Abstract
MULTIPRED is a web-based computational system for the prediction of peptide binding to multiple molecules (proteins) belonging to human leukocyte antigens (HLA) class I A2, A3 and class II DR supertypes. It uses hidden Markov models and artificial neural network methods as predictive engines. A novel data representation method enables MULTIPRED to predict peptides that promiscuously bind multiple HLA alleles within one HLA supertype. Extensive testing was performed for validation of the prediction models. Testing results show that MULTIPRED is both sensitive and specific and it has good predictive ability (area under the receiver operating characteristic curve AROC > 0.80). MULTIPRED can be used for the mapping of promiscuous T-cell epitopes as well as the regions of high concentration of these targets—termed T-cell epitope hotspots. MULTIPRED is available at http://antigen.i2r.a-star.edu.sg/multipred/.
INTRODUCTION
T-cells of the human immune system recognize antigens as short peptide fragments (T-cell epitopes) derived from the degradation of proteins. Major histocompatibility complex (MHC) proteins play a vital role in the initiation and regulation of immune responses (1–4). Their primary function is to bind and subsequently present antigenic peptides on the cell surface for recognition by T-cells of the immune system. The recognition of T-cell epitopes is critical for the immune response to infectious, autoimmune, allergic and neoplastic disease. T-cell epitopes are important for the development of peptide-based vaccines (5). There is a great diversity of human leukocyte antigens (HLAs; human MHC) genes with some 2000 known variants characterized to date (6). HLA proteins share 3D structure with main differences observed in residues that form the peptide-binding groove. HLA proteins that have small differences in their peptide-binding grooves and share similar peptide-binding specificities are grouped into HLA supertypes (7,8). Promiscuous peptides—those that bind more than one HLA variant—are prime targets for vaccine and immunotherapy development because they are relevant to higher proportions of the human population. Because of the large number of HLA proteins, experimental approaches for identifying T-cell epitopes (from overlapping peptides that span the length of protein antigens) are time-consuming and costly, and thus not applicable for large-scale screening. Computer modeling methods can help to simulate the biological process of antigen presentation, minimize the number of experiments required, enable a systematic scanning for candidate MHC-binding peptides and thus speed up vaccine development (9).
MULTIPRED is a web-based system for the prediction of peptides that bind multiple HLA alleles. Current implementation can predict peptides that bind HLA proteins belonging to supertypes A2 and A3 (HLA class I) as well as DR (HLA class II) and in future will be extended to other supertypes. The predictive engines implemented in MULTIPRED are hidden Markov models (HMMs) and artificial neural networks (ANNs). A novel data representation method enables MULTIPRED to predict peptides that bind to multiple HLA alleles belonging to one HLA supertype by a single prediction model per supertype.
SYSTEM DESCRIPTION
The predominant length of peptides that bind HLA-A2 and-A3 (class I) proteins is nine amino acids (10). HLA-DR (class II) proteins bind longer peptides through the core binding region, which is nine amino acids long (11). The training data comprise 3050 9mer peptide sequences (664 binders and 2386 non-binders) related to 15 variants of the HLA-A2 supertype, 2216 9mer peptide sequences (680 binders and 1536 non-binders) related to eight variants of the HLA-A3 supertype and 2396 9mer peptides (448 binders and 1948 non-binders) related to six HLA-DR variants. These data are mainly from three sources, the MHCPEP database (12), published articles and a set of HLA non-binding peptides (V. Brusic, unpublished data). For both training and prediction the data representation includes both the peptide and its binding environment (HLA contact residues). This ‘virtual peptide’ representation comprises both peptide residues and the environment for each residue of the 9mer peptides (13,14). To simplify the data representation and eliminate redundant information, for each HLA supertype, we considered only those contact residues that vary across various HLA variants and discarded the residues, which are conserved.
In MULTIPRED, a three-layer backpropagation network with sigmoid activation functions was built for HLA-A2 and -A3 supertype and a four-layer backpropagation network with a hyperbolic tangent sigmoid activation function between the two hidden layers and a sigmoid activation function between the second hidden layer and the output for HLA-DR supertype. Various techniques, including optimization of ANN architecture and balancing datasets, were explored to improve the prediction accuracy of the ANN models (14). MULTIPRED also has a first-order HMM as an alternative prediction engine (13). The user can select either the ANN or the HMM model for prediction—both methods have been optimized and show similar performance. The AROC is >0.8 in all cases, indicating good prediction capability [see (13,14) for details on HLA-A2 models, (15) for HLA -A3 models, and V. Brusic, A. Sette, G. L. Zhang, K. N. Srinivasan, J. T. August and V. Brusic, manuscript in preparation for HLA-DR models.
