Abstract
Foldalign is a Sankoff-based algorithm for making structural alignments of RNA sequences. Here, we present a web server for making pairwise alignments between two RNA sequences, using the recently updated version of foldalign. The server can be used to scan two sequences for a common structural RNA motif of limited size, or the entire sequences can be aligned locally or globally. The web server offers a graphical interface, which makes it simple to make alignments and manually browse the results. The web server can be accessed at http://foldalign.kvl.dk.
INTRODUCTION
As transcriptional high-throughput sequence data are being generated, it is becoming clear that a large fraction of the data cannot be annotated by comparison with existing genes using conventional methods, such as BLAST (1). For example, a study of 10 human chromosomes shows that 15.4% of the nucleotides are transcribed, which is ∼10 times as many as expected from the annotation (2). Clearly, phenomena, such as junk transcription, are expected to account for some fraction of this transcription, but the same study also found that there are twice as many transcripts without a poly(A) tail as transcripts with a poly(A) tail in the cytosol. These results indicate that a significant portion of the existing transcription could be non-coding RNAs.
Searches for novel non-coding RNAs by comparative genomics are often highly dependent on a substantial amount of sequence similarity (3). Hence, genomic regions with low sequence similarity between related organisms remain to be systematically compared.
Foldalign makes alignments of sequences containing RNA secondary structures (4–6). The newly updated version uses a combination of a light weight energy model and sequence similarity to find common folds and alignments between two sequences (4). The method is based on the Sankoff algorithm (7). Other methods based on the work of Sankoff have also been introduced (8–10).
The foldalign software can make three different types of comparisons. Local, where a single local fold and alignment between the two input sequences is produced. Global, where the sequences are folded and aligned globally. Scan is used when the sequences have lengths that make the folding and aligning of the entire sequences prohibitive. The sequences can then be aligned by limiting the length of the resulting folds and alignments, i.e. a mutual scan for structural similarities between the two sequences can be carried out.
Here, we present a web server which provides a graphical output for the different types of comparisons. This graphical output enables the non-informatics user to navigate quickly to desired parts of the results. The web server (and foldalign) is especially suited for comparing sequences expected to be functionally related when the sequences are too diverged for similarity-based methods to work. The algorithm was previously tested on sequences with <40% identity (see Supplementary Material) (4). Supplementary Figure S2 shows novel performance results for global alignments, with similarity up to 70% identity. These results also show as expected that foldalign can be used when the sequences are >40% identical.
INPUT
Here, we present the options of the web server. The first choice is the Comparison type. The default value Scan compares the two sequences and reports a ranked list of the local folds and alignments. The length of each local motif is limited (see below). The other possible values are Local which reports just a single local fold and alignment, and Global which reports a single global fold and alignment.
All types of comparisons require two sequences in FASTA format. The maximum sequence length is 200 for global and local comparisons and 500 for scanning. For scanning, the maximum length of the motif searched for is limited to 200. An Email address can be provided for reporting when the results are ready. For scans, the score matrix found to be optimal for scanning in (4) is used. For local and global alignments, a novel score matrix optimized for global structure prediction is used (see Supplementary Material).
All types of comparisons use three parameters: Maximum length difference (delta—δ), Gap opening cost and Gap elongation cost. δ is the maximum difference between two subsequences being compared. It is a heuristic which limits the computational complexity (5). Obviously, for global alignments δ has to be longer than the length difference between the two sequences. This is not required for the other two types of comparisons, but setting δ to low will affect the quality of the alignment. The maximum value of δ is 15 for Scan and 25 for Local and Global. Which gap values to choose depend on the problem at hand. When scanning, the cost must be high enough to quench spurious alignments. Empirically, a gap opening cost of −50 has given good results. For Local and Global alignment the gap opening cost depends on the RNAs being aligned as observed by us and others (4,8). Testing a few values in the range −10 to −100 can be necessary. Supplementary Figure S1 shows the average performance as a function of gap opening penalty for four different types of RNA structures. The gap elongation cost can often be fixed at half the gap opening cost. An extra Comment/ID (id) field is provided for the user's convenience. This can be used to mark different submissions.
