Abstract
The three most abundant extracellular proteins of Mycobacterium tuberculosis, the 30-, 32-, and 16-kDa major extracellular proteins, are particularly promising vaccine candidates. We have mapped T-cell epitopes of these three proteins in outbred guinea pigs by immunizing the animals with each protein and assaying splenic lymphocyte proliferation against a series of overlapping synthetic peptides covering the entire length of the mature proteins. The 30-kDa protein contained nine immunodominant epitopes, the 32-kDa protein contained two immunodominant epitopes, and the 16-kDa protein contained a highly immunodominant region at its N terminus. The immunodominant epitopes of the 30- and 32-kDa proteins in outbred guinea pigs were frequently identified in healthy purified-protein-derivative-positive or BCG-vaccinated individuals in previous studies. The immunodominant epitopes of these major extracellular proteins have potential utility in an epitope-based vaccine against tuberculosis.
Tuberculosis continues to ravage humanity, and an improved vaccine is urgently needed. The major extracellular proteins of Mycobacterium tuberculosis are prime candidates for a subunit vaccine against tuberculosis (1, 8, 15, 16). Purified major extracellular proteins have been shown to induce protective immunity against aerosol challenge in the outbred guinea pig model of pulmonary tuberculosis (8). Tuberculosis in the highly sensitive guinea pig closely mimics the human disease immunologically, clinically, and pathologically, and hence the guinea pig is the most relevant small animal model of tuberculosis.
The three most abundant extracellular proteins of M. tuberculosis, the 30-kDa (also called antigen 85B), the 32-kDa (also called antigen 85A), and the 16-kDa (also called MPT 63) proteins are particularly promising vaccine candidates. Together, they comprise approximately one-half of the total proteins exported by M. tuberculosis into broth culture (8). They are also among the most abundant M. tuberculosis proteins expressed in macrophages (10). The 30- and 32-kDa proteins of M. tuberculosis are present throughout the genus and function as mycolyltransferases (2, 19). The 16-kDa protein is specific to the M. tuberculosis complex but its function is not known.
Proteins often contain immunodominant epitopes that cross major histocompatibility complex types within and between species (3, 13, 14). One strategy for enhancing the protective efficacy of a protein vaccine is to immunize with selective immunodominant epitopes of the protein. In theory, an epitope vaccine may be more effective than a whole protein vaccine because it would contain a high proportion of the most relevant peptide components and a low proportion of irrelevant or potentially deleterious components, e.g., immunosuppressive components. Consistent with this idea, in one report, mice immunized with a peptide of hen eggwhite lysozyme produced a vigorous T-cell proliferative response to the peptide that did not occur after immunization with the whole protein (20).
In this study, we sought to identify immunodominant epitopes of the 30-, 32-, and 16-kDa major extracellular proteins of M. tuberculosis. To do so, we immunized outbred guinea pigs with one of each of the three proteins and assayed the proliferative responses of their splenic lymphocytes to synthetic peptides overlapping the entire mature protein to which the animal was immunized.