In addition to individual 9mer predictions, MULTIPRED also predicts immunological hotspots (regions of high concentration of 9mer promiscuous binders). We have developed two scoring schemes to identify immunological hotspots within antigens for HLA classes I and II supertype. The scheme for HLA class I supertype is based on high-scoring individual 9mers within a window of 30 amino acids (15) and the scheme for HLA class II supertype is based on average scores of individual 9mers within a window of 15 amino acids. The selection of window lengths was based on a trial-and-error process. Window lengths of 15, 20, 25 and 30, were explored and the results were compared with the representative experimental results. The window length 30 was found to suit class I predictions and window length 15 to class II predictions. The lengths outside these ranges are considered too short or too long as targets for experimental validation. The prediction performance of MULTIPRED for HLA-A2 and -A3 hotspots was validated using experimental results from a systematic study of human papillomavirus type 16 E6 (P03126) and E7 (P03129) proteins (16). The prediction performance of MULTIPRED for HLA-DR hotspots was validated using experimental results from systematic binding studies of overlapping peptides from Myelin Oligodendrocyte glycoprotein (MOG) (CAA88109), bee venom protein (1POC) and hepatitis C virus 1B protein (AAB00216).
USING THE SYSTEM
The web interface of MULTIPRED uses a set of graphical user interface forms with a combination of Perl, CGI and C background programs. Development of MULTIPRED was carried out in SunOS 5.9 UNIX environment. The functions provided by MULTIPRED include (i) running predictions, (ii) model building, (iii) prediction accuracy evaluation and (iv) identifying consensus predictions among up to three sets of predictions on the same input protein sequence.
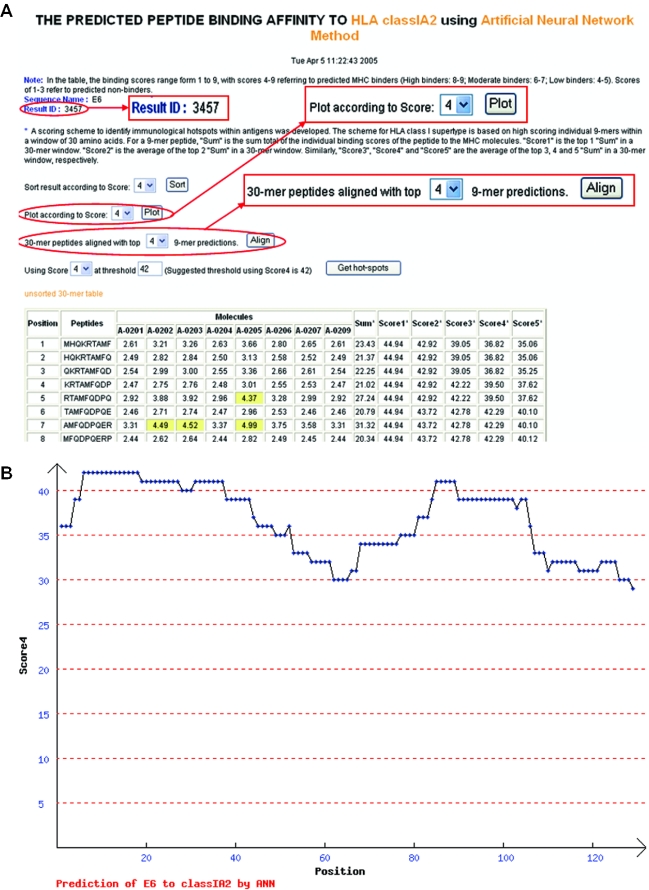
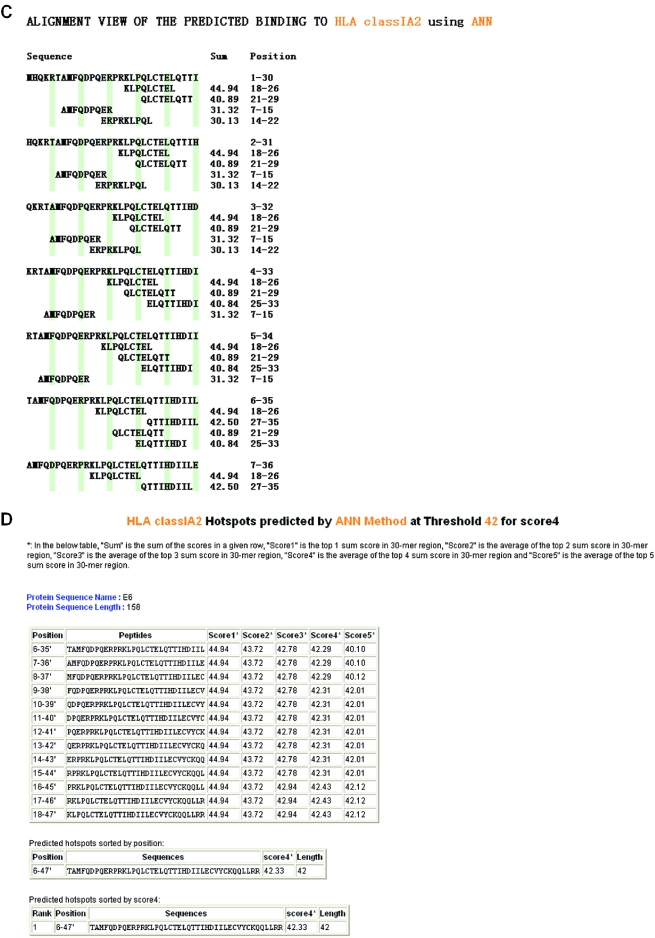
To predict peptides binding to a supertype, users must first select ‘Run prediction’. The required input is the selection of supertype and prediction method (pre-defined ANN or HMM). Alternatively, users can select a pre-defined model (built by model building function). By selecting the ‘Submit’ button users get to a sequence input page where the required input is a protein sequence and its name. The length of the input sequence must be between 9 and 2000 amino acids. If the input sequence contains symbols other than amino acids (space and carriage returns are allowed) or if the sequence is outside the length limits, an error message will be displayed. The input can either be a protein sequence or a list of peptides. The default selection on the webpage is ‘Protein sequence’, which means the input sequence is treated as one single protein sequence and carriage returns are ignored. If users changed the sequence