Abstract
InterProScan [E. M. Zdobnov and R. Apweiler (2001) Bioinformatics, 17, 847–848] is a tool that combines different protein signature recognition methods from the InterPro [N. J. Mulder, R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bradley, P. Bork, P. Bucher, L. Cerutti et al. (2005) Nucleic Acids Res., 33, D201–D205] consortium member databases into one resource. At the time of writing there are 10 distinct publicly available databases in the application. Protein as well as DNA sequences can be analysed. A web-based version is accessible for academic and commercial organizations from the EBI (http://www.ebi.ac.uk/InterProScan/). In addition, a standalone Perl version and a SOAP Web Service [J. Snell, D. Tidwell and P. Kulchenko (2001) Programming Web Services with SOAP, 1st edn. O'Reilly Publishers, Sebastopol, CA, http://www.w3.org/TR/soap/] are also available to the users. Various output formats are supported and include text tables, XML documents, as well as various graphs to help interpret the results.
INTRODUCTION
When carrying out analysis of protein sequences, the aim is to find out as much information as possible about potential relationships with other sequences as well as characterizing their physiochemical properties. The first step usually involves comparing the protein sequence against a non-redundant protein sequence database by using Blast (1) or Fasta (2), which will reveal which sequence(s) are similar to the query sequence alone. To obtain further information about a protein's specific function, searches against secondary databases (also known as pattern or signature databases) are necessary. When such searches return significant matches or hits, these results help in the assignment of a particular function or functional domain to the query protein. The InterPro (http://www.ebi.ac.uk/interpro) (3) database was created to unite secondary databases that contain overlapping information on protein families, domains and functional sites (Table 1). InterPro entries are divided into groups based on the protein families or the domains that the signatures represent. If the structure and function of a protein family is well characterized, searches of the secondary databases offer a fast track into inferring biological function. Searching individually against each of these databases to get the most information is repetitive, time consuming and labour intensive. To search InterPro with a novel protein sequence, a tool, InterProScan (4), has been developed (http://www.ebi.ac.uk/InterProScan) that combines the protein function recognition methods of the member databases of InterPro into one application. Since its creation, there have been several releases to improve it, adding functionality as well as new databases to the system.
Table 1.
Database members and their applications
| Database | Application |
|---|---|
| ProDom (6) | BlastProDom (Blastall) (4) |
| PRINTS (7) | FingerPrintScan (8) |
| SMART (9) | Hmmpfam (http://hmmer.wustl.edu/) |
| TIGRFAMs (10) | Hmmpfam (http://hmmer.wustl.edu/) |
| Pfam (11) | Hmmpfam (http://hmmer.wustl.edu/) |
| PROSITE (12) | ScanRegExp + ProfileScan (13) |
| PIRSF (14) | Hmmpfam (http://hmmer.wustl.edu/) |
| SUPERFAMILY (15) | Hmmpfam (http://hmmer.wustl.edu/) |
| CATH (16) | Hmmpfam (http://hmmer.wustl.edu/) |
| PANTHER (17) | Hmmsearch (http://hmmer.wustl.edu/) |
| SignalPHMM | SignalPHMM (18) |
| Transmembrane | TMHMM2.0 (19) |
InterProScan TOOL
Here, we describe the use of the web browser based version of InterProScan available from the EBI at http://www.ebi.ac.uk/InterProScan (Figure 1). This service is free to all academic and commercial organizations and offers interactive as well as email job submission. Direct email submissions should be directed to interproscan@ebi.ac.uk. Instructions and documentation are available when sending an email to the above address that contains the word ‘help’ in the message body. Users requiring high-throughput use of the application or who wish to carry out analysis using other databases can download a standalone version from ftp://ftp.ebi.ac.uk/pub/databases/interpro/iprscan. Users requiring programmatic access to the InterProScan can do so using a SOAP-based Web Service called WSInterProScan (5), which is described at http://www.ebi.ac.uk/Tools/webservices/WSInterProScan.html. All of these make use of a centrally maintained core version of InterProScan version 4.0.
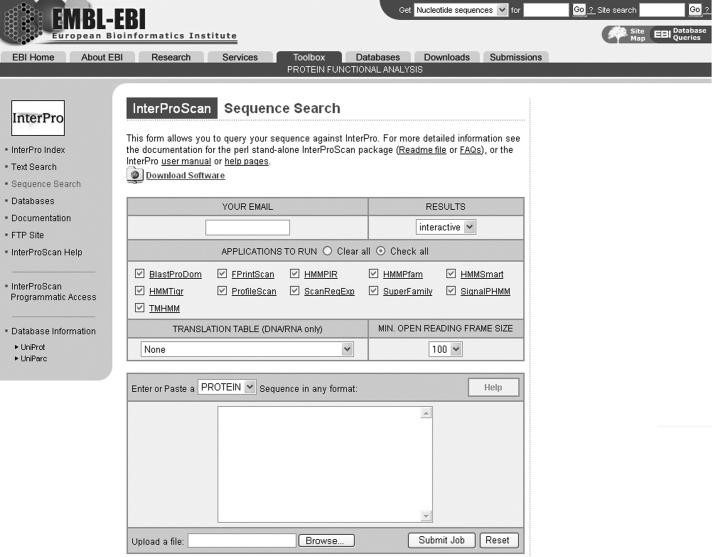
Figure 1.