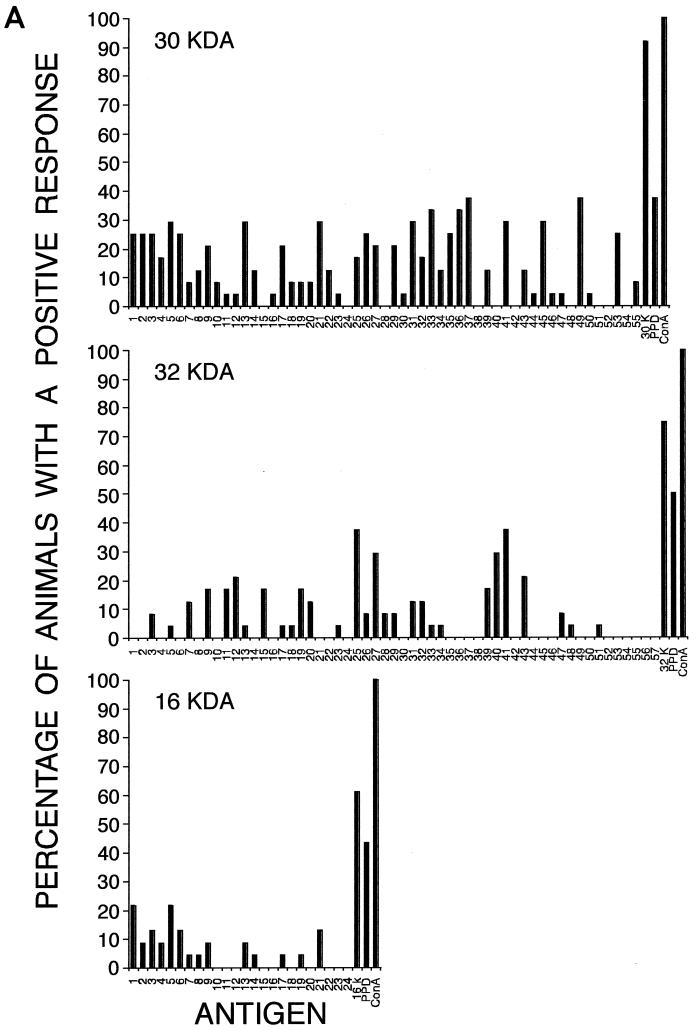
Native 30-, 32-, and 16-kDa proteins of M. tuberculosis used in this study were purified from culture filtrates as described previously (8). Outbred male Hartley strain guinea pigs (Charles River Breeding Laboratories) were immunized three or four times with 100 μg of a purified protein in Syntex adjuvant formulation (SAF) every 3 weeks, also as described previously (8). A skin test for the proteins was performed at the end of the immunization protocol (8). Control guinea pigs were sham immunized with phosphate buffer only in SAF. For each protein, a series of synthetic peptides (15-mers) covering the entire mature protein and overlapping by 10 amino acids was used in splenic lymphocyte proliferation assays. Synthetic peptides were purified by high-pressure liquid chromatography, and the accuracy of the peptide sequence for the 32- and 16-kDa proteins was confirmed by mass spectrometry. The purity of the synthetic peptides was typically greater than 80%, with few exceptions. Lymphocyte proliferation assays were performed as described earlier (15). A positive lymphocyte proliferative response to a synthetic peptide or control antigen was defined as (i) a stimulation index (SI) that is greater than 1.2 and (ii) a value of (decays per minute [dpm] with antigen) − (dpm without antigen) that is greater than 1,500. By these criteria, at least 60% of the immunized animals had a positive response to the protein used for immunization. Four separate experiments were conducted for each protein. The data from the four experiments (23 or 24 animals for each protein) were combined in Fig. 1, which shows the percentage of animals with a positive response to individual peptides of each protein.
FIG. 1.
Lymphocyte proliferative responses of immunized (A) and control (B) guinea pigs to overlapping synthetic peptides of the M. tuberculosis 30-, 32-, and 16-kDa major extracellular proteins. Peptides (15-mers) covering the entire mature protein and overlapping by 10 amino acids were synthesized for use in the lymphocyte proliferation assays. Splenic lymphocytes (106) from guinea pigs immunized with the 30-, 32-, or 16-kDa protein or from sham-immunized control guinea pigs were incubated with peptides (10 μg), purified protein (1 μg), PPD (1 μg), or concanavalin A (0.5 μg) in the presence of polymyxin B (10 U) for 48 h and then pulsed with [3H]thymidine (1 μCi) for 16 h. Each of the three proteins was studied in four independent experiments, each of which had five or six immunized and six control animals (12 experiments in total). For each protein, the data represent the combined results of the four experiments (23 to 24 immunized and 24 control animals per protein). The definition of a positive response was as follows: ([dpm with antigen] − [dpm without antigen] > 1,500) and ([dpm with antigen]/[dpm without antigen] > 1.2).
Peptide mapping of the 30-kDa protein.
