Abstract
YASS is a DNA local alignment tool based on an efficient and sensitive filtering algorithm. It applies transition-constrained seeds to specify the most probable conserved motifs between homologous sequences, combined with a flexible hit criterion used to identify groups of seeds that are likely to exhibit significant alignments. A web interface (http://www.loria.fr/projects/YASS/) is available to upload input sequences in fasta format, query the program and visualize the results obtained in several forms (dot-plot, tabular output and others). A standalone version is available for download from the web page.
INTRODUCTION
Modern bioinformatics relies heavily on alignment programs and motif discovery tools, and numerous comparative genomics projects need ever more precise and faster tools for comparing two or several genomic sequences with different resolutions.
Except for small sequences, the exact local alignment algorithm of Smith and Waterman (1) is not frequently used, and most alignments are obtained using heuristic alignment tools such as FASTA (2), FLASH (3), BLAST (4,5), BLASTZ (6) and PatternHunter (7,8). All these methods introduce a trade-off between two competing parameters: selectivity (or specificity) directly affecting the speed of the algorithm and sensitivity affecting its precision (i.e. the number of relevant alignments missed). Achieving a good trade-off between sensitivity and selectivity is the key issue in local alignment tools. The recently introduced spaced seeds technique (7,8) allows an increase in sensitivity without loss in selectivity. This innovation triggered various studies (9–15) related to the usage, design and generalizations of spaced seeds.
In this note, we present YASS (Yet Another Similarity Searcher)—a new software for computing local alignments of two DNA sequences—and its web server (http://yass.loria.fr/interface.php). Compared with other tools, YASS is based on two innovations. The first is a new spaced seed model called transition-constrained seeds that takes advantage of statistical properties of real genomic sequences. The second feature is a new statistically founded hit criterion that controls the formation of groups of closely located seeds that are likely to belong to the same alignment. An implementation of these improvements, reported here, provides a fast and sensitive tool for local alignment of large genomic sequences.
DESCRIPTION
Web interface
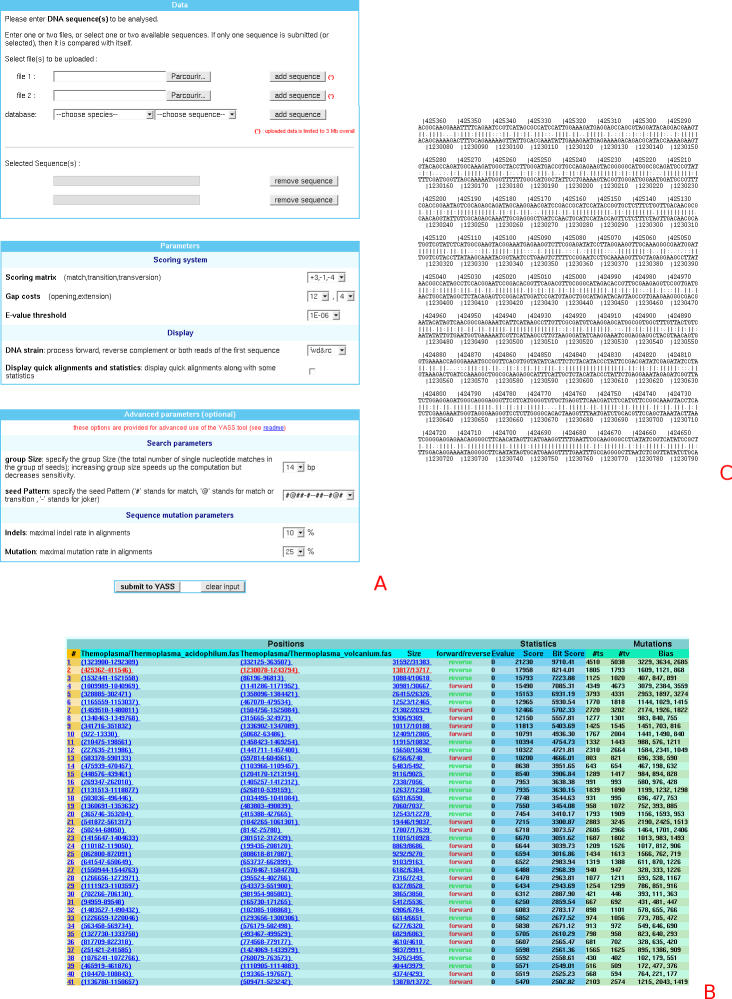
The main user input (Figure 1A) consists of one or two sequences in fasta format either chosen from a predefined database or uploaded to the web server.
Figure 1.
The input window (A) allows users to control most YASS parameters, from most basic to more advanced. Results are output in tabular format (B), with the possibility of displaying each sequence alignment (C).
