Abstract
Using the Distributed Annotation System (DAS) we have created a protein annotation resource available at our web page: http://www.cbs.dtu.dk, as a part of the BioSapiens Network of Excellence EU FP6 project. The DAS protocol allows us to gather layers of annotation data for a given sequence and thereby gain an overview of the sequence's features. A user-friendly graphical client has also been developed (http://www.cbs.dtu.dk/cgi-bin/das), which demonstrates the possibility of integrating DAS annotation data from multiple sources into a simple graphical view. The client displays protein feature annotations from the Center for Biological Sequence Analysis as well as from the BioSapiens reference UniProt server (http://www.ebi.ac.uk/das-srv/uniprot/das) at the European Bioinformatics Institute. Other DAS data sources for protein annotation will be added as they become available.
INTRODUCTION
In recent years, numerous computational tools for gene and protein analysis have been constructed by various laboratories. Several such analysis tools have been created and published by the Center for Biological Sequence Analysis (CBS), many of which are available online for all users at the CBS web page: http://www.cbs.dtu.dk. The analysis results of such tools have led to an explosion in the amount of data in biological databases and available information that exists for biological sequences. Today, one of the major tasks of systems biology is to integrate as much of the experimental and computational information as possible and thereby gain biological insight into the properties and function of the macromolecules under observation. This means the integration of several types of data, in various formats, dispersed around the face of the globe into a unified structure. This integration of online annotations is greatly simplified if the annotation services follow accepted standards. One such standard is the Distributed Annotation System (DAS) (1).
DAS services have existed for several years now. Version 1.0 of the DAS specification was released in 2001 and version 2 is under development. The DAS protocol is a simple http-based client-server system. A query in the form of a URL is made to the server, which replies with annotations for the sequence entry specified in the URL query. The reply from the server is in XML format. The DAS web page (http://www.biodas.org) has both Perl- and Java-based server software for download. Client libraries in Perl and Java are also available.
The DAS specification was originally written with genomic sequences in mind, but the standard has proven itself flexible enough to handle protein data as well. Several annotation databases are now serving annotations using the DAS system, including Ensembl (2), FlyBase (3), UniProt (4) and WormBase (5).
The flexibility and success of the DAS protocol has made it the annotation method of choice for the BioSapiens Network of Excellence, of which the CBS DAS server detailed here is a part. The various consortium members will in the near future deploy several DAS servers, which will serve protein annotations for the same UniProt sequences as the DAS server at CBS and all the data can therefore be easily integrated in a coherent manner.
The full list of query types that the DAS specification supports is beyond the scope of this document. We refer readers to the DAS web page and specification for detailed information and it suffices to say that for queries on protein sequences, the most important queries are probably the ‘sequence’ to which a reference DAS server responds with the full sequence and ‘features’ to which reference and annotation servers respond with feature annotations they store for a specified sequence identifier. An example query to the CBS DAS server is shown below.
SERVER INFRASTRUCTURE
At CBS, we have implemented a Perl-based DAS server, ProServer (http://www.sanger.ac.uk/Software/analysis/proserver), which accepts queries at the address: http://genome.cbs.dtu.dk:9000/das. We provide annotations for several of CBS's protein sequence annotation servers, which predict protein sorting [LipoP (6), NetNES (7), SignalP (8), SecretomeP (9), TargetP (10)], protein post-translational modification [NetAcet (11), NetPhos (12), NetOGlyc (13), NetNGlyc, ProP (14)] and protein structure and function [TMHMM (15)]. Statistics and data source names (DSNs) for the individual methods are shown in Table 1. The annotations provided by the DAS server include: the start and end position of the feature annotated; the score from the prediction method that assigned the feature; a hyperlink to the web page of the prediction method with sequence information preloaded in the form input and possibly some further information.
Table 1.
