Abstract
RPBS (Ressource Parisienne en Bioinformatique Structurale) is a resource dedicated primarily to structural bioinformatics. It is the result of a joint effort by several teams to set up an interface that offers original and powerful methods in the field. As an illustration, we focus here on three such methods uniquely available at RPBS: AUTOMAT for sequence databank scanning, YAKUSA for structure databank scanning and WLOOP for homology loop modelling. The RPBS server can be accessed at http://bioserv.rpbs.jussieu.fr/ and the specific services at http://bioserv.rpbs.jussieu.fr/SpecificServices.html.
INTRODUCTION
Recent years have seen the development of an increasing number of bioinformatics methods. Although reference servers such as the NCBI server (http://www.ncbi.nlm.nih.gov) or the EBI server (http://www.ebi.ac.uk/services/) provide access to the most established methods, numerous new approaches are being continuously developed by many research teams. Although servers such as eva (http://cubic.bioc.columbia.edu/eva/) or the CAFASP server (http://bioinfo.pl/cafasp/) help in the identification and performance comparison of well established applications, many new specific tools remain barely visible on the Internet.
RPBS (Ressource Parisienne en Bioinformatique Structurale) is the result of the joint effort of several teams and aims at making available, at a unique entry point, original services devoted to structural bioinformatics. The expertise ranges from sequence/structure analysis to protein modelling and drug design, although not all these topics are yet tackled on the RPBS server. The server consists of a web portal for many tools, with the ultimate purpose of addressing the many areas of structural bioinformatics in an integrated manner. At the present time, this section (P-server) is only partially functional. RPBS also offers an interface to original software (specific services) developed by our teams. These services cover topics from the sequence field to the structure field. Several tools are structure-oriented sequence tools, such as CysState for the prediction of cysteine oxidation state (1), COUDES for the prediction of turns (2), HCA for secondary structure prediction and alignment (3), JPBS for local structure prediction using a structural alphabet (4) and PredAcc for the prediction of solvent accessibility (5). Other RPBS tools deal directly with 3D structures, such as SA-Search for finding structural similarities based on a structural alphabet (6) and Scit for comparing side chain conformations (7). RPBS also maintains several collections of commercially available organic compounds for structure-based in silico screening experiments.
As an illustration of RPBS specific services, we present in this article three methods covering the fields of sequence and structure analysis: AUTOMAT, YAKUSA and WLOOP (8). AUTOMAT is devoted to scanning databanks for sequence similarity. Its conception is different from that of FASTA and BLAST, which leads to complementary results. AUTOMAT is the first program to automatically eliminate local redundant alignments. YAKUSA is devoted to the screening of structural databanks in order to produce local structural alignments based on a description of protein conformations using their α angles. WLOOP is a backbone loop builder for loops of 3–12 residue length.
AUTOMAT: A SEQUENCE DATABASE SCANNING PROGRAM
AUTOMAT (http://bioserv.rpbs.jussieu.fr/Automat/index.html) was developed in the early 1990s (9) to search for similarities between a query sequence (DNA or protein) and a databank. It is based on an automaton scanning the database. Its design makes it more systematic than BLAST. AUTOMAT and BLAST lead to complementary results in terms of analysing protein sequences (10). In brief, the query sequence is split into words of minimal length w chosen by the user, who can also define an alphabet of classes of equivalent amino acids. Hits of the words derived from the query (called triggers) against identical words in the target database sequence are collected. In order to generate alignments, the program gathers neighbouring triggers, if they are separated by a distance of fewer than k mismatching residues, with the same offset between the query and the target. Alignments of neighbouring triggers adding up to n or more matching residues are qualified for scoring by means of a substitution matrix. The score corresponds to the maximum cumulative cost of aligned symbols found within the generated alignment. The numbers w, n and k are the first class parameters for AUTOMAT, the latter two having negligible effect on computer time. These parameters allow the accommodation of various types of query sequences.