EBI's InterProScan job submission page.
The job input form
The first section of the input form consists of the users email address and how the results are to be displayed. The first thing a user needs to decide when using the InterProScan submission form (Figure 1) is how he/she wants to see the results. This is carried out by making a selection on the RESULTS menu. Two options are available: ‘interactive’, which will return the results to the browser once the job is completed, and ‘email’, which will return the results to the email specified in the YOUR EMAIL text dialog.
The next section has a set of check boxes that either choose all or clears all the methods available. Each method can be ticked on or off, according to the user's requirements. For example, users interested only in signal peptide cleavage sites or the transmembrane domains described in InterPro entries may choose the corresponding methods individually.
The third section of the submission form is specific for DNA as the sequence input. DNA sequences will be translated to protein according to the translation rules specified in the TRANSLATION TABLE menu. The default is the standard code. Each translation will generate peptide sequences in six frames and all will be searched. The minimum length of an open reading frame produced after translation can be specified in the MIN. OPEN READING FRAME SIZE menu. This dictates that only peptides above the selected value will be searched by the methods chosen in the second section.
The fourth section of the input form consists of the sequence input panel. The components of this panel include a selection menu for the molecule type. This one can be DNA or protein. The default is protein. When DNA is selected it enables the TRANSLATION TABLE menu in the third section of the form. Help is available by clicking on the HELP image. This will open a new browser window that contains comprehensive information about InterProScan. There is also an UPLOAD dialog that can be used instead of cutting and pasting sequences into the input window. Finally, there are the Submit and Reset buttons. The sequence input text dialog will accept protein or DNA sequence in any of the standard sequence formats in use today. These include EMBL, SWISS, GenBank, NBRF/PIR, CODATA, Fasta, GCG and RAW text. Primary or secondary identifiers (accession number or identifier) of a protein sequence in the databases can also be used. In this case, the user will type a database name followed by a colon and the identifier. For example, ‘UNIPROT:INSR_HUMAN’. It is not possible to submit more than 10 protein input sequences at the same time. Each protein sequence must be at least five amino acids long. Only one nucleic acid sequence may be used at a time and the length for this sequence must be ≤5000 bases.
InterProScan output
Before InterProScan launches each of the protein sequence analysis applications, it takes advantage of pre-computed results whenever possible. It calculates a checksum (CRC64) for the query sequence and compares it with the checksums of the protein sequences that are present in a database called IPRMATCHES. This is a database that lists all the entries from UniProt/Swiss-Prot and UniProt/TrEMBL that match one or more InterPro entries. If the checksum calculated for the query sequence does not match any checksums found in the IPRMATCHES database, the protein sequence analysis applications are launched in parallel; otherwise the IPRMATCHES entry is returned.
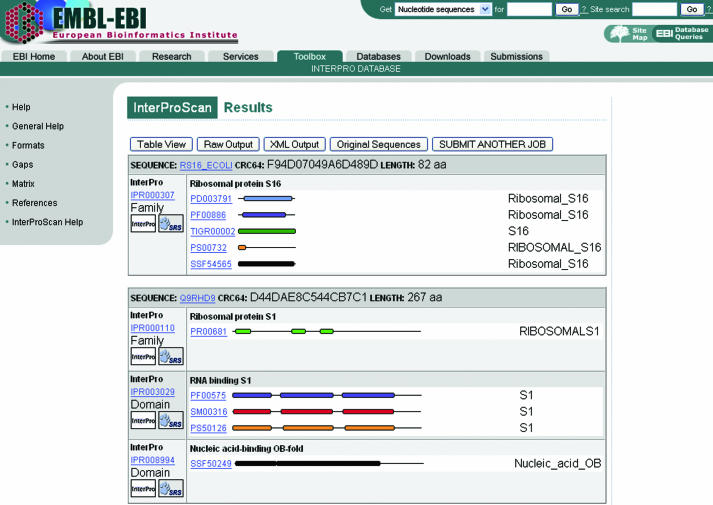
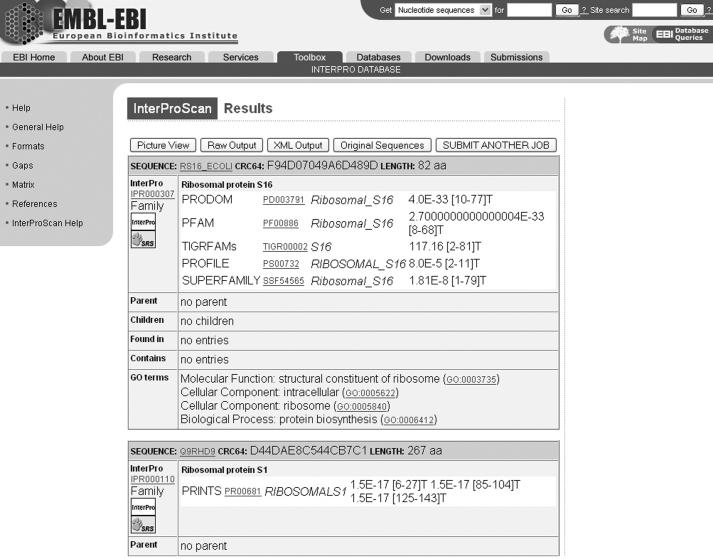
Once a job is completed, the output of each of the applications is individually parsed to produce a merged results file. This file is in the tab-delimited format. A converter is called onto generate, on the fly, an XML document, which is used to generate the HTML output. This consists of two views: a picture or a graphical view (Figure 2) that displays a cartoon of the sequence with highlighted domains or functional sites corresponding to the matches in the InterPro databases. Each match contains hypertext links to the InterPro database main web resource as well as to the individual member databases' websites where the matches are further described. A table view (Figure 3) is also available by clicking on the ‘table view’ button. This one consists of complete database names, hyperlinked match identifiers, the sequence coordinates (start–stop pairs) where the match occurs, E-values and the status of the match in InterPro (e.g. ‘T’ for true or ‘?’ for unknown). Parent–child relationships are displayed if they exist in an InterPro entry. GO annotation is also shown if available. Other options in the HTML results page include the raw output in the tab-delimited format, the XML document and the sequences used as input (original sequences). The results for each job are stored at the EBI for at least 24 h.
