Abstract
SEARCHGTr is a web-based software for the analysis of glycosyltransferases (GTrs) involved in the biosynthesis of a variety of pharmaceutically important compounds like adriamycin, erythromycin, vancomycin etc. This software has been developed based on a comprehensive analysis of sequence/structural features of 102 GTrs of known specificity from 52 natural product biosynthetic gene clusters. SEARCHGTr is a powerful tool that correlates sequences of GTrs to the chemical structures of their corresponding substrates. This software indicates the donor/acceptor specificity and also identifies putative substrate binding residues. In addition, it provides interfaces to other public databases like GENBANK, SWISS-PROT, CAZY, PDB, PDBSum and PUBMED for extracting various information on GTrs homologous to the query sequence. SEARCHGTr would provide new dimension to our previously developed bioinformatics tool NRPS-PKS. Together, these tools facilitate comprehensive computational analysis of proteins involved in biosynthesis of aglycone core and its downstream glycosylations. Apart from presenting opportunities for rational design of novel natural products, these tools would assist in the identification of biosynthetic products of secondary metabolite gene clusters found in newly sequenced genomes. SEARCHGTr can be accessed at http://www.nii.res.in/searchgtr.html.
INTRODUCTION
Glycosyltransferases (GTrs) are enzymes that carry out diverse biological functions by catalyzing transfer of activated sugars to varied acceptor molecules, like proteins, nucleic acids, saccharides, lipids or small molecules (1–3). Among the various classes of GTrs, one group of particular importance is the family of GTrs that differentially glycosylate the secondary metabolite aglycone during the late stages of biosynthesis to produce biologically active antibiotics. The site of glycosylation, nature of the sugar and the number of sugars are known to affect the efficacy of the antibiotic (4,5). The NDP-sugar substrates for antibiotic GTrs are typically TDP-hexoses, with the hexoses in either D- or L-configuration (6). A variety of functional modifications of the deoxyhexoses can result in enormous variations in donor substrates. Similarly there can be a vast repertoire of acceptor substrates, as these aglycones can be polyketides or nonribosomal peptides with enormous variations in their chemical structure. Therefore, understanding the donor/acceptor specificity of GTrs is crucial for the rational design of novel antibiotics by the reprogramming of their biosynthetic process.
The relaxed substrate specificity of some GTrs from vancomycin, chloroeremomycin and elloramycin pathway and mutational experiments on GTrs from urdamycin pathway have been successfully exploited to generate new compounds by using unnatural substrates (7–12). However, for the successful engineering of GTrs with altered recognition properties, it is essential to have powerful bioinformatics tools that can correlate the sequence of the GTrs to the chemical structures of their substrates and provide guidelines for various genetic manipulation studies. Such tools can also help in predicting substrate specificities of a large number of uncharacterized GTrs found in newly sequenced genomes.
We have recently used a knowledge-based approach to develop such in silico tools (13–15) for analyzing polyketide synthases (PKS) and nonribosomal peptide synthetases (NRPS). Apart from helping in experimental characterization of several proteins in Mycobacterium tuberculosis from PKS/NRPS family (16,17), these tools have also been successfully used to reprogram the PDIM biosynthesis pathway to produce an altered metabolite (18). In principle, a similar knowledge-based approach can be used for identifying specificity determining residues of GTrs involved in the biosynthesis of secondary metabolites. Recently available crystal structures of few antibiotic GTrs (19–21) indicate that, despite the divergence in their primary sequence they adopt the same structural fold. In view of the conserved structural fold, the structures for various GTrs with known substrates can be modeled using threading approach and putative substrate binding residues can give insights into the structural basis of their substrate recognition. Hence, we have carried out a comprehensive analysis of the sequence and structural features of various experimentally characterized GTrs, and based on the results of this analysis, we have developed, SEARCHGTr, software for correlating sequences of GTrs to their substrate specificity. In this work, we describe methods for developing SEARCHGTr, its various features and results from benchmarking.