Once sequences have been selected, the user can run the program right away with all other parameters set by default. Alternatively, the user can set other parameters such as the scoring matrix or gap penalties (preselected matrices are proposed), and specify the DNA strain to be processed (direct, complementary or both). The user can also choose to display complete alignments rather than only alignment positions.
More advanced parameters are available for expert users. For example, the right choice of the seed pattern can increase the search sensitivity considerably provided that some knowledge of target alignments is available (10–14). The web interface provides a preselection of seeds including three transition-constrained seeds, one providing a good performance compromise between coding and non-coding sequences, and the other two tuned respectively for non-coding and coding regions. The accompanying Hedera program (http://www.loria.fr/projects/YASS/hedera.html) is also provided for advanced users in order to design new seed patterns according to different probabilistic models of alignments (15).
Finally, the user can specify some statistical parameters of target alignments, such as the assumed substitution rate or indel rate. These parameters control the hit criterion, i.e. the rules for grouping together closely located seeds to detect similarities.
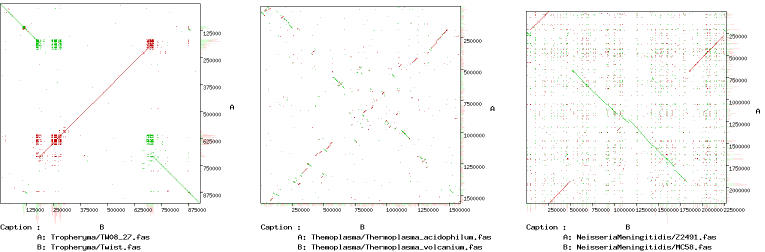
Once the results are obtained, it is possible to generate a clickable dot-plot (Figure 2) where each alignment is linked to a URL with its text representation (Figure 1C). A tabular output (Figure 1B) is also available: alignments are sorted according to their E-value and linked to their text representation. Finally, the YASS output can also be downloaded in text format for further analysis.
Figure 2.
Three YASS dot-plots are shown, each obtained from pairs of closely related bacterial sequences. Green segments represent alignments of forward reads and red segments correspond to alignments between the reverse complement of one sequence and the forward read of the other.
Technical issues
The YASS server available at http://yass.loria.fr/interface.php currently runs Apache 2.0.47 (PHP and Perl-CGI modules) on a Linux Mandrake 9.2. Dot-plots are obtained with the GD graphical library interfaced to PHP. The YASS program has been developed in C and is distributed under the Gnu General Public License.
Owing to limitations of computational resources, some restrictions have been made on the web interface. For example, uploaded files are currently limited to 3 Mb, scoring systems can be chosen only among preselected ones and for each parameter a fixed range of possible values has been settled.
Standalone version
The standalone version is recommended for frequent users or those who need specific parameters to be set outside preselected values. It provides access to two other output formats, including a BLAST-like tabular format that can be used by existing postprocessing parsers. Note that YASS does not need one of the sequences to be preprocessed (formatdb command of BLAST), rather, it treats both sequences on the fly.
METHODS
Here we briefly outline the underlying principles of the YASS algorithm, including some novel features. For a more detailed presentation the reader is referred to (16) (http://www.biomedcentral.com/1471-2105/5/149/).
Seed model
Seeds are specified using a seed pattern built over a three-letter alphabet #, @ and –, where # stands for a nucleotide match, – for a don't care symbol and @ for a match or a transition (mutation A↔G or C↔T). The weight of a pattern is defined as the number of # plus half the number of @. The weight is the main characteristic of seed selectivity.
The advantage of transition-constrained seeds stems from the biological observation that transition mutations are relatively more frequent than transversions, in both coding and non-coding regions. Typically, biologically relevant alignments contain about the same number of transitions and transversions, whereas transitions are half as frequent in independently and identically distributed random sequences. Transition-constrained seeds increase the possible number of transitions in a hit relative to spaced seeds without the transition constraint, and this is done without loss of sensitivity or efficiency.
The sensitivity of a given seed has been estimated using the algorithm of (15), which is a generalization of the one proposed in (11). Two main alignment models have been considered: a Bernoulli model (13) assumed to simulate alignments of non-coding DNA and a hidden Markov model (10) assumed to simulate alignments of coding DNA. By default, YASS currently uses the seed #@# –– ## –– # – ##@# of weight 9, which provides a good compromise in detecting similarities in both coding and non-coding sequences. The standalone version of YASS allows users to specify their own seeds. Several preselected seeds are provided by the YASS web interface.