Annotation methods provided by the CBS DAS system
| Method | Data source name | Organism coverage | Number of records | Reference |
|---|---|---|---|---|
| LipoP-1.0 | lipop | Gneg | 7 597 | (6) |
| NetAcet-1.0 | netacet | E | 122 664 | (7) |
| NetNES-1.1 | netnes | E | 1 945 054 | (11) |
| NetNGlyc-1.0 | netnglyc | H | 137 800 | |
| NetOGlyc-3.1 | netoglyc | M | 81 310 | (13) |
| NetPhos-2.0 | netphos | E | 8 940 654 | (12) |
| ProP-1.0 | prop | E | 127 553 | (14) |
| SecretomeP-1.0 | secretomep | E | 58 318 | (9) |
| SignalP-3.0 | signalp | E, Gpos, Gneg | 1 189 706 | (8) |
| TargetP-1.01 | targetp | E | 750 111 | (10) |
| TMHMM-2.0 | tmhmm | A | 5 086 476 | (15) |
| All the above combined | cbs_total | 18 447 243 |
The annotation methods are specific to the following phylogenetic groups: ‘A’ stands for all proteins, ‘E’ for eukaryotes, ‘Gpos’ for Gram-positive bacteria, ‘Gneg’ for Gram-negative bacteria, ‘H’ for human and ‘M’ for mammals. The data source name is the name of the particular annotation method on the DAS server.
In general, the annotations span all of UniProt (4), but are limited to phylogenetic subsets of the database, as the annotation methods are usually constructed with a specific phylogenetic group as a target (see the reference for each server for details). Currently, the CBS DAS servers provide over 18 million protein annotations for over 1.5 million protein sequences from the UniProt database and we hope that this wide coverage makes our services of general interest to the scientific community. The predicted annotations include several highly cited methods, e.g. SignalP and NetPhos, which are among the top 1% of the most cited papers in the scientific literature according to the Institute for Scientific Information.
The annotations are precalculated and the results stored in a relational database, allowing for fast retrieval and update of data.
Regarding the terminology of the predicted features, we have generally used the nomenclature of the original prediction method. In some cases, we have modified the feature names to mimic the UniProt feature table, thus reflecting the reference database structure, allowing for easy comparison between the reference UniProt server and other annotation resources. It is quite conceivable that the vocabulary will be updated at a later point to make use of standard ontologies such as the Gene Ontology (GO) (16), so that post-translational modifications would be mapped onto GO ‘biological process’, etc. The concept of the Sequence Ontology (SO) (http://song.sourceforge.net/) is highly relevant to this project, however the SO does not yet provide sufficient coverage of protein sequence attributes, such as post-translational modification, to be useful for our purposes.
A query example
When querying a DAS server for annotation, one must append the DSN, along with a query type and a sequence identifier to the address of the server. For example, if we wish to ask for annotations from the SignalP signal peptide prediction method (8) for the protein EGFR_HUMAN we first append the DSN for that method (‘signalp’, Table 1). Then we use the ‘features’ query to ask for feature annotations and identify the sequence as a ‘segment’. The whole query string thus looks like this: http://genome.cbs.dtu.dk:9000/das/signalp/features?segment=EGFR_HUMAN.