METHODS
Compilation of GTrs with known specificity
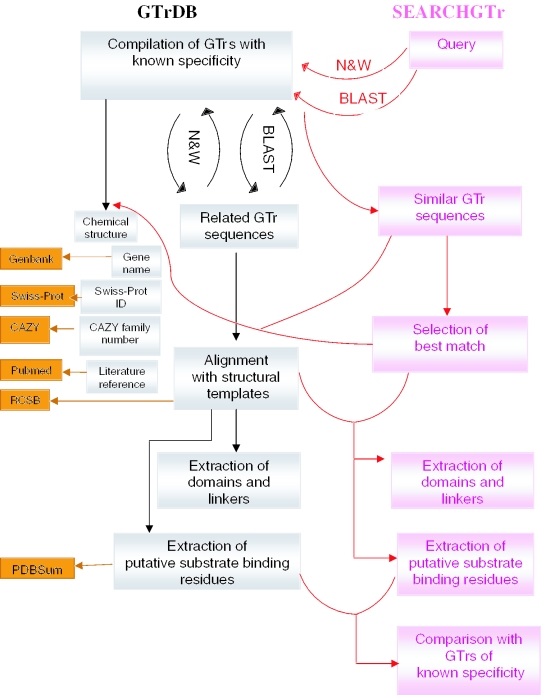
SEARCHGTr uses a knowledge-based approach to carry out various predictions by comparing the query sequence with GTrs of known specificity. Hence, an essential task in the development of this tool involved compilation of sequences of experimentally characterized GTrs in the form of a searchable database, GTrDB. Figure 1 illustrates the organization of the database. The compilation of GTrDB involved extracting sequences of GTrs glycosylating various antibiotics and identifying their donor and acceptor specificity through exhaustive literature survey. GTrDB is a compilation of 102 annotated GTr sequences from 52 different natural product biosynthetic gene clusters. The database gives the chemical structure of the antibiotic and a variety of information on the GTrs involved in its biosynthesis. For each GTr, GTrDB stores only the primary sequence in FASTA format, name of the source organism, identifiers for other databases like GenBank (22), Swiss-Prot (23), CAZY (3) etc. Using these identifiers, links are provided to respective databases for additional details on these proteins. GTrDB also provides link to the literature which describes the experimental characterization of the corresponding GTr. For each GTr, the natural donor and acceptor specificity is given along with their chemical structures. If unnatural substrates are known for a specific GTr, they are listed and a link is also provided to the corresponding literature. For deriving the homology relationships between various GTrs in the database, GTrDB stores both global and local alignments of each GTr with all other GTrs. In order to derive information about the structural features of various GTrs, each GTr sequence has been aligned, using a local version of THREADER program (24), with structural templates from the antibiotic GTr family available in PDB (25), namely 1IIR, 1RRV and 1PN3. The structures of antibiotic GTrs show bi-domain architecture with the N- and C-terminal domains containing a majority of residues binding to acceptor and donor respectively. Hence, the N- and C-terminal domains and linkers for each GTr have been identified from their threading alignments with 1RRV.
Figure 1.
A flowchart depicting the organization of SEARCHGTr and its backend database GTrDB. The links to other databases are shown in orange.
Out of the three crystal structures of antibiotic GTrs, 1RRV and 1PN3 represent GTrs in complex with donor as well as acceptor substrates. Using the LIGPLOT interactions from PDBSum database (26), 23 acceptor binding residues (ABR) and 15 donor binding residues (DBR) were identified for each GTr from their respective threading alignments with 1RRV. Similarly, 18 ABR and 16 DBR were identified for each GTr using 1PN3 as the template. It may be noted that despite the high degree of sequence and structural similarity between 1RRV and 1PN3, identical acceptor substrate binds to them in different orientations and only a very small number of amino acids are common between the ABR of 1RRV and 1PN3. It has been proposed that vancomycin GTrs achieve their regioselectivity for glycosylation by employing these two different binding modes (20,21). In view of the limited number of structures for GTrs in the complex with the substrates, prediction of the SBR for the GTrs in GTrDB remains a difficult task. Thus, the SBR identified by using 1RRV and 1PN3 as templates only help to give a consensus view of the approximate location of the substrate binding pocket. A more accurate prediction of the substrate binding pocket is possible only if additional crystal structures for members of other antibiotic GTr families are available.
Development of query interface
The program consists of two major components, the backend database GTrDB, which have well-annotated GTr sequences and the query module SEARCHGTr, which allows analysis of an uncharacterized GTr sequence. The workflow of the program is shown schematically in the Figure 1.