For trigger detection, AUTOMAT relies on a finite state automaton and triggers are stored in an offset-hashed chained-list structure. Once the target sequence has been scanned, triggers are clustered and stored in a heap structure (semi-sorted binary tree), the dimension of which is the maximum number of reported alignments per sequence. Matching sequences are themselves stored in a larger heap structure according to their maximum alignment score, with the dimension of that heap being the maximum number of reported sequences.
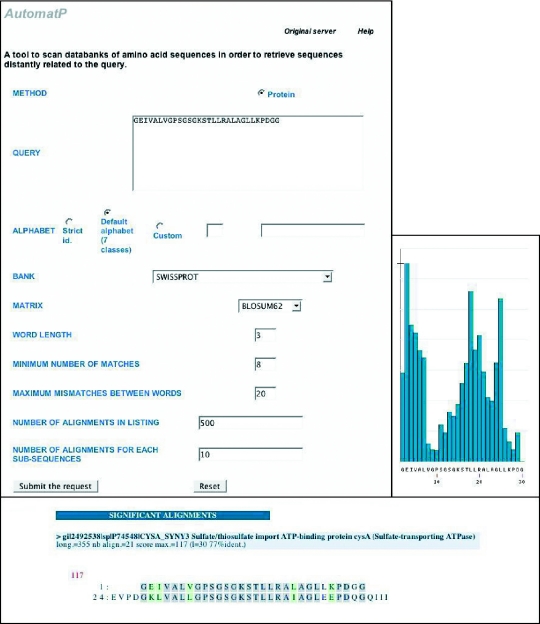
AUTOMAT has been the first software program to introduce histograms presenting the frequencies in the database of all the words of the query larger than w and to delete automatically redundant alignments originating from different sequences in the output listing (10,11). AUTOMAT and BLAST lead to coherent results for protein sequences in comparable computer time, but for nucleic acids AUTOMAT allows triggers w shorter (5 characters by default) than BLAST (words of 11 characters), thus leading to more sensitivity than BLAST. In the example given in Figure 1, a short peptide of 30 amino acids length is retrieved by both programs from SwissProt. With BLAST, two hits are recorded from CYSA_YERPS and CYSA_YERPE, ranked at positions 70 and 71 in the output listing, with 19 identities over 30 positions for both. These two complete sequences contain 363 residues and differ by only one single residue outside the region of alignment with the query. Owing to redundancy elimination by AUTOMAT, only CYSA_YERPS is listed in the output, at position 48, thus rendering human use of the listing easier. More detailed comparisons between AUTOMAT and BLAST have been presented elsewhere and have demonstrated the complementarity of the results produced by these two programs on proteins such as the RED (reductase-epimerase-dehydrogenase), HMP (haemoglobin-like protein of Escherichia coli) and HIV-1 Env protein families (10).
Figure 1.
Front page of RPBS for AutomatP, with an example of a histogram and one alignment for a short peptide 30 amino acids long.
Input
Two different services are available: AutomatP and AutomatN, for proteins or nucleic acids, respectively. Their interfaces are similar (see Figure 1), the parameters are:
A query sequence. Numerous input formats are accepted.
An alphabet, which can be provided by the user. By default a seven class alphabet (VILM, FYW, ACT, RQEK, HSD, GN and P) is used for proteins and four classes are used for nucleic acids.
The target databank. By default SwissProt is selected for proteins and human DNA sequences provided by Ensembl (12) for nucleic acids.
The substitution matrix, by default BLOSUM62 for proteins, and the identity matrix for nucleic acids. Other matrices can be selected by the user.
The parameter w, the minimal size of the triggers, can be chosen by the user. The default value is 3 for proteins and 5 for nucleic acids.
The parameter k, the maximal space between two consecutive triggers, can be chosen by the user. The default value is 20 for proteins and nucleic acids.