CBS DAS VIEWER
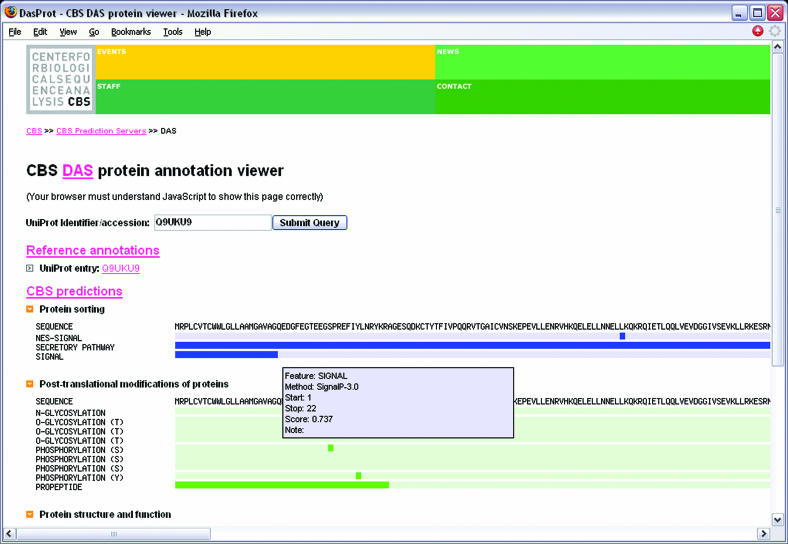
As the raw XML output of DAS servers is not very suitable for browsing of feature annotations, we have developed a client viewer to allow visualization of CBS DAS annotations in a simple graphical way. This viewer is publicly available at http://www.cbs.dtu.dk/cgi-bin/das. All the user is required to do is to input a UniProt accession number or identifier. The viewer then collects the annotations provided by the CBS DAS servers, along with annotations from a UniProt reference DAS server at the European Bioinformatics Institute (http://www.ebi.ac.uk/das-srv/uniprot/das) for that particular sequence. All the annotations are then displayed as aligned graphical tracks, allowing for easy inspection of features along the length of the protein. Additional information about the annotations is shown in a pop-up window when the user points the mouse to an annotation track. This is the first time CBS has provided a composite graphical display of several of its protein prediction methods simultaneously, which the users of CBS prediction services may find interesting. Some types of feature annotations carry a hyperlink in the XML payload. When the user clicks on a graphical track for such an annotation, the CBS DAS protein viewer will open a new browser window, following the hyperlink. The graphical tracks can also be folded and expanded to allow simplified overview. A screenshot of the client in action can be seen in Figure 1. The client demonstrates how easily different data sources can be integrated using the DAS. We plan to incorporate relevant DAS protein annotation resources into the graphical client as they appear. At the time of writing, only one external DAS source was incorporated in the view; a resource where RCSB Protein Data Bank (17) structures are aligned upon UniProt entries, provided by the Sanger Institute (http://www.sanger.ac.uk).
Figure 1.
The CBS protein DAS viewer. The browser interface is very simple, it has only one form field and the graphical tracks show the annotations for a given UniProt protein. Additional information for individual features is shown in a pop up help window when the mouse is pointed at the feature.
Acknowledgments
The author wishes to thank Søren Brunak, Kristoffer Rapacki and Hans-Henrik Stæfreldt at CBS, as well as the anonymous reviewers for helpful comments and suggestions. This work was supported by a grant from the BioSapiens Network of Excellence (NoE), FP6, contract no. LSHG-CT-2003-503265, the Danish Platform for Integrative Biology with Focus on Systemic Proteomics—DPIB, funded by the Danish National Research Foundation and the Danish Center for Scientific Computing. Funding to pay the Open Access publication charges for this article was provided by the Danish National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dowell R.D., Jokerst R.M., Day A., Eddy S.R., Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drysdale R.A., Crosby M.A., Gelbart W., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., et al. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N., Harris T.W., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Bradnam K., Canaran P., Chan J., Chen C.-K., et al. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juncker A.S., Willenbrock H., von Heijne G., Brunak S., Nielsen H., Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.la Cour T., Kiemer L., Molgaard A., Gupta R., Skriver K., Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 8.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 9.Bendtsen J.D., Jensen L.J., Blom N., von Heijne G., Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 10.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 11.Kiemer L., Bendtsen J.D., Blom N. Netacet: prediction of N-terminal acetylation sites. Bioinformatics. 2005;21:1269–1270. doi: 10.1093/bioinformatics/bti130. [DOI] [PubMed] [Google Scholar]
- 12.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 13.Julenius K., Molgaard A., Gupta R., Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 14.Duckert P., Brunak S., Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 15.Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 16.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology the gene ontology consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., Feng Z., et al. The RCSB protein data bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]