Hit criterion
YASS is based on a multi-seed hit criterion that defines a hit as a group of closely located and possibly overlapping seeds. Two seeds belong to the same group if they occur within a bounded distance or, on the other hand, are located at close dot-plot diagonals. Distance threshold parameters are computed according to probabilistic sequence models taking into account substitution and indel rates, similarly to models used in (17). Note that seeds of a group are allowed to overlap. An additional group size parameter sets a lower bound on the total number of individual matches and transitions of the group. Using the group size results in a flexible criterion that combines a guaranteed selectivity with a good sensitivity on both short and long similarities. More details on the hit criterion can be found in (16).
Comparative tests
To validate the better performance of transition-constrained seeds compared with ordinary spaced seeds, several comparative experiments have been presented in (16). Transition-constrained seeds have been shown to be more sensitive with respect to some Bernoulli and hidden Markov models of alignments of coding and non-coding DNA [Tables 1 and 2 in (16)]. Moreover, transition-constrained seeds have been shown to be more sensitive in detecting alignments of real genomic sequences [Table 3 in (16)].
YASS has been compared with bl2seq (NCBI BLAST 2.2.6) according to several criteria: running time, number of significant alignments found (with E-value ≤ 10−6) and number of significant alignments found exclusively by one program and their total length [Table 4 in (16)]. The results show that YASS detects more significant alignments than bl2seq, within a smaller time for large DNA sequences.
CONCLUSIONS
In this paper, we have described YASS—a new DNA local alignment tool. The proposed web interface features several output formats suitable for a coup d'oeil analysis as well as for a deeper analysis of alignments. An upcoming release of YASS will include multi-seed indexing strategies and an optimized processor-cache algorithm.
Acknowledgments
The authors wish to thank Marie-Pierre Etienne and Pierre Valois for their help during early stages of this work, Mikhail Roytberg for his contribution to the seed design problem and Christophe Valmir for improving the YASS code. The authors have been supported by the ACI IMPBio of the French Ministry of Research. They thank the reviewers for their helpful remarks. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smith T., Waterman M. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 2.Lipman D., Pearson W. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Califano A., Rigoutsos I. Flash: a fast look-up algorithm for string homology. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1993;1:56–64. [PubMed] [Google Scholar]
- 4.Altschul S., Gish W., Miller W., Myers E., Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S., Madden T., Schäffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S., Kent J., Smit A., Zhang Z., Baertsch R., Hardison R., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma B., Tromp J., Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- 8.Burkhardt S., Kärkkäinen J. Better filtering with gapped q-grams. Proceedings of the 12th Symposium on Combinatorial Pattern Matching (CPM'01); 1–4 July; Jerusalem, Israel. Springer; 2001. pp. 73–85. LNCS 2089. [Google Scholar]
- 9.Li M., Ma B., Kisman D., Tromp J. PatternHunter II: highly sensitive and fast homology search. J. Bioinform. Comput. Biol. 2004;2:417–439. doi: 10.1142/s0219720004000661. [DOI] [PubMed] [Google Scholar]
- 10.Brejova B., Brown D., Vinar T. Optimal spaced seeds for hidden Markov models, with application to homologous coding regions. In: Baeza-Yates R., Chavez E., Crochemore M., editors. Proceedings of the 14th Symposium on Combinatorial Pattern Matching; 25–27 June; Morelia, Mexico. Springer; 2003. pp. 42–54. LNCS 2676. [Google Scholar]
- 11.Buhler J., Keich U., Sun Y. Designing seeds for similarity search in genomic DNA. Proceedings of the 7th Annual International Conference on Computational Molecular Biology (RECOMB'03); 10–13 April; Berlin, Germany. ACM Press; 2003. pp. 67–75. [Google Scholar]
- 12.Choi K.P., Zeng F., Zhang L. Good spaced seeds for homology search. Bioinformatics. 2004;20:1053–1059. doi: 10.1093/bioinformatics/bth037. [DOI] [PubMed] [Google Scholar]
- 13.Keich U., Li M., Ma B., Tromp J. On spaced seeds for similarity search. Discrete Appl. Math. 2004;138:253–263. [Google Scholar]
- 14.Kucherov G., Noé L., Ponty Y. Estimating seed sensitivity on homogeneous alignments. Proceedings of the IEEE 4th Symposium on Bioinformatics and Bioengineering (BIBE2004); May 19–21; Taichung, Taiwan. IEEE Computer Society Press; 2004. pp. 387–394. [Google Scholar]
- 15.Kucherov G., Noé L., Roytberg M. A unifying framework for seed sensitivity and its application to subset seeds. 2004. Rapport de recherche INRIA RR-5374. http://www.inria.fr/rrrt/rr-5374.html. [DOI] [PMC free article] [PubMed]
- 16.Noé L., Kucherov G. Improved hit criteria for DNA local alignment. BMC Bioinformatics. 2004;5:149. doi: 10.1186/1471-2105-5-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson G. Tandem repeats finder: a program to analyse DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]