The parameter n, the minimal number of identical residues according to the alphabet, can be chosen by the user. The default value is 8 for proteins and 25 for nucleic acids.
The number of alignments in the listing. The default value is 500, but this can be changed at will.
A list of advanced parameters can be given, concerning the suppression of redundancies (default is on) and the visualization of local alignments for an entry, i.e. including alignments whose score is <50% of the maximum scored obtained for that entry (default is on). The histogram can be shown at will (default is off). In addition, the user can decide for the output listing to discard all the alignments with a score below a certain threshold (by default all are shown). An option (default is on) takes into account the identities for computing n in the regions separating consecutive triggers.
For a more thorough search, w can be taken as 2 with AutomatP or as ≤4 for AutomatN, but at the cost of a much longer processing time. The other two parameters, k and n, although important for the quality of sequence retrieval, are less critical to the computing time. Depending on the goal of the search, the user will keep the 20 class alphabet (all amino acids are distinguished) or use a degenerated alphabet of <20 classes. In our experience, the default alphabet with 7 classes is the best for AutomatP and the default identity matrix is the best for AutomatN.
Output
The output is a classical list of entries in the databank hyperlinked to the alignments between the query and these targets. The alignments are sorted according to their score computed through the homology matrix chosen in the input.
YAKUSA: FAST STRUCTURAL DATABANK SCANNING
YAKUSA (http://bioserv.rpbs.jussieu.fr/Yakusa/index.html) is designed for rapid real-time scanning of a structural databank with a user query protein structure. It finds the longest common substructures, called SHSPs (structural high-scoring pairs), between the query structure and every structure in the databank. On RPBS, the structural databanks available are the ASTRAL databanks (13) (at 45% or 90% identity), several PISCES databanks (14), non-redundant ‘clusters50’, ‘clusters70’ and ‘clusters90’ PDB databanks and the overall PDB databank (15).
YAKUSA describes protein structures as sequences of their α angles' internal coordinates. The α angles are dihedral angles between four consecutive Cα (16) and are discretized in order to be used as symbols. The query is split into overlapping patterns of α angle symbols with equal length (usually 5). These patterns and generated neighbour patterns are stored in a deterministic finite state automaton, as in the Aho–Corasick method (17). The neighbour patterns generated are simply the original query patterns with limited ‘errors’ within given local and global thresholds. The latter are structurally similar to the original patterns and the use of these neighbour patterns permits a more sensitive search, as structural similarities are not ‘exact’. The automaton scans every structure from the databank to detect seeds sharing structural similarities with the query. The retrieved seeds are extended into longer matching substructures (i.e. SHSPs). First, overlapping or neighbouring seeds are gathered, then the query and databank structures are aligned according to each seed in turn and the seed is extended on both sides to an aligned fragment with maximal similarity score. The similarity score of an aligned fragment is the sum of the similarity score of its corresponding residue pairs, which is related to the α angles of the two residues. For each query/databank pair a best path is computed between all SHSPs, and only the SHSPs belonging to this path are kept. The spatial compatibility between two SHSPs is established by measuring the distance between the two fragments forming one SHSP when the other two fragments of the other SHSP are superimposed. If this distance is short, the two SHSP are spatially compatible (i.e. they share the same relative configuration in the two structures). Groups of spatially compatible SHSPs can be established.
Statistics for occurrences of α angle runs (i.e. structural fragment) can be computed from all PDB structures. These statistics must take into account the high correlation between successive α angles in protein structures. As structures in the PDB are not numerous enough to estimate the parameters of high-order Markov chain models, YAKUSA uses mixture transition distribution models (18) that are pair approximations for such high-order Markov chains. As SHSPs are defined at the α angle level, a probability of occurrence can be assigned to each databank fragment of an SHSP, giving a measure of the commonness of this SHSP. The databank structures are ranked according to the score of their spatially compatible SHSPs with the query structure (a Z-score is also computed as YAKUSA gets one score for each query/databank pair).