type to ‘a list of peptide sequences’, then sequences divided by carriage returns are treated as separate peptides. The processing steps and result pages for the two types of inputs are different. The detailed description on processing steps involved when the input sequence is a protein sequence or a list of peptides are available at http://antigen.i2r.a-star.edu.sg/multipred/HTML/faq.html#Q3 and http://antigen.i2r.a-star.edu.sg/multipred/HTML/faq.html#Q4, respectively. The 9mer binding scores range from 1 to 9 (Figure 1A), with scores 4–9 referring to predicted binders (8 or 9 referring to high, 6 or 7 to moderate, and 4 or 5 to low confidence of peptide binding). Scores 1–3 refer to predicted non-binders. MULTIPRED saves the prediction result and the users may note down the ID number of the saved jobs for the comparison of prediction results generated by different prediction models (Figure 1A). Two scoring schemes to identify immunological hotspots within antigens were developed for HLA classes I and II supertype. The scheme for HLA class I supertypes is based on high-scoring individual 9mers within a window of 30 amino acids (15). In the result table (Figure 1A), ‘Sum’ is the sum total of the individual binding scores of a peptide to the MHC proteins, ‘Score 1’ is the top 1 ‘Sum’ in a 30mer window (A 30mer window comprises 22 consecutive 9mer peptides). ‘Score 2’ is the average of the top 2 ‘Sum’ in a 30mer window. Similarly, ‘Score 3’, ‘Score 4’ and ‘Score 5’ are the average of the top 3, 4 and 5 ‘Sum’, respectively, in a 30mer window. To show the user a clear view of the binding capacity of an input protein, Scores 1–5 of all 30mer peptides of the input protein can be displayed as graphs, in which x-axis represents the starting position of a 30mer window and the y-axis represents Score 1 (2/3/4/5) of the 30mer window. For example, in Figure 1B, which is the graph of Score 4 of the protein E6, the first three 30mer windows (starting at positions 1, 2 or 3) are 36.82 and the next two windows (starting at positions 4 or 5) have scores 39.50. The following 13 30mer windows (starting at positions 6–19) have scores >42, the recommended threshold for Score 4 for HLA-A2 ANN models (Figure 1A), indicating a predicted hotspot, which corresponds to an experimentally determined HLA-A2 hotspot in E6 protein (16). To locate the individual 9mers with top binding scores in each 30mer window, the ‘align’ function can be used. Figure 1C shows an example of the alignment view of the top four 9mers in each 30mer window. The user can also identify hotspots at a certain threshold by using the ‘Get hotspots’ function (Figure 1D). The default values on the web page are the recommended thresholds for Score 4. In a HLA-DR prediction result table, ‘Average’ was calculated as the average of the ‘Sum’ within a 15mer window (seven consecutive 9mers make a 15mer window).