There are two additional parameters for Scan. Maximum motif length (lambda—λ) and Maximum number of structures. λ is the maximum length of an alignment. This parameter greatly affects the time needed to do the alignment. As mentioned, λ is limited to a maximum of 200 nt. The parameter Maximum number of structures controls the maximum number of hits to be realigned and backtracked to produce a structure. If only the structure of the best hit is of interest, then this value should be set to one. A maximum of 10 structures can be produced.
The time needed to do an alignment varies from seconds (short sequences and a small δ) to several hours (scan of 500 nt long sequences with λ = 200 and δ = 15). Examples of run times for different sets of parameters are available in the online documentation. When a job is submitted, its number in the server queue is reported.
OUTPUT
Upon completion of a job, the web server produces a web page where the results are displayed and can be downloaded. The main parts of the outputs from the Scan, Global and Local comparisons are shown in Figures 1 and 2.
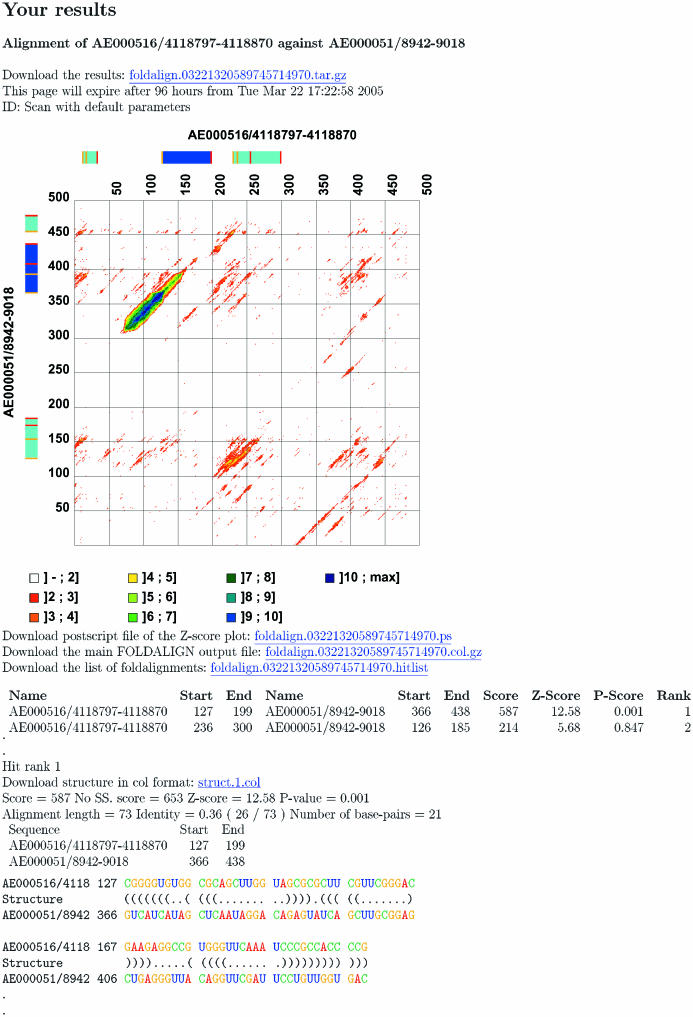
Figure 1.
An example of the output from a scan comparison. The sequences contain one tRNA each. The tRNA structures were taken from the tRNA database and the surrounding sequences from GenBank (14,15). Default parameters were used for the alignment. At the top of the output, there is a plot of the Z-scores. It is followed by a ranked list of non-overlapping local alignments. In the example the two best alignments have been included. The locations of the best hits are marked with bars on the sides of the Z-score plot. The bars of the best hit have a darker blue color than the rest. The final section shows the structures of the best hits.
Figure 2.
An example of the output from Local and Global comparisons. The two tRNA sequences were aligned using the Local comparison type with default parameters. The sequences were taken from the tRNA database (14).
The typical output from a scan alignment can be seen in Figure 1. There are three main sections. The figure at the top shows the Z-score for the best local alignment starting at each pair of positions along the two sequences. Correct alignments will often show up as big blotches. The plot is made using MatrixPlot (11). The bars at the top of the plot and on the left side indicate the location of the best alignments. The best alignment has a darker blue color than the others. To distinguish between alignments overlapping in one of the sequences, start and stop positions are colored yellow and red. A set of bars is drawn for each of the alignments for which a structure is produced and reported.