The mature 30-kDa protein of M. tuberculosis consists of 285 amino acids (5, 7). It is the most abundant protein secreted by the bacterium and is apparently the most immunogenic of the three proteins tested in this study. Splenic lymphocytes from 22 of 24 guinea pigs (92%) immunized with purified 30-kDa protein proliferated in response to the protein. Lymphocytes from nine of the immunized animals (38%) proliferated against M. tuberculosis purified protein derivative (PPD). The mean SIs of the immunized animals were 2.8 ± 1.6 in response to the 30-kDa protein and 1.4 ± 0.5 in response to PPD. Peptides recognized by 25% or more of the animals were as follows: peptide numbers 1 to 3 (amino acids 1 to 25), 5 to 6 (amino acids 21 to 40), 13 (amino acids 61 to 75), 21 (amino acids 101 to 115), 26 (amino acids 126 to 140), 31 (amino acids 151 to 165), 33 (amino acids 161 to 175), 35 to 37 (amino acids 171 to 195), 41 (amino acids 201 to 215), 45 (amino acids 221 to 235), 49 (amino acids 241 to 255), and 53 (amino acids 261 to 275). Immunized animals recognized an average of eight peptides (range, 0 to 28).
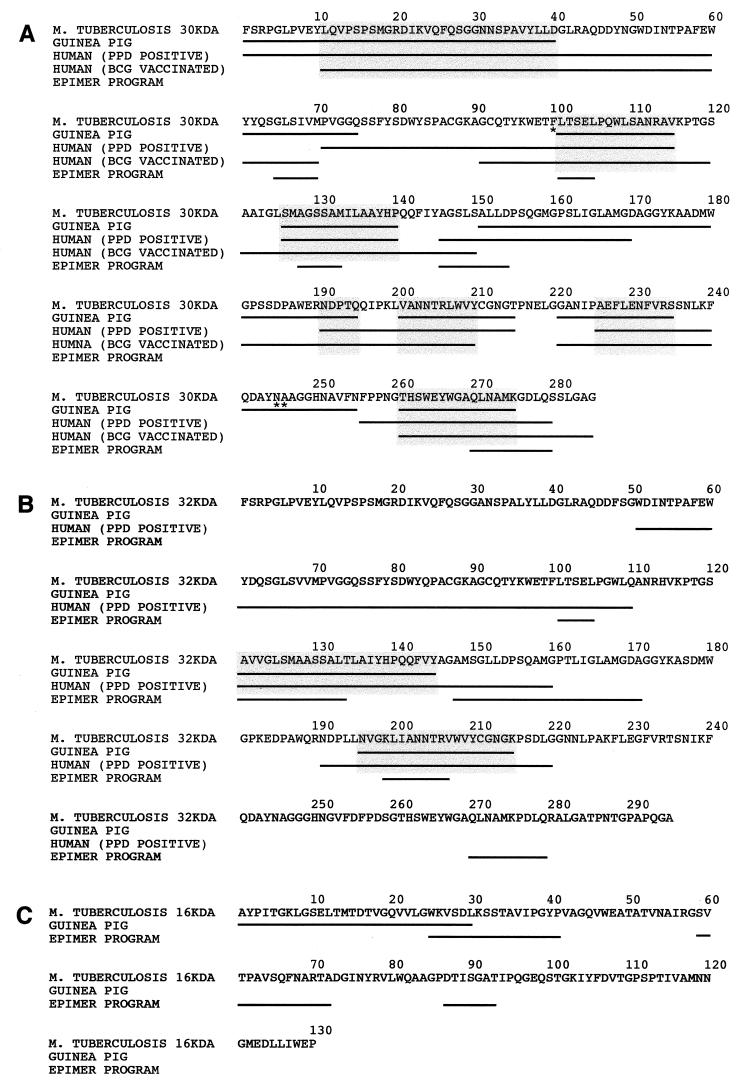
The M. tuberculosis 30-kDa protein is identical to the M. bovis BCG strain Tokyo homolog except for three amino acid residues at positions 100, 245, and 246 (i.e., Phe, Asn, Ala and Leu, Lys, Pro for M. tuberculosis and M. bovis BCG Tokyo, respectively) (5). Immunoreactive epitopes of the M. tuberculosis 30-kDa protein identified in this study on the basis of mapping in the guinea pig frequently overlapped with immunoreactive epitopes of the M. bovis BCG 30-kDa protein identified in two previous studies on the basis of mapping in healthy PPD-positive and BCG-vaccinated individuals (17, 18) (Fig. 2A). Regions on the 30-kDa mature protein recognized by at least 25% of the immunized guinea pigs and healthy PPD-positive or BCG-vaccinated individuals were amino acids 11 to 40, 101 to 115, 126 to 140, 191 to 195, 201 to 210, 226 to 235, and 261 to 275 (Fig. 2A). Moreover, three of these seven immunoreactive regions of guinea pigs and humans (amino acids 101 to 115, 126 to 140, and 261 to 275) overlap with T-cell epitopes of the 30-kDa protein predicted by a newly developed computer program (EpiMer) that predicts putative T-cell epitopes by searching the HLA-binding motifs on a given protein sequence (12) (Fig. 2A). Amino acids 241 to 255 were identified as an immunoreactive region in guinea pigs but not in healthy PPD-positive or BCG-vaccinated individuals. It is possible that this discrepancy between the studies is due to the two consecutive amino acid differences at positions 245 and 246 in the 30-kDa protein of M. tuberculosis, mapped in guinea pigs, and the 30-kDa protein of M. bovis BCG Tokyo, mapped in humans.
