Abstract
The Onto-Tools suite is composed of an annotation database and six seamlessly integrated, web-accessible data mining tools: Onto-Express, Onto-Compare, Onto-Design, Onto-Translate, Onto-Miner and Pathway-Express. The Onto-Tools database has been expanded to include various types of data from 12 new databases. Our database now integrates different types of genomic data from 19 sequence, gene, protein and annotation databases. Additionally, our database is also expanded to include complete Gene Ontology (GO) annotations. Using the enhanced database and GO annotations, Onto-Express now allows functional profiling for 24 organisms and supports 17 different types of input IDs. Onto-Translate is also enhanced to fully utilize the capabilities of the new Onto-Tools database with an ultimate goal of providing the users with a non-redundant and complete mapping from any type of identification system to any other type. Currently, Onto-Translate allows arbitrary mappings between 29 types of IDs. Pathway-Express is a new tool that helps the users find the most interesting pathways for their input list of genes. Onto-Tools are freely available at http://vortex.cs.wayne.edu/Projects.html.
INTRODUCTION
Molecular biology and genetics are currently at the center of an information revolution. High-throughput techniques generate very large amounts of heterogeneous data. There is a large gap between our ability to collect data and our ability to interpret it. The challenge faced by today's researchers is to develop effective ways to analyze the vast amount of data that has been and will continue to be collected. First released in 2001, Onto-Tools is an open access software suite that partially addresses this problem (1–5). This is achieved by using a probabilistic functional analysis that bridges the gap between low-level, high-throughput gene expression data and high-level functional knowledge. This analysis approach has become de facto standard in the second-stage analysis of microarray experiments (6–18). The Onto-Tools suite includes: (i) Onto-Express, which can be used to translate lists of differentially regulated genes into a better understanding of the underlying biological phenomena through the use of Gene Ontology (GO); (ii) Onto-Design, which can be used to select the best set of genes to be included on a custom microarray designed for the study of a given biological phenomenon; (iii) Onto-Compare, which can be used to analyze the functional bias of various focused commercial microarrays and select the one that is most appropriate for a given biological hypothesis; (iv) Onto-Translate, which can be used to translate lists of genes from one reference system to another (e.g. from GenBank accession numbers to UniGene cluster IDs to Affymetrix probe IDs, etc.); (v) Onto-Miner, which provides a unified access point and an application programming interface allowing queries for various information, such as the gene name, official symbol, reference accession number, coded protein, etc.; and (vi) Pathway-Express, which helps the users find most interesting pathway(s) involving their genes of interest. Previous publications have described in detail the motivation, implementation and validation of these tools (1–5). This paper describes a new tool added to the ensemble and discusses various other additions and enhancements made to the existing tools and the database.
ENHANCEMENTS AND ADDITIONS
The back-end annotation database
In February 2005, NCBI phased out its LocusLink database and replaced it with the Entrez Gene database (19). All data previously stored in LocusLink (20) have been migrated to the new Gene database. However, the structure of the new database and the format of the files used to export this data have changed. Previously, the Onto-Tools database used the LocusLink gene as its backbone data structure to link the functional annotations imported from GO (21,22) with the sequence data imported from dbEST and UniGene. Hence, the LocusLink phase-out has had a dramatic impact on the schema of the Onto-Tools back-end annotation database, as well as on the tools themselves. The Onto-Tools back-end database has been re-designed to use a new data structure based on the Gene data model. In addition, the database download and parser modules are also modified to download and parse the Entrez Gene database.
Over the past year, Onto-Tools (OT) database has been expanded dramatically. Now, the database contains various types of data from 13 new databases which include: Swiss-Prot (23), TrEMBL (24), PIR (25), UniProt (26), Eukaryotic Promoter Database (EPD) (27), Human Genome Nomenclature Committee (HGNC) (28), GenPept, Online Mendelian Inheritance in Man (OMIM) (28), Protein Data Bank (PDB) (29), iProClass (24), HomoloGene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?DB=homologene), RefSeq (20) and GO (21,22). The Onto-Tools database now integrates information from 21 databases. The other databases previously integrated in the Onto-Tools database include dbEST (30), GenBank (31), UniGene (32), KEGG (33), WormBase (http://www.wormbase.org), NetAffx, dbEST library (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html) and eVOC (34).