Figure 1.
An example of the output pages of MULTIPRED when the input is a single protein sequence. The input protein sequence is a human papillomavirus type 16 E6, the prediction method used is ANN and the HLA supertype of interest is HLA-A2. (A) The main result page. The input sequence is truncated into overlapping 9mers for the prediction of binding scores to multiple HLA-A2 variants, *0201, *0202, *0203, *0204, *0205, *0206, *0207 and *0209. The red ovals are added by the authors for the clarity of viewing. (B) Example graph of Score 4. (C) Alignment view of the top four 9mers in the 30mer windows. (D) The prediction hotspot region is 6–47 at threshold 42.
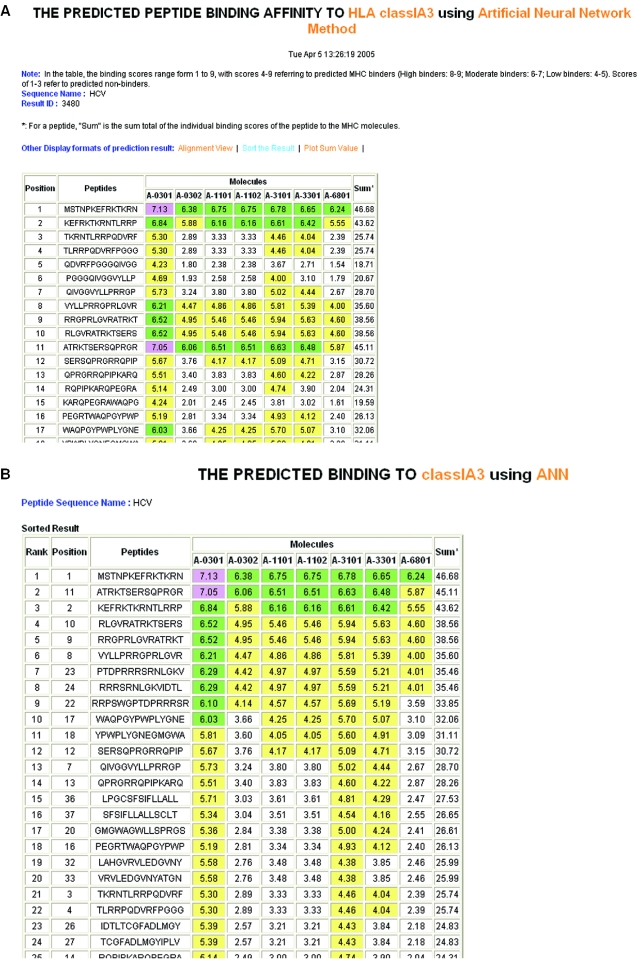
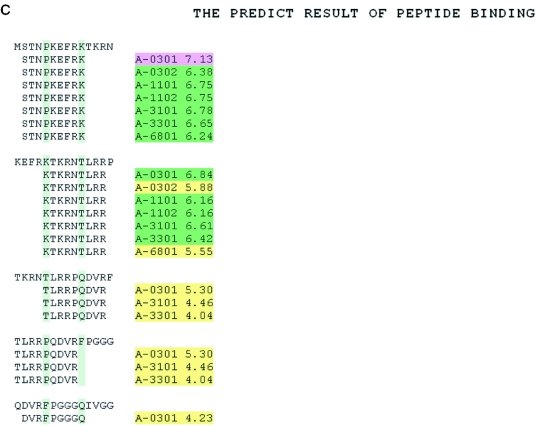
When users select the input sequence as ‘a list of peptide sequences’, the input sequences separated by carriage returns or line breaks are treated as different peptides. All overlapping 9mers in each peptide are submitted for prediction. In the result tables, predicted binding scores are represented by the highest individual binding score of each input peptide. The predicted binding scores of individual 9mers in each peptide in the list are data not shown (Figure 2A). To display the input peptides in the order of their binding scores, the user can use the function ‘Sort the Result’. In the result page (Figure 2B), the input peptides are listed in descending order of their binding scores. To display the predicted 9mer binders from each input peptide, the user can use the function ‘Alignment View’. In the result page (Figure 2C), the 9mers with binding scores ≥4 are aligned with the input peptides. The predicted 9mer binders are displayed with the names of the HLA alleles, which produced binding scores above the selected threshold.