With regard to sensitivity and selectivity of the structural matches, YAKUSA does as well as the best related programs such as DALI and CE, although it is by far faster (∼1 min), depending on the choice of the databank being used. We ran YAKUSA with the test cases of the experiment of Novotny et al. (19) and noticed that the three best performing programs for the overall test are YAKUSA, CE and DALI (20). Fisher et al. (21) selected 68 protein pairs that have similar folds but are not easy to detect. We selected the 14 most difficult pairs distributed among the four CATH main classes and submitted the 14 query structures to DALI, CE and YAKUSA. YAKUSA and CE showed the same overall performances (50% success), but the success distribution is different and DALI failed in more cases (29% success). In fact YAKUSA seems particularly suited for finding similarities in αβ and ‘few SSEs’ protein structures and, in a way, it is complementary to CE (20).
Input
The query can be uploaded as a PDB-formatted protein structure (it can be a new fold) or specified using the PDB code and chain identifier (in the case of a known fold).
Output
The output result for a run with a query structure is a listing of ranked structurally similar proteins in the databank; for each similar protein, its score, Z-score and a list of its SHSPs is given with their scores, probabilities, RMS and sequences. A simple visualization tool in Java (the Jmol applet from http://jmol.sourceforge.net/) allows the user to see the query/databank structures (superimposed according to their longest SHSP) and highlights all SHSPs between the two structures. Rasmol can also be used as a browser helper for this purpose as Rasmol scripts are generated by the YAKUSA server (Figure 2).
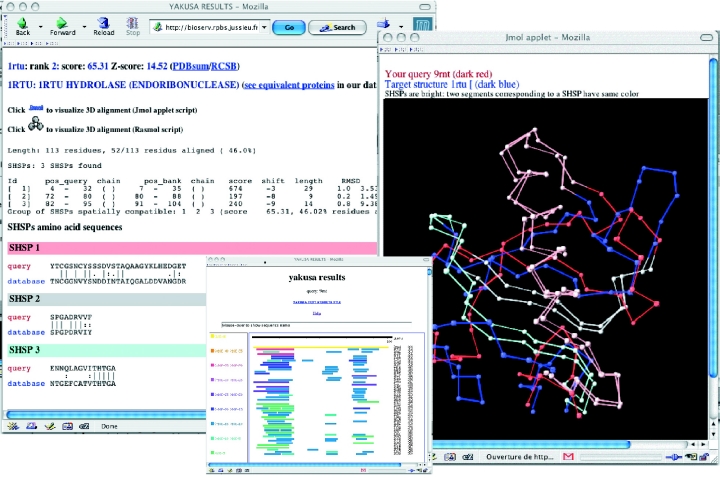
Figure 2.
YAKUSA result pages. Front window: visual listing of synthetic results of the scan of the databank with PDB structure 9rnt. For each entry (one line), SHSPs are coloured according to their probabilities of occurrence. Back window: part of a listing showing the SHSPs between the query (9rnt) and a databank entry (1rtu). Middle window: superimposition in the Jmol applet of the two structures (9rnt: dark red and 1rtu: dark blue) according to their longest SHSP (coloured in pink). The two others SHSPs are coloured in light grey and light blue.
WLOOP: A SERVER TO MODEL PROTEIN LOOP BACKBONES
Comparative modelling or homology modelling usually involves building the coordinates for structurally conserved regions (often the secondary structure elements) from selected experimental template(s). In the second step, the backbone of the connecting loops is built, and in the final step the side chains are located. At present, although several methods have been proposed to model the conformation of loops, there are, to our knowledge, only two other web servers offering such a service. CODA, from the group of Tom Blundell (22), combines ab initio prediction (from 3 to 5 amino acids) and a knowledge-based approach (from 3 to 16 residues) where loops are selected from an experimental databank, fitted and sorted using an energy function. The output produced is a set of PDB files containing only the fragments concerned, i.e. the loops themselves and their regular flanks (limited to 5 residues). ModLoop, from Sali lab (23), is based on the optimization by molecular dynamics with simulated annealing of an energy function. It accepts requests that can correspond to several loops, but can solve only up to 20 residues at one time. Thus, a complete modelling requires several requests.