The query interface prompts the user to provide the query sequence in FASTA format. It is aligned with all the 102 GTrs in the GTrDB using both local alignment program BLAST as well as Needleman and Wunsch global alignment programs. Sequence alignments are carried out using BLOSUM62 scoring matrix, E-value cut off of 0.000001 and default values of gap initiation and extension penalties. The program gives a list of GTr sequences homologous to the query. After selecting a particular homologous sequence, the user can view its local/global alignment with the query. The page showing the alignment also provides a link to the GTrDB, which gives other details on the aligned GTr sequence, specifically the chemical structures of its donor and acceptor. Unnatural donor substrates for the matching GTr, if known, are also listed. This information on donor and acceptor substrates of the best matching GTr can provide important clues about possible substrates for the query GTr.
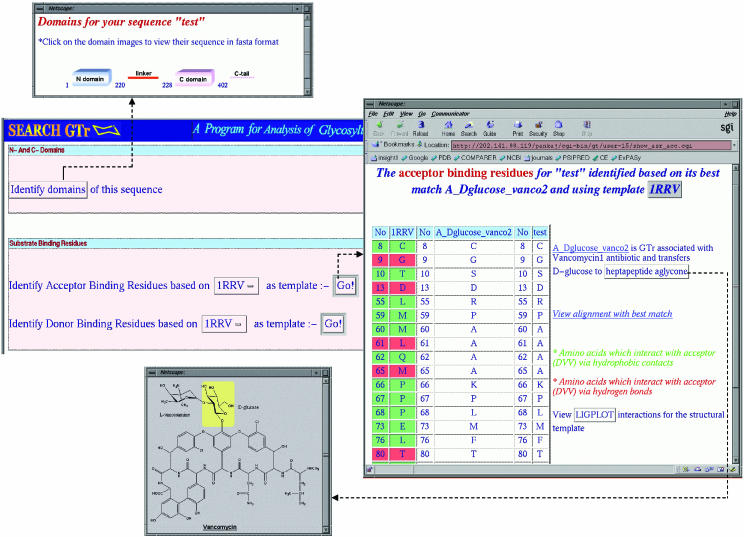
Using the best matching GTr from GTrDB and its precomputed threading alignment with 1RRV, SEARCHGTr identifies the N- and C-terminal domains and linkers in the query sequence. These are then depicted in a pictorial format in a pop up window (Figure 2). The numbers below the image of the domain specify its start and end positions on the query sequence. If there are parts of GTr which are overhangs on the N-terminal or C-terminal domains, they are shown as N-tail and C-tail, respectively. The FASTA sequence of the domain or linker or tail can be viewed by clicking on the respective images.
Figure 2.
A screenshot from SEARCHGTr showing extraction of domains and linker and identification of putative acceptor binding residues (ABR) using 1RRV as template. The ABR of the structural template, best match and query are depicted in tabular format. The page provides link to the chemical structure of the donor/acceptor of best match and LIGPLOT interactions for structural template.
The program identifies putative SBR in the query sequence from the threading alignment of its best match with 1RRV or 1PN3 (Figure 2). The user is given the option to choose either 1RRV or 1PN3 from a pull down menu as template for identifying the SBR. Links are provided to the PDBSum site for viewing the interactions between the protein and the ligand in the template structure using the program LIGPLOT. As can be seen from Figure 2, for the structural template 1RRV/1PN3, the amino acids interacting with the ligand via hydrogen bonds and hydrophobic contacts are highlighted in different colors. However, no such coloring scheme is used for the query or the best match as equivalent residues are expected to have dissimilar amino acids depending on the chemical structure of the donor/acceptor substrates. In order to facilitate correlation between the residues in the substrate binding pocket and substrate specificity, the program provides a link to the chemical structure of the acceptor/donor substrate of the best matching GTr.
Additionally, SEARCHGTr provides an option for comparing the query sequence with the experimentally characterized GTrs in terms of their substrate (acceptor/donor) binding pocket residues alone. The SBR of the query, identified based on either 1RRV or 1PN3 as the template, is compared with the corresponding library of SBR from GTrs with known donor and acceptor. The query SBR are pair-wise compared with SBR of all GTrs in GTrDB and each position is scored using the BLOSUM62 matrix. The results are displayed as a sorted list of the best matching acceptor/donor-binding residues from GTrs in GTrDB along with their donor/acceptor specificities. This allows a one to one comparison of amino acids at each position of donor/acceptor binding pocket.
Furthermore, the program also has options for the alignment of the query sequence with any specific GTr in GTrDB by selecting them based on the type of donor substrate, acceptor substrate or type of antibiotic.
Web server description
Our web interface is implemented using Perl, CGI scripts and Apache web server. Global alignments are carried out using a local version of Needleman and Wunsch programs from the EMBOSS package (27). BLAST program downloaded from NCBI is used for local alignments (28).
