sequence logos are constructed from a set of aligned sequences and graphed as columns of stacked symbols. The height of each column corresponds to the information content (IC) (2) of the corresponding position in the alignment, and the size of the individual symbols within each column reflects the frequency of the corresponding nucleotide at this position. Typically, the IC is calculated from the frequencies of the nucleotides (or amino acids) in each position. Logos have been extensively used for the representation of transcription factor binding site (TFBS) preferences. Typically, a collection of known binding sites of a particular transcription factor (TF) is used to estimate the position-specific probabilities of the 4 nt as frequencies in the alignment. However, it is also possible to directly determine the interaction energies between a TF and its binding site (3–5). Currently, there is one web resource that can generate a PostScript or PNG logos from a set of aligned sequences (Weblogo, http://weblogo.berkeley.edu/), but not from other types of data.
In this paper, we describe enoLOGOS, a tool developed primarily for converting energy or log-probability DNA weight matrices to normalized sequence logos (energy normalized logos). In addition, enoLOGOS can utilize a wide variety of input formats to generate sequence logos, including sequence alignments (in plain or FASTA format), frequency or alignment matrices (as generated by most motif-finding programs) and TRANSFAC matrices. We also describe the C2H2-enoLOGOS web version of this program that has been previously used to graphically represent predicted DNA-binding preferences of zinc-finger proteins (6). C2H2-enoLOGOS can generate sequence logos from predicted energies of Cys2His2 zinc-finger DNA-binding proteins selected from the Pfam protein family (7) or from a user-defined alignment of ‘contacting’ amino acid residues for this DNA-binding domain (6). Finally, when provided a DNA (or RNA) alignment, enoLOGOS can display the mutual information among all pairs of alignment positions implicating correlation between positions in a binding site or secondary structure in the case of RNA alignments (8).
METHODS
Following the notation of Benos et al. (6), given a matrix of binding energy contributions for each base at each position of a TFBS, we can obtain the probability of observing any sequence (Di) bound to the corresponding TF using a derivative of the Boltzmann distribution:
| 1 |
where Pref(Di) is the prior (background) probability of the sequence Di (e.g. in the genome), and Z is the partition function (the sum of the numerator over all possible binding sequences). The inclusion of Pref(Di) is essential, as it is obvious that for a non-specific protein the probabilities of the bound DNA will be equal to the prior probabilities. E(Di) is the binding energy of the protein to the sequence Di (in kBT units), which is obtained from the matrix of binding energy contributions, summing those values that correspond to the sequence (9). Note that the absolute values of the energy contributions do not matter to the probability calculations, only the difference in energy between different bases. If we add any constant to all of the energy terms, the probability values will remain unchanged (Equation 1). We have often defined the ‘specific binding energy’ contributions of base b at position i, as (9), but one can also use the convention of Berg and von Hippel (10) of setting the best binding base to have zero energy as long as the differences with the other bases are maintained. It is important to specify the units of the energy as they will affect the binding probabilities. We offer the user the choice of several typical energy units (kBT, kcal/mol, log-odds scores using log base 2 or e).
The energy matrix for a TF is an intrinsic property of the protein, and does not depend on the probabilities of different sequences that it has the opportunity to bind (it will depend on the binding conditions, such as pH, temperature and ionic strength of the buffer, but we consider those to be fixed quantities that are not the subject of this analysis). In the generated logo the total height of bases at each position is the ‘IC’ of the position, and the height of each base is the proportion of that base in the total. The IC of DNA position i is defined as:
| 2 |
where P(b, i) is the probability of base b at position i in the bound set of sites. IC is the average specific binding energy, as defined above, and will be zero if the bound probabilities are the same as the prior probabilities (i.e. a non-specific protein has zero IC). Regardless of how the binding energies are determined, the bound probabilities can be obtained for any set of prior probabilities, and these are provided in the text output (probability_matrix). The IC depends on the prior base probabilities, as one would expect from the relationship between the IC and the expected frequency of sites in a genome (2); the higher the IC the less likely a site is expected to occur by chance. Hence sites that are GC-rich, for example, would be expected to occur less often in AT-rich genomes (and thus have a higher total IC) than in GC-rich genomes. By setting equiprobable prior probabilities (default setting of 0.25 for each base), the bound frequencies will be those obtained if all sequences are equally available for binding, and under this assumption the ‘intrinsic’ IC for the protein will be estimated. IC calculated with equiprobable priors is equivalent to the reduction in Shannon entropy (11) from its maximum value.
IMPLEMENTATION
Algorithm
The enoLOGOS program is written in C++ and includes refactored PostScript functions from the Delila package (12). The web tools enoLOGOS and C2H2-enoLOGOS are implemented in Perl (CGI) and provide the interface to the C++ program. The algorithm offers all the standard options for plotting logos and extend the alignment-based outputs with probability normalized and energy normalized logos.