The second main section is a list of the best scoring non-overlapping alignments between the two sequences. A maximum of 100 hits is included in the list on the web page, but the file with the entire list is one of the files available for download. Hits can overlap in one of the sequences, but not in both. The format of each line is: the name of sequence one, its start position, its end position, the name of sequence two, its start position, its end position, the foldalign score, the Z-score, the P-value and the rank. Start and end are the start and end positions of the alignment. The P-value is calculated using the island method, (12,13), using the scores of the non-overlapping hits as the scores used for estimating the extreme value parameters. The distribution parameters can be found at the bottom of the page (not shown in the figure). The P-value estimate is very crude since the distribution is estimated from very few alignment scores, and any non-random alignments will bias the estimate. The rank is simply the hit's position in the list. The final main section is the predicted structures of the best hits. The structures are in parentheses notation. The NS score is the foldalign score without the contribution from single strand sequence similarity. This score can be used to separate alignments that have a high score due to conserved structure from alignments that have a high score due to sequence conservation.
The output from both local and global alignment shows the alignment score, the score without the contribution from the single strand substitution costs, the positions, the local identity of the sequences, the number of base pairs in the predicted structure, the sequences and the common structure (Figure 2).
DISCUSSION
Foldalign performs structural alignment of two RNA sequences or local structural alignment between structural similar regions in two sequences. The algorithm uses a combination of a light weight energy model and sequence similarity (4).
A foldalign web server is now available, which predicts alignments and structures for pairs of sequences. The minimum input to the server is two sequences in FASTA format. It can make three types of comparisons: Scan makes a local alignment and reports a ranked list of the best local alignments. The input sequences can be long, but the length of the motif searched for is limited. The Local comparison type makes a local alignment where the motif can be as long as the input sequence. The Global comparison type folds and aligns the sequences from end-to-end.
Even though the sequence length, λ, and δ are limited on the web server, arbitrarily long sequences can in principle be scanned by using the foldalign software itself. λ and δ are then limited by the amount of memory available on the local machine.
The foldalign software is also available for download at http://foldalign.kvl.dk.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank Paul Gardner for turning our attention to global alignments, and Gary Stormo for useful discussions. This work was supported by the Danish Technical Research Council, the Ministry of Food, Agriculture and Fisheries and the Danish Center for Scientific Computing. Funding to pay the Open Access publication charges for this article was provided by Danish Technical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005 doi: 10.1126/science.1108625. doi:10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havgaard J.H., Lyngsø R., Stormo G.D., Gorodkin J. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40% Bioinformatics. 2005;21:1815–1824. doi: 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- 5.Gorodkin J., Heyer L.J., Stormo G.D. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorodkin J., Stricklin S.L., Stormo G.D. Discovering common stem-loop motifs in unaligned RNA sequences. Nucleic Acids Res. 2001;29:2135–2144. doi: 10.1093/nar/29.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- 8.Mathews D.H., Turner D.H. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- 9.Hofacker I.L., Bernhart S.H., Stadler P.F. Alignment of RNA base pairing probability matrices. Bioinformatics. 2004;20:2222–2227. doi: 10.1093/bioinformatics/bth229. [DOI] [PubMed] [Google Scholar]
- 10.Holmes I. A probabilistic model for the evolution of RNA structure. BMC Bioinformatics. 2004;5:166. doi: 10.1186/1471-2105-5-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorodkin J., Stærfeldt H.H., Lund O., Brunak S. MatrixPlot: visualizing sequence constraints. Bioinformatics. 1999;15:769–770. doi: 10.1093/bioinformatics/15.9.769. [DOI] [PubMed] [Google Scholar]
- 12.Olsen R., Bundschuh R., Hwa T. Rapid assessment of extremal statistics for gapped local alignment. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:211–222. [PubMed] [Google Scholar]
- 13.Altschul S.F., Bundschuh R., Olsen R., Hwa T. The estimation of statistical parameters for local alignment score distributions. Nucleic Acids Res. 2001;29:351–361. doi: 10.1093/nar/29.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.