RESULTS
In order to benchmark the SEARCHGTr program, predictions were carried out for the 102 GTrs of known specificity using a jackknife-type approach. The 52 antibiotics in GTrDB were grouped into 20 acceptor families based on the structural similarity of the acceptor aglycone core, e.g. the vancomycin group, the anthracycline group, the orthosomycin group, etc. Substrate specificity was assigned to the query GTr based on the best match predicted by SEARCHGTr. If the best match was from the same acceptor family as the query GTr, it was considered a correct prediction. For example, if the GTr from erythromycin transferring mycarose is used as query sequence, its closest homolog is the GTr which transfers mycarose in analogous position in megalomicin. In view of the very high degree of structural similarity of the acceptor cores of erythromycin and megalomicin, it can be considered a correct prediction. This type of jackknife test was carried out for all 102 GTrs in GTrDB and the correct acceptor family could be predicted for 72 GTrs. However, 9 out of 102 GTrs belong to acceptor families containing single members only; thus their acceptor family cannot be predicted by our knowledge-based approach. Hence, the accuracy of SEARCHGTr for the prediction of acceptor family is 77%. Similar analysis also indicated that SEARCHGTr can correctly predict donor group in 45 out of 74 GTrs of known donor specificity, thereby giving a prediction accuracy of 61%.
In a separate analysis, the nr database of NCBI was searched to identify experimentally uncharacterized GTrs, whose specificity could be predicted with a reasonable confidence level by our program. Out of the 806 proteins extracted from the nr database, 19 sequences showed high sequence similarity (>40% identity) to the known GTrs in the GTrDB. However, seven of them had been annotated as hypothetical proteins or putative GTrs. Therefore, the results from in silico analysis by SEARCHGTr program can aid experimental characterization of these proteins. The other proteins from this set of 806 show a relatively lower level of sequence similarity with known GTrs in GTrDB, so the prediction of substrate specificity for them may not be reliable. However, out of these 806 sequences, 111 proteins have been annotated as hypothetical proteins in the nr database, even though they show statistically significant sequence similarity with known GTrs in GTrDB. Thus, SEARCHGTr could also aid in the annotation of GTrs.
In order to benchmark the prediction accuracy of SBR by SEARCHGTr, we attempted to predict the DBR for 1RRV using 1PN3 as template and vice versa. It was found that 14 of the 15 known DBR could be predicted for 1RRV using 1PN3 as template. On the other hand, by using 1RRV as template 14 out 16 DBR could be identified for 1PN3. These two GTrs belong to the same antibiotic family and share a sequence identity of 55%. Hence, the prediction accuracy of DBR was very high. We also tested DBR prediction accuracy of SEARCHGTr using MurG structure, which adopts a GT-B fold but is not an antibiotic glycosyltransferase. For the structure of MurG-donor substrate complex (1NLM), SEARCHGTr could identify 6 out of the 12 donor binding residues correctly, using 1RRV or 1PN3 as template. It may be noted that the sequence identity between 1NLM and 1RRV (or 1PN3) is only 20%. Even by the structural superposition of the donor binding domains of 1RRV (or 1PN3) and 1NLM only 7 of the 12 donor binding residues can be identified. Hence, even for a difficult test case like MurG, the performance of SEARCHGTr is reasonably good. The eventual inclusion of additional structural templates would help in further improving the prediction accuracy.
DISCUSSION
SEARCHGTr is an interactive web server for the analysis of GTrs involved in natural product biosynthesis. It has options for carrying out a variety of detailed analyses on GTrs of known substrate specificity. It allows identification of homologous sequences, depiction of domains and linkers and extraction of putative donor/acceptor binding residues. As the program allows comparison of amino acids lining the substrate binding pocket, it can provide clues for altering donor/acceptor selectivity of GTrs by site- directed mutagenesis experiments. Apart from its utility in the rational design of novel antibiotics, SEARCHGTr can also help in in silico identification of substrates for various uncharacterized GTrs in newly sequenced genomes. Benchmarking a set of GTrs of known substrate specificity indicate that the program can predict the acceptor specificity of antibiotic GTrs with an accuracy of 77% based on whole sequence comparisons. In view of the enormous diversity in the chemical structure of the antibiotic GTrs, even at this level of accuracy, SEARCHGTr would be a valuable tool for identifying substrates for uncharacterized GTrs. Our analysis indicates that SEARCHGTr fails to identify the correct substrates by comparison of whole sequences if the sequence similarity between the query sequence and the GTrs of known specificity is very low. It is possible that the prediction accuracy in such cases of low homology may be improved if active site residues alone are used for substrate prediction instead of whole sequences. Even though SEARCHGTr has options for comparing GTrs using only active site residues, the current version of the program extracts the putative active site residues using crystal structures of two GTrs (1RRV/1PN3) belonging to the vancomycin group of antibiotics. In view of the structural diversities of the acceptor group, it is possible that the mode of acceptor binding may be different for different groups. Elucidation of crystal structures of acceptor complexes for members of other antibiotic families will help in better identification of their binding pocket.