Figure 2.
An example of the output pages of MULTIPRED when input is a list of peptides. The input protein peptides are from hepatitis C virus, the prediction method used is ANN and the HLA supertype of interest is HLA-A3. (A) The main result page. As can be seen here, the input sequence is truncated into overlapping 9mers for the prediction of binding scores to multiple HLA-A3 variants, *0301, *0302, *1101, *1102, *3101, *3301 and *6801. (B) Input peptides displayed in the descending order of binding scores. (C) Alignment view of the predicted 9mer binders.
If the user has 9mer peptides with known binding affinities to proteins belonging to HLA-A2, -A3 or -DR supertypes and wants to build his own prediction models, the user can use the ‘Model build’ function in MULTIPRED. Only 9mer peptides can be used as training data. The users have the option to use their data only, or combine their data with the existing MULTIPRED data and build the model on the server. Currently, users can expect to train an HMM model within 1 min while training of ANN models may take up to 50 min (depending on the size of the training dataset)—there are actually four ANNs trained in the background. The ANN models trained by the same dataset are usually slightly different because the initial weights of networks are assigned randomly (14). To make the trained models more stable, the training is repeated four times, and four sets of weights are trained—the predictions are the averages of these four predictions. When the model building request is submitted, an intermediate page (Figure 3) will be displayed providing the result URL can be bookmarked for later model retrieval.
Figure 3.
When the model building request is submitted, an intermediate page will be displayed providing the result URL that can be bookmarked for later model retrieval.
If the user has 9mer peptides with known binding affinities and would like to evaluate the prediction accuracy of a model with these peptides, the user can use the ‘accuracy evaluation’ function of MULTIPRED. The system predicts the binding affinities of the input 9mers and calculates AROC of the predictions. For each supertype, there are two built-in prediction models available in MULTIPRED. Predictions can also be performed by user-built models. Therefore, MULTIPRED may produce several sets of predictions for the same sequence. The comparison of predictions helps identify the most promising peptides picked up as predicted binders by multiple models. The comparison is facilitated by the ‘Consensus predictions’ function. The user needs to input the individual Result IDs (up to three) to the system. The Result IDs must be predictions of the same protein and to the same HLA supertype, if the Result IDs belong to predictions on different proteins, an error message will be displayed. The user can select the analysis of top 5 or 10% of the predicted binders. In the output table, top 5 or 10% predictions are displayed in the descending order of their binding scores. The peptides selected by multiple models are highlighted in blue or red.
DISCUSSION
Several web-based systems have been developed and widely used for the prediction of MHC binders, such as SYFPEITHI (17), BIMAS (18), SMM (19), MHCPred (20), RANKPEP (21), TEPITOPE (22), NetMHC (23) and SVMHC (24). Although MULTIPRED is similar to them in its overall goal of predicting MHC-binding peptides, there are significant differences in both functionality and methodology. SYFPEITHI uses binding motifs. BIMAS, MHCpred, RANKPEP and TEPITOPE use quantitative matrices, and SMM is based on an improved matrix-based algorithm called stabilized matrix method. SVMHC uses support vector machines (SVMs) and NetMHC uses ANNs. Each of these methods uses one prediction model per MHC proteins, making them difficult to maintain and assess accuracy. TEPITOPE allows prediction of peptides to many different Class II proteins (using multiple prediction models), but it is not available through the Web. MULTIPRED predicts peptide binding to multiple HLA proteins with one model per HLA supertype. It can also identify promiscuous peptides and T-cell epitope hotspots. Since HLA proteins are highly polymorphic, promiscuous peptides that bind more than one HLA protein are prime targets for vaccine and immunotherapy development because they are relevant to higher proportions of the human population. T-cell epitope hotspots are highly promising regions as targets of T-cell immune responses, which are of interest for experimental validation as potential vaccine targets. In addition, MULTIPRED provides several functions which are not available in other prediction systems, such as model building by user function, accuracy evaluation function and consensus prediction function. The pathway from epitopes to vaccine development is lengthy and cost-intensive, involving exhaustive experiments. The main utility of MULTIPRED is in the selection of key antigenic regions to minimize the number of experiments required for mapping of promiscuous T-cell epitopes and T-cell epitope hotspots.
SUPPLEMENTARY MATERIAL
Supplementary material is available at NAR online.