Onto-Express
Over the past year, we have added support for the functional profiling of 19 new organisms by integrating annotations from GO database. Onto-Express (OE) now supports functional profiling for a total of 24 organisms, including Homo sapiens, Saccharomyces cerevisiae, Drosophila melanogaster, Mus musculus, Arabidopsis thaliana, Caenorhabditis elegans, Rattus norvegicus, Oryza sativa, Danio rerio, Dictyostelium discoideum, Candida albicans, Bacillus anthracis Ames, Coxiella burnetii RSA 493, Geobacter sulfurreducens PCA, Listeria monocytogenes 4b F2365, Methylococcus capsulatus Bath, Pseudomonas syringae DC3000, Shewanella oneidensis MR-1, Vibrio cholerae, Leishmania major, Plasmodium falciparum, Schizosaccharomyces pombe, Trypanosoma brucei and Glossina morsitans.
In addition to broadening its scope by adding new organisms, OE is now able to support 11 more types of input data. Previously, OE allowed the users to submit a list of GenBank accession numbers, UniGene cluster IDs, Affymetrix probe IDs, LocusLink IDs, WormBase accession IDs and gene symbols. Now OE allows the users to submit any of the database IDs used in the GO annotations. These ID types include Saccharomyces Genome Database (SGD) IDs, FlyBase IDs, Mouse Genome Informatics (MGI) IDs, The Arabidopsis Information Resource (TAIR) IDs, The Institute for Genomic Research (TIGR) IDs, Rat Genome Database (RGD) IDs, Gramene IDs, Zebrafish Information Network (ZFIN) IDs, DictyBase IDs, Candida Genome Database (CGD) IDs and Sanger GeneDB IDs.
Onto-Translate
In order to correctly interpret the results of any experiment, the researchers need to build a complete picture of the biological phenomenon under study, to the extent possible. Most often, one needs to take into account various types of data: DNA sequence, mRNA sequence and expression, protein expression and structure, gene interactions, protein–protein interactions, etc. However, various databases are rather specialized (e.g. GenBank focuses on sequence data, PDB focuses on protein structure data, Swiss-Prot focuses on protein annotations, etc.) and no public or private resource contains all available data. In order to allow the users to navigate from one resource to another, as often required, all public databases cross-reference to some of the other databases. For example, an mRNA or expressed sequence tag (EST) database (e.g. UniGene) will provide links to a gene database (e.g. Entrez Gene), indicating the gene from which the mRNA is transcribed and to a protein database (e.g. GenPept) that will provide information about the protein which is translated from the mRNA. Such a gene database will also be cross-linked to a genome map database (e.g. NCBI genome assembly) to help with the location of the gene on its genome. Similarly, a protein database (e.g. Swiss-Prot) will provide links to a gene database (e.g. Entrez Gene) and a protein structure database (e.g. Pfam or InterPro). More recently, this cross-linking has started to be extended to include clinical aspects. For example, disease databases, such as OMIM, can provide a link to a gene database (e.g. Entrez Gene) where one can obtain functional annotations for those genes involved in specific diseases. In a parallel movement, organism-specific databases, such as MGI, RGD, etc. are geared toward providing a one-stop information resource for a specific organism. These organism-specific databases also provide links to other large-scale, organism-independent, generic databases, such as Entrez Gene, Swiss-Prot, etc.
The major problem in the field is that each of these databases has its own schema, which is designed independently of any other. In consequence, each of these resources will use its own main identifier (i.e. will have independent name-spaces). For example, GenBank uses accession numbers, UniGene uses cluster IDs, Entrez Gene uses gene symbols and gene IDs, Swiss-Prot uses Swiss-Prot accession IDs, TrEMBL uses TrEMBL IDs, etc. The net result is that each database refers to the same information differently. For example, the gene beta actin in mouse is referred to as MGI:87904 in MGI, Actb (Gene ID: 11461) in Entrez Gene, Mm.297 in UniGene, ACTB_MOUSE (primary accession number: P60710) in UniProt and TC1242885 in TIGR gene index. In addition, the beta actin gene in mouse is referred to by 29 mRNA sequences and 4552 ESTs in dbEST, 5 secondary accession numbers in UniProt, 4 other accession IDs in MGI and 5 probe IDs on 4 different Affymetrix mouse arrays. The burden of mapping various types of ID on each other is left entirely on the shoulders of the researchers, who often have to revert to cutting and pasting lists of IDs from one database to another.