WLOOP (http://bioserv.rpbs.jussieu.fr/WLoop/index.html) is a service to model loops of 3–12 residues. It relies on a bank of loops extracted from the PDB as fragments connecting two consecutive secondary structures of at least 4 residues. Loops of equivalent length are classified into families and sequence signatures specific to each class have been detected (24). The different steps of WLOOP (8) are as follows. First, the atomic coordinates of the backbone of the regular secondary structures surrounding the loop to be modelled are extracted, and the distance between the extremities of the fragments flanking the loop is calculated. This is compared with the average values of the distance between the first and last Cα of the loops of the different families. The loop family used for modelling is selected on the basis of the best agreement between these distances. Then, given the loop family, the sequence of the loop to model is considered in order to select the most appropriate template, using a substitution matrix taking into account the specific propensity of amino acids for loops. Finally, given the template, loop closure is performed using a simple procedure that scans three dihedral angles at the junction between the upstream flank and the loop. This is achieved with the X-Plor (25) molecular mechanics program, which checks stereochemistry and Ramachandran plot compatibility. As the side chains are not considered at this step, a crude energetic expression is considered (bonds, angles, dihedral and improper), and no solvation is considered. In the present version of the service, this elementary procedure is called iteratively for each loop to model.
To assess the performance of WLOOP, Table 1 present results for a test set of loops initially proposed by van Vlijmen and Karplus (26) and used by CODA (22) or ModLoop (23). Loops belonging to proteins included in the present WLOOP databank of loops have been removed. The choice of a criterion best adapted to quantifying the quality of the structural prediction of a fragment has been the subject of many discussions in the literature, e.g. (27). Here, we present both the local root mean square distance (RMSD), i.e. the RMSD between the model and the actual conformations taking into account only the atoms of the region to model, and the global RMSD, which includes the flanks in the comparison, and thus gives some assessment of the loop closure procedure. Looking at the local RMSDs, it is clear that WLOOP is able to propose candidates that are close to the native structure: the average deviation on the set of loops is 1.8 Å. This is somewhat less accurate than ModLoop (average accuracy of 0.87 Å), even if for 3grs_83–89 and 3dfr_20–23 WLOOP proposes better solutions. In terms of the flanks, WLOOP appears less accurate. These results suggest that the local conformation retrieval from the databank is reasonable, but that the loop closure should be improved. Work is in progress to address this point. It is important to note that the present comparison is not really fair as ModLoop and CODA involve time-consuming refinement procedures whereas WLOOP is a very fast approach with no refinement. The WLOOP server provides at present only a fast method to model loops (computing times being on the order of only a few minutes versus several hours) and users need to perform additional refinements (the choice is left to them) at a later stage. Yet WLOOP remains the only server that builds in one run all the loops of a homology model.
Table 1.
Comparison of predictions for a test loop used by several groups expressed as the root mean square distance (RMSD, Å) between the model and the actual structure.