FIG. 2.
Alignment of T-cell epitopes of the 30-kDa (A), 32-kDa (B), and 16-kDa (C) proteins mapped in guinea pigs and in humans. T-cell epitopes that were recognized by at least 25% of the immunized guinea pigs or healthy PPD-positive or BCG-vaccinated individuals are indicated by underlining. Common epitopes of guinea pigs and humans are highlighted with shaded boxes. Epitopes of the 30-, 32-, and 16-kDa protein sequences of M. tuberculosis predicted by the EpiMer computer-based program that are at least five amino acids long are underlined. Epitope mapping in guinea pigs was performed with overlapping peptides derived from the sequence of the 30-, 32-, and 16-kDa proteins of M. tuberculosis shown in the figure. Synthetic peptides used to map the T-cell epitopes of the 30-kDa protein in humans were based on the sequence from M. bovis BCG Tokyo (17, 18). An asterisk indicates a difference in amino acid residue between the 30-kDa protein sequence from M. tuberculosis and that from M. bovis BCG Tokyo (i.e., F100, N245, A246 and L100, K245, P246 for M. tuberculosis and M. bovis BCG Tokyo, respectively). Synthetic peptides used to map the T-cell epitopes of the 32-kDa protein in humans were based on the sequence from M. tuberculosis (9).
Peptide mapping of the 32-kDa protein.
The mature 32-kDa protein, consisting of 295 amino acids (4, 6), is closely related to the 30-kDa protein in both primary sequence and physiological function. It is also a major target of host immune response to mycobacteria. Among 24 guinea pigs immunized with the 32-kDa protein, splenic lymphocytes from 18 (75%) animals had a positive proliferative response to the purified 32-kDa protein; splenic lymphocytes from 12 (50%) animals proliferated in response to PPD. The mean SIs were 2.1 ± 1.2 to the 32-kDa protein and 1.5 ± 0.7 to PPD. Immunized animals recognized an average of four peptides (range, 0 to 14). The most immunostimulatory peptides were peptide numbers 25 and 27 (amino acids 121 to 145) and 40 to 41 (amino acids 196 to 215). These two epitopes, recognized by 30 to 40% of the immunized guinea pigs, overlap with regions (i.e., amino acids 121 to 160 and 191 to 220) preferentially recognized by the peripheral blood mononuclear cells of PPD-positive individuals (9) (Fig. 2B). These two epitopes also overlap with the T-cell epitope predictions of the EpiMer program. In contrast, the immunoreactivity of amino acids 51 to 110 and 146 to 160 are human specific since these regions were preferentially recognized by PPD-positive individuals and predicted by the EpiMer program but were apparently not immunoreactive in outbred guinea pigs. Although the 30- and 32-kDa proteins of M. tuberculosis are highly homologous, the majority of 15-mer peptides that correspond between these two proteins are not identical. Corresponding peptides of these two proteins have from 1 to 6 different residues and these are frequently nonconservative changes. The only peptides of the 30- and 32-kDa proteins of M. tuberculosis that are identical are peptides 1 to 4, 18 to 19, and 53. However, peptides 1 to 3 and 53 were recognized by 25% of the animals immunized with the 30-kDa protein but not by animals immunized with the 32-kDa protein. This suggests that the processing and presentation of these peptides is influenced by the context in which they reside in the protein.