As a side effect of the efforts to link genomics, proteomics and annotation databases, some information is replicated in multiple databases. An immediate problem created by this duplication is data coherency and consistency. The same record stored in different databases will be updated at different times, which means that different versions will co-exists for a while. For example, one can obtain functional annotations for a gene from either the GO database or Entrez Gene database. Note that the annotations in Entrez Gene and GO database are obtained from the same source, the GO Annotation project at EBI. An illustrative example is the gene erythrocyte membrane protein band 4.1-like 3 [H.sapiens] (Entrez Gene ID:23136, EPB41L3). A search for this gene in the GO annotation database returned only one GO term, plasma membrane. However, the same gene, EPB41L3, is annotated with 6 GO terms: actin binding, structural molecule activity, cortical actin cytoskeleton organization and biogenesis, cytoplasm, cytoskeleton and plasma membrane in the Entrez Gene database.
Onto-Translate (OT) is designed to address these name-space issues and help the user with the problem of mapping various types of IDs on each other. The ultimate goal of OT is to provide the users with a non-redundant and complete mapping from any type of identification system to any other type. Currently, OT allows such arbitrary mappings among 29 types of IDs. OT uses the custom design of Onto-Tools database that integrates 19 sequence, gene, protein and annotation databases.
Pathway-Express
The automated functional profiling approach of OE helps the researchers to better understand the biological phenomenon under study by pointing out statistically significant cellular functions. However, graphical representations of gene interactions (pathways) have been shown to be very useful (33,35,36). As more data become available, the question ‘is there a known pathway containing my gene(s) of interest?’ will gradually transform into ‘how do I find the most interesting pathway(s) involving my gene(s)?’
Pathway-Express (PE) is a new tool in the Onto-Tools ensemble that was designed to answer such questions. Our goal is to provide a system that will automatically find such interesting pathways. When the user submits a list of genes, the system performs a search and builds a list of all associated pathways. This is illustrated in Figure 1. After generating a list of pathways for the input list of genes from the Onto-Tools database, PE first calculates a perturbation factor PF(g) for each input gene. This perturbation factor takes into account the (i) normalized fold change of the gene and (ii) the number and amount of perturbation of genes downstream from it. This gene perturbation factor reflects the relative importance of each differentially regulated gene. The impact factor of the entire pathway includes a probabilistic term that takes into consideration the proportion of differentially regulated genes on the pathway and gene perturbation factors of all genes in the pathway. The impact factors of all pathways are used to rank the pathways before presenting them to the user. Note that all pathways affected are presented regardless of their impact factors. In Figure 1, for example, the pathway selected as the most important had only three differentially regulated genes out of 209 genes. However, this pathway is dramatically impacted if the two transmembrane receptors and the ligand shown are affected.
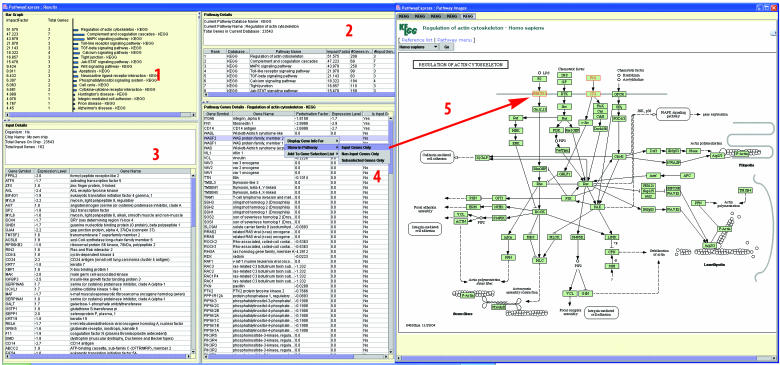
Figure 1.
Pathway-Express (PE) performs a pathway level impact analysis and orders the affected pathways in the decreasing order of their expected importance for the given condition (panel 1). PE displays details about each pathway including: the source database, the total number of genes, etc. (panel 2). Panel 3 shows the gene symbols, expression values (or fold changes), and gene names for the differentially regulated genes. Panel 4 contains details about each pathway, including all genes in the pathway with their perturbation factors and expression levels. The users can also visualize the input genes in a pathway diagram. The input genes are highlighted in red color on the pathway diagram (panel 5).