| Loop pedigree | Length | ModLoop | CODA | WLOOP | ||
|---|---|---|---|---|---|---|
| Global RMSD | Local RMSD | Global RMSD | Global RMSD | Local RMSD | ||
| 8abp_203–208 | 6 | 0.37 | 0.24 | 0.8 | 2.7 | 1.53 |
| 2act_198–205 | 8 | 2.21 | 1.6 | 3.1 | 3.9 | 3.1 |
| 3grs_83–89 | 7 | 0.58 | 0.47 | 1.4 | 2.7 | 0.18 |
| 5cpa_231–237 | 7 | 1.23 | 1.06 | 0.2 | 3.3 | 2.39 |
| 8tln_E32–E38 | 7 | 2.26 | 1.82 | 1.9 | 3.11 | 2.29 |
| 8tln_E248–E255 | 8 | 0.98 | 0.84 | 1.8 | 3.57 | 2.47 |
| 3dfr_20–23 | 4 | 1.59 | 1.51 | 0.4 | 0.93 | 0.99 |
| 3dfr_89–93 | 5 | 1.14 | 0.85 | 0.6 | 1.65 | 2.41 |
| 3dfr_120–124 | 5 | 0.28 | 0.2 | 0.7 | 1.55 | 0.66 |
| 3blm_131–135 | 5 | 0.22 | 0.14 | 0.2 | 3.38 | 1.9 |
Input
The following data are required:
A PDB file from which the regions flanking the loops will be extracted. At the moment, only a file containing one single chain can be processed.
The sequence of the complete protein must be specified, using the one-letter code, and using lower and uppercase. Lowercase letters are used to specify rigid parts of the molecules containing the loop flanks. Regions specified using uppercase letters will be considered as regions to model. This convention was chosen to be consistent with programs such as SCWRL and Scit, which address side chain modelling. Loops <3 or >12 residues will be automatically discarded before further processing.
The secondary structure assignment must be provided, matching the amino acid sequence, with the following code: H for helices, E for strands; all other letters (such as T for turn) will be considered as coils. A utility for secondary structure assignment and sequence extraction from a PDB file is provided.
Output
A PDB file is returned, with the amino acids renumbered from 1. It contains the atomic coordinates of the backbone of the modelled loops, along with the coordinates of remaining parts of the structure. Logs related to the modelling of each loop are also returned.
CONCLUSIONS AND FUTURE DIRECTIONS
In this work, we have chosen to describe three original services available at RPBS that cover various aspects of structural bioinformatics: sequence similarity search, structural similarity search and loop modelling. RPBS offers many other programs dealing with structure, such as secondary structure assignment, solvent accessibility and side chain modelling. One outcome of the joint effort of the different groups is clearly to make available to the community original services that are grouped at one unique entry point and cover a large range of topics.
In addition to offering original services, work is under way to implement a collection of basic tools clearly missing, such as facilities for structure edition or comparison. An important concern is also the integration of the different services to evolve towards their chaining as a pipeline or a workflow. Hence, RPBS has contributed to the biomoby project by developing a Python API and has started to offer some options in the form of a web service. Finally, work is also under way to implement a generic way of organizing the interface of the different services to chain them on the web.
Acknowledgments
The development of RPBS was supported by a grant from ‘Action conjointe CNRS INSERM Bioinformatique 2002’. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mucchielli-Giorgi M.H., Hazout S., Tuffery P. Predicting the disulfide bonding state of cysteines using protein descriptors. Proteins. 2002;46:243–249. doi: 10.1002/prot.10047. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs P.F.J., Alix A.J.P. High accuracy prediction of beta-turns and their types using propensities and multiple alignments. Proteins. 2005;59:828–839. doi: 10.1002/prot.20461. [DOI] [PubMed] [Google Scholar]
- 3.Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Henrissat B., Mornon J.P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Brevern A.G., Etchebest C., Hazout S. Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Proteins. 2000;41:271–287. doi: 10.1002/1097-0134(20001115)41:3<271::aid-prot10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 5.Mucchielli-Giorgi M.H., Hazout S., Tuffery P. PredAcc: prediction of solvent accessibility. Bioinformatics. 1999;15:176–177. doi: 10.1093/bioinformatics/15.2.176. [DOI] [PubMed] [Google Scholar]