Peptide mapping of the 16-kDa protein.
The gene for the 16-kDa protein of M. tuberculosis has recently been cloned and sequenced (6, 11). The mature protein contains 130 amino acids. Of 23 guinea pigs immunized with the 16-kDa protein, 14 (61%) recognized the purified protein versus 42% of the controls, and 10 (43%) recognized PPD versus 4% of the controls. The mean SIs for immunized animals were 1.7 ± 0.6 to the 16-kDa protein and 1.4 ± 0.4 to PPD. Immunized animals recognized an average of one peptide (range, 0 to 7). The most immunostimulatory region of the protein was its N terminus; peptide numbers 1 (amino acids 1 to 15) and 5 (amino acids 15 to 30), in particular, were recognized by 20% of the immunized animals. Three putative T-cell epitopes were predicted by the EpiMer program (Fig. 2C). Except for six amino acids (amino acids 25 to 30), these epitopes do not overlap with the immunoreactive region mapped in guinea pigs.
Assays of lymphocyte proliferation to whole proteins discriminated very well between immunized and control animals in the case of the 30-kDa protein (92 versus 4%) but less well in the cases of the 32- and 16-kDa proteins (75 versus 54% for the 32-kDa protein and 61 versus 42% for the 16-kDa protein). When the results of all of the studies are combined, it can be seen that ca. 33% of the control animals responded to purified proteins and 18% responded to PPD. Responses to purified proteins and PPD were highly correlated, i.e., animals that recognized PPD generally also recognized the purified proteins. Presumably, these responses of the control animals reflect cross-reactive immunity from exposure to environmental antigens, possibly including those from nonpathogenic mycobacteria.
Interestingly, in separate experiments in which both immunized and control animals were tested for cutaneous delayed-type hypersensitivity to the proteins (unpublished data), the skin tests to the 32- and 16-kDa proteins discriminated between immunized and control animals better than the lymphocyte proliferation assay did in the present study. In the separate study, 72, 78, and 72% of the animals had a positive response to the 30-, 32-, and 16-kDa proteins, respectively, versus 0% of the controls. Paradoxically, although immunized animals consistently recognized the whole proteins more frequently than did the sham-immunized controls, they did not consistently recognize individual epitopes better. In the case of the 30-kDa protein, immunized animals recognized virtually all immunoreactive epitopes more frequently than did the controls. However, in the case of the 32- and 16-kDa proteins, immunized animals recognized most immunoreactive epitopes less frequently than did the controls.
In this study, we have identified a total of 12 T-cell epitopes on the three most abundant extracellular proteins of M. tuberculosis in outbred guinea pigs. The T-cell epitopes of the 30- and 32-kDa proteins mapped in immunized guinea pigs were frequently identified in healthy PPD-positive or BCG-vaccinated individuals. (The 16-kDa protein has not been mapped in humans.) This suggests that outbred guinea pigs may be a useful model for identifying broadly immunodominant epitopes of microbial proteins in humans and that the broadly immunoreactive epitopes of the three major extracellular proteins of M. tuberculosis identified in this study may have utility in an epitope-based vaccine against tuberculosis.
Acknowledgments
We thank Barbara J. Dillon, Günter Harth, and Chai Chaloyphian for assistance with animal experimentation. We thank Joseph Reeve, Jr., of the UCLA Peptide Sequencing Facility for advice and for assistance with peptide synthesis and purification. We also thank Anne S. DeGroot of the TB/HIV Research Lab, Brown University, for providing the EpiMer program predictions of T-cell epitopes of the 30-, 32-, and 16-kDa proteins of M. tuberculosis.
Bai-Yu Lee was supported by National Institutes of Health training grant AI-07126. This work was supported by NIH grant AI31338.