Input data types
The enoLOGOS program accepts various types of input data. The default input data type is energies in which case the user can specify the units as kBT units (default), kcal/mol, J/mol or kJ/mol. The conversion rates we use are: 1 kBT unit (at 298 K) = 0.592 kcal/mol = 2.477 kJ/mol. Energies in kBT units are then used directly on Equation 1 to calculate the relative frequencies of the bases in each position. Other types of input data include alignment counts, probabilities (or KA values) and aligned sequences. All these data types are transformed into relative frequencies in a straightforward way. In addition, enoLOGOS can generate a logo from practically any type of matrix provided through its plot option ‘weights as entered’.
Input data format
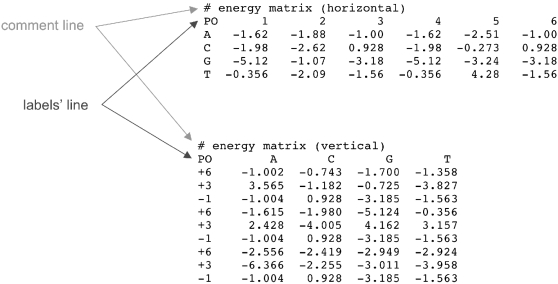
All numerical data can be entered in a weight matrix form. The weight matrix can be formatted horizontally or vertically, where the orientation refers to the length of the pattern. Our web tool can infer the orientation, provided that letters are used to identify nucleotide rows or columns and position labels (optional) are defined as integers. Examples of the horizontal and vertical format are presented in Figure 1. Lines that are preceded by ‘#’ are considered comment lines and are ignored. A single matrix header line starting with ‘PO’ [as used in TRANSFAC matrices (13)] can specify position labels (horizontal matrices) or base types (vertical matrices) of the logo columns. If a matrix header is found, then the first item on each subsequent line will be used as either the base type or the position label of the horizontal or vertical matrix, respectively (Figure 1). If the input data are aligned sequences (i.e. raw sequences or sequences in the FASTA format), an alignment will be inferred and an alignment matrix will be created. Any character other than white space in an alignment (e.g. ‘−’, ‘.’ or ‘*’) designates insertion. All white space characters are ignored.
Figure 1.
Examples of input weight matrices (horizontal and vertical).
LOGO calculation parameters
Independent of the type of data entered, the resulting logos can be plotted using option relative entropy, frequency or ‘weights as entered’. For the relative entropy option (default value), we use Equation 2, while frequency or ‘weights as entered’ options render logos as one would expect. The user may also choose how they want the individual bases to be scaled within a column. The available options are frequency (the default and most often used method) and relative entropy. If the user chooses to scale the bases in each position by their relative entropies, then the bases with negative relative entropy are plotted upside-down. This is an additional feature that helps users to visually identify those bases that are energetically favoured in each position taking into consideration the background base frequencies in the genome. In the case that the user selects relative entropy in either the calculation of column height or the base scaling, then a choice of the logarithm-base becomes important. The logarithm-base options are as follows: 2, giving information units in bits (default value); and e, giving information in nats and 10. In the case where the input consists of a number of aligned sequences, enoLOGOS can also calculate the mutual information and present it in a graphical way. Mutual information is the relative entropy between a joint distribution (in our case, the two columns under comparison) and the product distribution (of the independent columns). Finally, the user defines the expected (background) probabilities for the four bases in terms of %GC content. The default has been set to equiprobable background and in this case the relative entropy will be equivalent to the reduction in Shannon entropy (11) from its maximum value. In addition, %GC content values are provided as options for various model organisms, such as Escherichia coli, yeast, worm, fruitfly, mouse and human.
Output format parameters
In the output format parameters section, the user can select the colours for each base in the logo (in RGB [0..1] additive colour scheme), titles for the plot and for the x- and y-axes, and a weights scale factor that can be used to scale or change the sign of the weights. The default colours are green, blue, orange and red for each of the A, C, G and T bases, respectively. The enoLOGOS tool is accessible via the web at http://biodev.hgen.pitt.edu/enologos/.