- 6.Guyon F., Camproux A.C., Hochez J., Tuffery P. SA-Search: a web tool for protein structure mining based on a Structural Alphabet. Nucleic Acids Res. 2004;32:W545–W548. doi: 10.1093/nar/gkh467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautier R., Camproux A.C., Tuffery P. SCit: web tools for protein side chain conformation analysis. Nucleic Acid Res. 2004;32:W508–W511. doi: 10.1093/nar/gkh388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojcik J., Mornon J.P., Chomilier J. New efficient statistical sequence-dependent structure prediction of short to medium-sized protein loops based on an exhaustive loop classification. J. Mol. Biol. 1999;289:1469–1490. doi: 10.1006/jmbi.1999.2826. [DOI] [PubMed] [Google Scholar]
- 9.Cantalloube H., Nahum C., Achour A., Lehner T., Callebaut I., Burny A., Bizzini B., Mornon J.P., Zagury D., Zagury J.F. Automat: a novel software system for the systematic search for protein (or DNA) similarities with a notable application to autoimmune diseases and AIDS. Comput. Appl. Biosci. 1994;10:153–161. doi: 10.1093/bioinformatics/10.2.153. [DOI] [PubMed] [Google Scholar]
- 10.Cantalloube H., Labesse G., Chomilier J., Nahum C., Cho Y.Y., Chams V., Achour A., Lachgar A., Mbika J.P., Issing W., Zagury J-F. Automat and BLAST: comparison of two protein sequence similarity search programs. Comput. Appl. Biosci. 1995;11:261–272. doi: 10.1093/bioinformatics/11.3.261. [DOI] [PubMed] [Google Scholar]
- 11.Cantalloube H., Chomilier J., Chiusa S., Lonquety M., Spadoni J.L., Zagury J.F. Filtering redundancies for sequence similarity search programs. J. Biomol. Struct. Dyn. 2005;22:487–492. doi: 10.1080/07391102.2005.10507020. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner S.E., Koehl P., Levitt M. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Dunbrack R.L., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 15.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- 17.Aho A.V., Corasick H.J. Efficient string matching: an aid to bibliographic search. Commun. ACM. 1975;18:333–340. [Google Scholar]
- 18.Raftery A.E. A model for high-order markov chains. J. R. Stat. Soc. Ser B. 1985;47:528–539. [Google Scholar]
- 19.Novotny M., Madsen D., Kleywegt G.J. Evaluation of protein fold comparison servers. Proteins. 2004;54:260–270. doi: 10.1002/prot.10553. [DOI] [PubMed] [Google Scholar]
- 20.Carpentier M., Brouillet S., Pothier J. Yakusa: a fast structural database scanning method. Proteins. 2005 doi: 10.1002/prot.20517. in press. [DOI] [PubMed] [Google Scholar]
- 21.Fischer D., Elofsson A., Rice D., Eisenberg D. Assessing the performance of fold recognition methods by means of a comprehensive benchmark. Pac. Symp. Biocomput. 1996:300–318. [PubMed] [Google Scholar]
- 22.Deane C.M., Blundell T.L. CODA: a combined algorithm for predicting the structurally variable regions of protein models. Protein Sci. 2001;10:599–612. doi: 10.1110/ps.37601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiser A., Do R.K., Sali A. Modeling of loops in protein structures. Protein Sci. 2003;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwasigroch J.M., Chomilier J., Mornon J.P. A global taxonomy of loops in globular proteins. J. Mol. Biol. 1996;259:855–872. doi: 10.1006/jmbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 25.Brünger A.T. Crystallographic refinement by simulated annealing: application to a 2.8 Å resolution structure of aspartate aminotransferase. J. Mol. Biol. 1988;203:803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- 26.Van Vlijmen H.W., Karplus M. PDB-based protein loop prediction: parameters for selection and methods for optimization. J. Mol. Biol. 1997;267:975–1001. doi: 10.1006/jmbi.1996.0857. [DOI] [PubMed] [Google Scholar]
- 27.Rohl C.A., Strauss C.E., Chivian D., Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55:656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]