REFERENCES
- 1.Andersen P. Effective vaccination of mice against Mycobacterium tuberculosis infection with a soluble mixture of secreted mycobacterial proteins. Infect Immun. 1994;62:2536–2544. doi: 10.1128/iai.62.6.2536-2544.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 3.Berzofsky J A. Immunodominance in T lymphocyte recognition. Immunol Lett. 1988;18:83–92. doi: 10.1016/0165-2478(88)90046-6. [DOI] [PubMed] [Google Scholar]
- 4.Borremans M, De Wit L, Volckaert G, Doms T, De Bruyn J, Huygen K, Van Vodren J-P, Stelandre M, Verhofstadt R, Content J. Cloning, sequence determination, and expression of a 32-kilodalton-protein gene of Mycobacterium tuberculosis. Infect Immun. 1989;57:3123–3130. doi: 10.1128/iai.57.10.3123-3130.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Wit L, Palou M, Content J. Nucleotide sequence of the 85B-protein gene of Mycobacterium bovis BCG and Mycobacterium tuberculosis. DNA Sequence. 1994;4:267–270. [PubMed] [Google Scholar]
- 6.Harth G, Lee B-Y, Horwitz M A. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major Mycobacterium tuberculosis extracellular proteins considered to be leading vaccine candidates and drug targets. Infect Immun. 1997;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harth G, Lee B-Y, Wang J, Clemens D L, Horwitz M A. Novel insights into the genetics, biochemistry, and immunocytochemistry of the 30-kilodalton major extracellular protein of Mycobacterium tuberculosis. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz M A, Lee B-W E, Dillon B J, Harth G. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1995;92:1530–1534. doi: 10.1073/pnas.92.5.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Launois P, Deleys R, Niang M N, Drowart A, Andrien M, Dierckx P, Cartel J L, Sarthou J L, Van Vooren J P, Huygen K. T-cell-epitope mapping of the major secreted mycobacterial antigen Ag85A in tuberculosis and leprosy. Infect Immun. 1994;62:3679–3687. doi: 10.1128/iai.62.9.3679-3687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B-Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Invest. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manca C, Lyashchenko K, Wiker H G, Usai D, Colangeli R, Gennaro M L. Molecular cloning, purification, and serological characterization of MPT63, a novel antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1997;65:16–23. doi: 10.1128/iai.65.1.16-23.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister G E, Roberts C G P, Berzofsky J A, DeGroot A S. Two novel T cell epitope prediction algorithms based on MHC-binding motifs: comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13:581–591. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- 13.Moudgil K D, Sekiguchi D, Kim S Y, Sercarz E E. Immunodominance is independent of structural constraints: Each region within hen eggwhite lysozyme is potentially available upon processing of native antigen. J Immunol. 1997;159:2574–2579. [PubMed] [Google Scholar]
- 14.Ou D, Chong P, Gillam S. Immunogenicity study of a synthetic T-cell epitope of rubella virus capsid protein recognized by human T cells in different strains of mice. Viral Immunol. 1994;7:41–45. doi: 10.1089/vim.1994.7.41. [DOI] [PubMed] [Google Scholar]
- 15.Pal P G, Horwitz M A. Immunization with extracellular proteins of Mycobacterium tuberculosis induces cell-mediated immune responses and substantial protective immunity in a guinea pig model of pulmonary tuberculosis. Infect Immun. 1992;60:4781–4792. doi: 10.1128/iai.60.11.4781-4792.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts A D, Sonnenberg M G, Ordway D J, Furney S K, Brennan P J, Belisle J T, Orme I M. Characteristics of protective immunity engendered by vaccination of mice with purified culture filtrate protein antigens of Mycobacterium tuberculosis. Immunology. 1995;85:502–508. [PMC free article] [PubMed] [Google Scholar]
- 17.Roche P W, Peake P W, Billman-Jacobe H, Doran T, Britton W J. T-cell determinants and antibody binding sites on the major mycobacterial secretory protein MPB59 of Mycobacterium bovis. Infect Immun. 1994;62:5319–5326. doi: 10.1128/iai.62.12.5319-5326.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silver R F, Wallis R S, Ellner J J. Mapping of T cell epitopes of the 30-kDa alpha antigen of Mycobacterium bovis strain bacillus Calmette-Guerin in purified protein derivative (PPD)-positive individuals. J Immunol. 1995;154:4665–4674. [PubMed] [Google Scholar]
- 19.Wiker H G, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992;56:648–661. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanetti M, Sercarz E, Salk J. The immunology of new generation vaccines. Immunol Today. 1987;8:18–25. doi: 10.1016/0167-5699(87)90827-9. [DOI] [PubMed] [Google Scholar]