CONCLUSION
The Onto-Tools suite is composed of an annotation database and six seamlessly integrated, web-accessible, free data mining tools: Onto-Express, Onto-Compare, Onto-Design, Onto-Translate, Onto-Miner and Pathway-Express. Pathway-Express is a new tool that allows to find out most interesting pathways for the input list of genes in addition to visualizing the interactions among the genes in the pathways. Over the past year, our database has been dramatically expanded to include various types of data from 12 new databases, including Swiss-Prot, TrEMBL, PIR-iProClass, EPD, HGNC, GenPept, OMIM, PDB, HomoloGene, RefSeq and Entrez Gene. The Onto-Tools database now integrates various types of data from 19 genomics databases. In addition, our database is also enhanced with the integration of functional annotations of new organisms. Currently, OE supports functional profiling for 24 organisms and 17 types of input IDs. Onto-Translate was enhanced to provide a non-redundant and complete mapping from one type of identification system to any other type. Using the custom design of the Onto-Tools database, Onto-Translate currently allows arbitrary mappings among 29 types of IDs. Onto-Tools are freely available at http://vortex.cs.wayne.edu/Projects.html.
Acknowledgments
This work has been supported by the following grants: NSF DBI-0234806, DOD DAMD 17-03-02-0035, NIH(NCRR) 1S10 RR017857-01, MLSC MEDC-538 and MEDC GR-352, NIH 1R21 CA10074001, 1R21 EB00990-01 and 1R01 NS045207-01. The Onto-Tools currently runs on equipment provided by Sun Microsystems under the grant EDU 7824-02344-US. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Draghici S., Khatri P., Martins R.P., Ostermeier G.C., Krawetz S.A. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 2.Draghici S., Khatri P., Shah A., Tainsky M. Assessing the functional bias of commercial microarrays using the Onto-Compare database. Biotechniques. 2003;(Suppl.):55–61. [PubMed] [Google Scholar]
- 3.Drăghici S., Khatri P., Bhavsar P., Shah A., Krawetz S.A., Tainsky M.A. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khatri P., Bhavsar P., Bawa G., Drăghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W556. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatri P., Drăghici S., Ostermeier G.C., Krawetz S.A. Profiling gene expression with Onto-Express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 6.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 7.Beissbarth T., Speed T.P. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 8.Berriz G.F., King O.D., Bryant B., Sander C., Roth F.P. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 9.Castillo-Davis C.I., Hartl D.L. GeneMerge-post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2002;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 10.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 11.Hosack D.A., Dennis G., Jr, Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:P4. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin D., Brun C., Remy E., Mouren P., Thieffry D., Jacq B. GOToolBox: functional analysis of gene datasets based on gene ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasquier C., Girardot F., Jevardat de Fombelle K., Christen R. THEA: ontology-driven analysis of microarray data. Bioinformatics. 2004;20:2636–2643. doi: 10.1093/bioinformatics/bth295. [DOI] [PubMed] [Google Scholar]
- 14.Shah N.H., Fedoroff N.V. CLENCH: a program for calculating Cluster ENriCHment using the Gene Ontology. Bioinformatics. 2004;20:1196–1197. doi: 10.1093/bioinformatics/bth056. [DOI] [PubMed] [Google Scholar]
- 15.Young A., Whitehouse N., Cho J., Shaw C. OntologytTraverser: an R package for GO analysis. Bioinformatics. 2005;21:275–276. doi: 10.1093/bioinformatics/bth495. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B., Schmoyer D., Kirov S., Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S., Tian L., Li C., Storch K.-F., Wong W.H. Comparative analysis of gene sets in the Gene Ontology space under the multiple hypothesis testing framework. IEEE Computational Systems Bioinformatics Conference; August 14–19; 2004. pp. 425–435. [DOI] [PubMed] [Google Scholar]
- 19.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-oriented information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C.H., Yeh L-S.L., Huang H., Arminski L., Castro-Alvear J., Chen Y., Hu Z., Kourtesis P., Ledley R.S., Suzek B.E., et al. The Protein Information Resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann S.F.B., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M.J., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Périer R.C., Praz V., Junier T., Bonnard C., Bucher P. The Eukaryotic Promoter Database (EPD) Nucleic Acids Res. 2000;28:302–303. doi: 10.1093/nar/28.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wain H.M., Bruford E.A., Lovering R.C., Lush M.J., Wright M.W., Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- 28.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boguski M.S., Lowe T.M.J., Tolstoshev C.M. dbEST—database for expressed sequence tags. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 31.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. Genbank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuler G.D. Pieces of puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 33.Kanehisa M., Goto S., Kawashima S., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien-Kruger S., Smedley D., Otgaar D., Greyling G., Jongeneel V., McCarthy M.I., et al. eVOC: a controlled vocabulary for gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 36.Doniger S.W., Salomonis N., Dahlquist K.D., Vranizan K., Lawlor S.C., Conklin B.R. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]