Figure 2.
InterProScan graphical results view.
Figure 3.
InterProScan table results view.
THE STANDALONE VERSION OF InterProScan
For users who wish to run their own installation of InterProScan, there is a free standalone version available from the EBI's ftp server (ftp://ftp.ebi.ac.uk/pub/databases/interpro/iprscan/RELEASE/latest). This version can run from the command line or as a CGI through a web interface. Additional features that have been developed for this version include the use of the Perl indexing library, which indexes all data files, input sequences and the applications results for easy querying and retrieval of primary identifiers, names, job results and their status. From the results page, users can access each input sequence individually or the full input file, and also access the original outputs from each application.
The standalone version of InterProScan has been designed to run on either a single machine or a cluster of machines. It supports the use of various queuing systems such as LSF, OpenPBS and SGE.
The standalone version is composed of three different packages:
Perl core package containing all the scripts and modules to run InterProScan.
Data package, which contains all the data needed by each application to run (∼4 GB unzipped).
Binary package precompiled for six different platforms (Linux, OSF1, AIX, Sun, IRIX and MacOSX).
CONCLUSIONS
We describe here the current state of the EBI InterProScan server, along with much of its unique flexibility, which is freely available to the public. Questions, comments and suggestions from users are encouraged and may be addressed to http://www.ebi.ac.uk/support/.
Acknowledgments
InterPro is funded by the award of grant number QLRI-CT-2000-00517 and in part by grant number QLRI-CT-2001000015 from the European Union under the RTD program ‘Quality of Life and Management of Living Resources’. InterPro is a member database of the MRC-funded eFamily project. Funding to pay the Open Access publication charges for this article was provided by The European Molecular Biology Laboratory (EMBL).
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped Blast and Psi-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson W.R., Lipman D.J. Improved tools for biological sequence analysis. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zdobnov E.M., Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 5.Snell J., Tidwell D., Kulchenko P. Programming Web Services with SOAP, 1st edn. Sebastopol, CA: O'Reilly & Associates; 2001. [Google Scholar]
- 6.Bru C., Courcelle E., Carrère S., Beausse Y., Dalmar S., Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attwood T.K., Bradley P., Flower D.R., Gaulton A., Maudling N., Mitchell A.L., Moulton G., Nordle A., Paine K., Taylor P., et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scordis P., Flower D.R., Attwood T.K. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15:799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- 9.Letunic I., Goodstadt L., Dickens N.J., Doerks T., Schultz J., Mott R., Ciccarelli F., Copley R.R., Ponting C.P., Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins D.G., Gibson T.J. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci. 1994;10:19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- 14.Wu C.H., Nikolskaya A., Huang H., Yeh L.S., Natale D.A., Vinayaka C.R., Hu Z.Z., Mazumder R., Kumar S., Kourtesis P., et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 16.Pearl F.M., Lee D., Bray J.E., Sillitoe I., Todd A.E., Harrison A.P., Thornton J.M., Orengo C.A. Assigning genomic sequences to CATH. Nucleic Acids Res. 2000;28:277–282. doi: 10.1093/nar/28.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]