C2H2-enoLOGOS
Another variation of this tool, C2H2-enoLOGOS, allows users to generate the LOGO of the predicted DNA-binding preferences for any member of the Cys2His2 zinc-finger protein family of TFs. This is the largest TF family (Pfam version 16.0; http://pfam.wustl.edu/) with more than 5000 members, including 39 Saccharomyces cerevisiae and 1438 human TFs. The Cys2His2 motif is a modular motif (each helix contacts three bases in an antiparallel fashion) and composite sites can be deduced from the targets of the single helices. The co-crystal structure of a Cys2His2 factor EGR1, a member of this family, and its preferred binding site has revealed that three amino acid residues each contact one base and that these contacts are primarily responsible for the DNA-specificity (14). This simple pattern of contacts makes the modelling of the DNA-binding preferences of its members possible (6,15). The C2H2-enoLOGOS tool is using the energy estimates calculated previously for this family (6) to produce the requested logos. The user can either directly specify the amino acids in each of the three contact positions of each helix (positions −1, +3 and +6) or select a particular protein family member via a simple query form. We have restricted the predictions to proteins with a maximum of four helices, since some studies have shown that when additional helices are presented, some of them are not used to contact the DNA (16). The query capabilities of the tool support the use of ‘AND’ or ‘OR’ logic, hence key words can be joined for more specific searches of the alignment entries. All proteins that meet the specified search criteria are presented in a list and the user is asked for a choice. The Pfam alignment (7) is used for the identification of the contacting amino acids for the specified protein. Once the contacting amino acids have been specified for each α-helix, a log-probability position-specific weight matrix is generated from the model of this family (6). Then, a logo plot is produced as described above and the corresponding weight matrices are provided in the output. The C2H2-enoLOGOS web tool is accessible from the enoLOGOS web page.
RESULTS
The resulting sequence logo is displayed in the main page in the PNG format (e.g. see Figure 2). The intermediate PostScript file and PDF versions are also provided to the user and hyperlinked from the web page. A hyperlink is also provided to a text version of the logo matrix, input matrix and all its numeric transformations (e.g. energies, probabilities/frequencies and log-likelihood). This file also contains the numerical values for the position-specific relative entropies and IC.
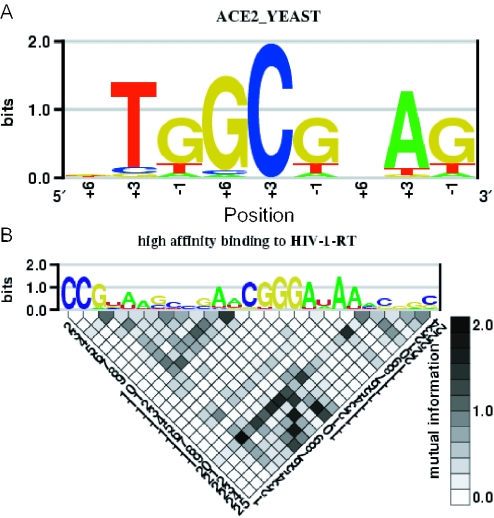
Figure 2.
Example of C2H2-enoLOGOS and enoLOGOS output. In this example, the x-axis of the C2H2-enoLOGOS output (top) represents the ‘contacting’ amino acid positions of fingers 3, 2 and 1, respectively. The output of web tool enoLOGOS (bottom) contains a grey-scale-coded matrix plot of the mutual information of each pair of positions of the alignment.
Acknowledgments
The authors would like to thank Claus A. Andersen for sharing code from his logo program. This work was supported by NSF grant MCB0316255. T.I. is funded by NCRR 018627. C.T.W. is funded by NIH 5T32HL07089-30. Funding to pay the Open Access publication charges for this article was provided by NSF grant MCB0316255.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider T.D., Stormo G.D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 3.Man T.K., Stormo G.D. Non-independence of Mnt repressor–operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulyk M.L., Gentalen E., Lockhart D.J., Church G.M. Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 5.Takeda Y., Sarai A., Rivera V.M. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc. Natl Acad. Sci. USA. 1989;86:439–443. doi: 10.1073/pnas.86.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benos P.V., Lapedes A.S., Stormo G.D. Probabilistic code for DNA recognition by proteins of the EGR family. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- 7.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorodkin J., Staerfeldt H.H., Lund O., Brunak S. MatrixPlot: visualizing sequence constraints. Bioinformatics. 1999;15:769–770. doi: 10.1093/bioinformatics/15.9.769. [DOI] [PubMed] [Google Scholar]
- 9.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 10.Berg O.G., von Hippel P.H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 11.Shannon C. The mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- 12.Schneider T.D., Stormo G.D., Yarus M.A., Gold L. Delila system tools. Nucleic Acids Res. 1984;12:129–140. doi: 10.1093/nar/12.1part1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wingender E. TRANSFAC, TRANSPATH and CYTOMER as starting points for an ontology of regulatory networks. In Silico Biol. 2004;4:55–61. [PubMed] [Google Scholar]
- 14.Pavletich N.P., Pabo C.O. Zinc finger-DNA recognition: crystal structure of a Zif268–DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 15.Benos P.V., Bulyk M.L., Stormo G.D. Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore M., Choo Y., Klug A. Design of polyzinc finger peptides with structured linkers. Proc. Natl Acad. Sci. USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]