Acknowledgments
Authors thank Dr Sandip K. Basu for his encouragement and support. P.K. is a recipient of Senior Research Fellowship from CSIR, India. R.S.G. is a Wellcome Trust International Senior Research Fellow for biomedical sciences in India. The work has been supported by grants to National Institute of Immunology from Department of Biotechnology, Government of India. Computational resources provided under BTIS project of DBT, India are gratefully acknowledged. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hu Y., Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem. Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 2.Unligil U.M., Rini J.M. Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 2000;10:510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 3.Coutinho P.M., Deleury E., Davies G.J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 4.Schlunzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez M.J., Snyder N.J., Zweifel M.J., Wilkie S.C., Stack D.R., Cooper R.D., Nicas T.I., Mullen D.L., Butler T.F., Thompson R.C. Novel glycopeptide antibiotics: N-alkylated derivatives active against vancomycin-resistant enterococci. J. Antibiot. (Tokyo) 1998;51:560–569. doi: 10.7164/antibiotics.51.560. [DOI] [PubMed] [Google Scholar]
- 6.Liu H.W., Thorson J.S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 7.Solenberg P.J., Matsushima P., Stack D.R., Wilkie S.C., Thompson R.C., Baltz R.H. Production of hybrid glycopeptide antibiotics in vitro and in Streptomyces toyocaensis. Chem. Biol. 1997;4:195–202. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- 8.Losey H.C., Peczuh M.W., Chen Z., Eggert U.S., Dong S.D., Pelczer I., Kahne D., Walsh C.T. Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: novel glycopeptides. Biochemistry. 2001;40:4745–4755. doi: 10.1021/bi010050w. [DOI] [PubMed] [Google Scholar]
- 9.Losey H.C., Jiang J., Biggins J.B., Oberthur M., Ye X.Y., Dong S.D., Kahne D., Thorson J.S., Walsh C.T. Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem. Biol. 2002;9:1305–1314. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]
- 10.Blanco G., Patallo E.P., Brana A.F., Trefzer A., Bechthold A., Rohr J., Mendez C., Salas J.A. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem. Biol. 2001;8:253–263. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmeister D., Ichinose K., Bechthold A. Two sequence elements of glycosyltransferases involved in urdamycin biosynthesis are responsible for substrate specificity and enzymatic activity. Chem. Biol. 2001;8:557–567. doi: 10.1016/s1074-5521(01)00039-4. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmeister D., Wilkinson B., Foster G., Sidebottom P.J., Ichinose K., Bechthold A. Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem. Biol. 2002;9:287–295. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 13.Yadav G., Gokhale R.S., Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003;215:403–410. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 14.Yadav G., Gokhale R.S., Mohanty D. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003;31:3654–3658. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari M.Z., Yadav G., Gokhale R.S., Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32:W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena P., Yadav G., Mohanty D., Gokhale R.S. A new family of type III polyketide synthases in Mycobacterium tuberculosis. J. Biol. Chem. 2003;278:44780–44790. doi: 10.1074/jbc.M306714200. [DOI] [PubMed] [Google Scholar]
- 17.Trivedi O.A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 18.Trivedi O.A., Arora P., Vats A., Ansari M.Z., Tickoo R., Mohanty D., Gokhale R.S. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Mulichak A.M., Losey H.C., Walsh C.T., Garavito R.M. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 20.Mulichak A.M., Losey H.C., Lu W., Wawrzak Z., Walsh C.T., Garavito R.M. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc. Natl Acad. Sci. USA. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulichak A.M., Lu W., Losey H.C., Walsh C.T., Garavito R.M. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43:5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 22.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2003;31:23–27. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D.T., Taylor W.R., Thornton J.M. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 25.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]