Supplementary Material
Acknowledgments
Authors thank Seng Hong Seah and Olivo Miotto for their valuable suggestions. This project has been funded in part with the USA Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Grant No. 5 U19 AI56541 and Contract No. HHSN2662-00400085C. Funding to pay the Open Access publication charges for this article was provided by the Institute for Infocomm Research, Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pamer E., Cresswell P. Mechanisms of MHC class I—restricted antigen processing. Annu. Rev. Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 2.Villadangos J.A., Bryant R.A., Deussing J., Driessen C. Proteases involved in MHC class II antigen presentation. Immunol. Rev. 1999;172:109–120. doi: 10.1111/j.1600-065x.1999.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 3.Yewdell J.W., Bennink J.R. Cut and trim: generating MHC class I peptide ligands. Curr. Opin. Immunol. 2001;13:13–18. doi: 10.1016/s0952-7915(00)00175-8. [DOI] [PubMed] [Google Scholar]
- 4.Bryant P., Ploegh H. Class II MHC peptide loading by the professionals. Curr. Opin. Immunol. 2004;16:96–102. doi: 10.1016/j.coi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Zhong W., Reche P.A., Lai C.C., Reinhold B., Reinherz E.L. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J. Biol. Chem. 2003;278:45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 6.Marsh S.G.E. Nomenclature for factors of the HLA system, update September 2003. Tissue Antigens. 2004;63:190–191. doi: 10.1111/j.1399-0039.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 7.Sette A., Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 8.Lund O., Nielsen M., Kesmir C., Petersen A.G., Lundegaard C., Worning P., Sylvester-Hvid C., Lamberth K., Røder G., Justesen S., et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- 9.Brusic V., Bajic V.B., Petrovsky N. Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods. 2004;34:436–443. doi: 10.1016/j.ymeth.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Rammensee H.G., Falk K., Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu. Rev. Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 11.Stern L.J., Brown J.H., Jardetzky T.S., Gorga J.C., Urban R.G., Strominger J.L., Wiley D.C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 12.Brusic V., Rudy G., Harrison L.C. MHCPEP, a database of MHC-binding peptides. Nucleic Acids Res. 1994;22:3663–3665. doi: 10.1093/nar/22.17.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brusic V., Petrovsky N., Zhang G.L., Bajic V.B. Prediction of promiscuous peptides that bind HLA class I molecules. Immunol. Cell Biol. 2002;80:280–285. doi: 10.1046/j.1440-1711.2002.01088.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G.L., Khan A.M., Srinivasan K.N., August J.T., Brusic V. Neural models for predicting viral vaccine targets. J. Bioinform. Comput. Biol. 2005 doi: 10.1142/s0219720005001466. (in press) [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan K.N., Zhang G.L., Khan A.M., August J.T., Brusic V. Predictions of Class I T-cell epitopes: evidence of presence of immunological hotspots inside antigens. Bioinformatics. 2004;20(Suppl. 1):i297–i302. doi: 10.1093/bioinformatics/bth943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kast W.M., Brandt R.M., Sidney J., Drijfhout J.W., Kubo R.T., Grey H.M., Melief C.J., Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- 17.Rammensee H.G., Bachmann J., Emmerich N.P., Bachor O.A., Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 18.Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 19.Peters B., Tong W., Sidney J., Sette A., Weng Z. Examining the independent binding assumption for binding of peptide epitopes to MHC-I molecules. Bioinformatics. 2003;19:1765–1772. doi: 10.1093/bioinformatics/btg247. [DOI] [PubMed] [Google Scholar]
- 20.Hattotuwagama C.K., Guan P., Doytchinova I.A., Zygouri C., Flower D.R. Quantitative online prediction of peptide binding to the major histocompatibility complex. J. Mol. Graph. Model. 2004;22:195–207. doi: 10.1016/S1093-3263(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 21.Reche P.A., Glutting J.P., Reinherz E.L. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 2002;63:701–709. doi: 10.1016/s0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
- 22.Bian H., Hammer J. Discovery of promiscuous HLA-II-restricted T cell epitopes with TEPITOPE. Methods. 2004;34:468–475. doi: 10.1016/j.ymeth.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Buus S., Lauemoller S.L., Worning P., Kesmir C., Frimurer T., Corbet S., Fomsgaard A., Hilden J., Holm A., Brunak S. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens. 2003;62:378–384. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 24.Donnes P., Elofsson A. Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics. 2002;3:25–38. doi: 10.1186/1471-2105-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.