Abstract
PREDBALB/c is a computational system that predicts peptides binding to the major histocompatibility complex-2 (H2d) of the BALB/c mouse, an important laboratory model organism. The predictions include the complete set of H2d class I (H2-Kd, H2-Ld and H2-Dd) and class II (I-Ed and I-Ad) molecules. The prediction system utilizes quantitative matrices, which were rigorously validated using experimentally determined binders and non-binders and also by in vivo studies using viral proteins. The prediction performance of PREDBALB/c is of very high accuracy. To our knowledge, this is the first online server for the prediction of peptides binding to a complete set of major histocompatibility complex molecules in a model organism (H2d haplotype). PREDBALB/c is available at http://antigen.i2r.a-star.edu.sg/predBalbc/.
INTRODUCTION
The T cells of the immune system recognize antigens as short peptide fragments (T-cell epitopes) derived from self or foreign proteins. Self proteins include all proteins produced by the cells of the host. Foreign peptides are derived from pathogens, environmental antigens, tumor cells and transplanted tissue. Immune recognition of both self and foreign antigens involves proteolytic processing of antigens, binding of the peptide epitopes by major histocompatibility complex (MHC) molecules and presentation of selected peptide epitopes on the cell surface to activate of T cells (1–4). Cytotoxic T cells (CD8+) recognize peptides bound to MHC class I molecules and helper T cells (CD4+) recognize antigen in the context of MHC class II molecules. MHC class I molecules are present in all cells and bind mainly endogenous peptides (those produced within the presenting cell), whereas class II molecules are present mainly in cells that recognize foreign proteins, such as macrophages and dendritic cells. T-cell epitopes are critical for the immune response to infectious, autoimmune, allergic and neoplastic disease. They have been studied for the development of peptide-based vaccines (5) and may also be important in the diagnosis of pathogens. It is estimated that between 1 and 5% of all peptides can bind a particular MHC molecule (6). Traditional approaches to the identification of T-cell epitopes that involve various biochemical and functional assays of overlapping peptides derived from proteins of interest are costly and not applicable to large-scale studies. Accurate predictions using computer models help speed up the identification of T-cell epitopes (7), minimize the number of experiments necessary and enable systematic scanning for candidate T-cell epitopes from larger sets of protein antigens, such as those encoded by complete viral genomes (8).
The BALB/c inbred laboratory mouse strain is one of the most commonly used animal models in immunological studies and has been used extensively in vaccine research (9,10). BALB/c mice express three class I (H2-Kd, H2-Ld and H2-Dd) and two class II (I-Ad and I-Ed) molecules. Several publicly available prediction systems for MHC class I and class II binding peptides provide the prediction models for (histocompatibility complex-2) H2d alleles. SYFPEITHI (11) has H2-Kd and H2-Ld models, BIMAS (12) has H2-Kd, H2-Dd and H2-Ld models, and RANKPEP (13) has models for all H2d molecules. SYFPEITHI uses binding motifs, whereas BIMAS and RANKPEP use binding matrices. These are general servers that contain prediction models for a range of MHC molecules in human and mouse, and several other mammalian organisms, but the accuracies of individual models have not been determined. On the other hand, quantitative matrices for H2-Kb, H2-Db, H2-Ld and H2-Kk (14,15) have been developed and validated.
PREDBALB/c is a computational system for the prediction of peptides binding to all five MHC molecules in BALB/c mice (H2d) class I (H2-Kd, H2-Ld and H2-Dd) and class II (I-Ad and I-Ed) that allows analysis of proteins for the presence of binding motifs to all five H2d molecules in parallel. We derived the initial quantitative matrices for PREDBALB/c using logarithmic equations based on the frequency of amino acids at specific positions within the training set of 9mer peptides as described previously (16). The initial matrices were refined by including information on the consensus (11) and other binding motifs, for example, H2-Kd binding peptides that have I, L or M at major anchor position p2 (17). The anchor positions (e.g. positions 2 and 9 in Kd binding peptides) were assigned higher weights than other positions. In addition, the prediction scores were inspected for all permissible amino acids at each of the anchor positions. All amino acids at the anchor positions other than the permissible ones were assigned low scores to exclude peptides with non-permissible amino acids from the list of predicted binders. The final binding scores were normalized to a scale of 1–9 and the final models were tested and validated rigorously. To our knowledge, PREDBALB/c is the first online server for the prediction of peptides binding to a complete set of MHC molecules in a model organism (H2d haplotype).
SYSTEM DESCRIPTION
The training data containing binding and non-binding peptides were extracted from MHCPEP (18), MHCBN (19), SYFPEITHI (11), JenPep (20) and a set of non-binders (V. Brusic, unpublished data). The 9mer peptides were used for deriving H2d class I matrices because the majority of peptides that bind these molecules are 9 amino acids long (21). Although the majority of H2d class II binding peptides are 12–20 amino acids long, their binding cores are 9 amino acids long (22,23). An iterative elimination method starting from I-Ad and I-Ed motifs in SYFPEITHI (11) was used to identify the core 9mer regions from long peptides (K.N. Srinivasan, G.L. Zhang, A. Veeramani, J.T. August and V. Brusic, manuscript in preparation). The number of peptides in the training sets is shown in Table 1. No one method of predicting peptide–MHC binding consistently outperforms the rest and the most appropriate predictive model depends on the amount of data available in Ref. (16). In our previous work, an artificial neural network method and hidden Markov models were applied to the prediction of human leukocyte antigens (HLAs) binding peptides (24,25), where more training data were available. Because relatively small training data sets are available for H2d, we adopted matrix models as the prediction method.
Table 1.
Number of peptides in the training sets for H2d matrices
| Class I | Class II | ||||
|---|---|---|---|---|---|
| Dd | Kd | Ld | Ad | Ed | |
| Binders (B) | 93 | 139 | 73 | 127 | 191 |
| Non-binders (NB) | 331 | 105 | 91 | 963 | 627 |
| Ratio B/NB | 28% | 132% | 87% | 13% | 30% |
Five 9 × 20 matrices were built, one for each of the five H2d alleles, and 10-fold cross-validations were performed to test the accuracy of the prediction models. The results show that PREDBALB/c predicts peptides binding to I-Ed, I-Ad and H2-Kd with excellent accuracy [area under the receiver operating characteristic (ROC) curve, AROC ≥ 0.90], and to H2-Dd and H2-Ld with good accuracy (AROC ≥ 0.85). The models were also rigorously tested using experimentally known peptides from viral, prokaryotic and eukaryotic origins (14,26–29) and validated by in vivo studies using severe acute respiratory syndrome (SARS) nucleocapsid and HIV GAG proteins. The H2d models accurately predicted 11 out of 12 ELISPOT positive regions from BALB/c mice splenocytes immunized with SARS nucleocapsid DNA vaccines (data not shown).
The web interface of PREDBALB/c uses a set of graphical user interface forms. The interface was built using a combination of Perl, CGI and C programs. PREDBALB/c has been implemented in a SunOS 5.9 UNIX environment.
USING PREDBALB/C
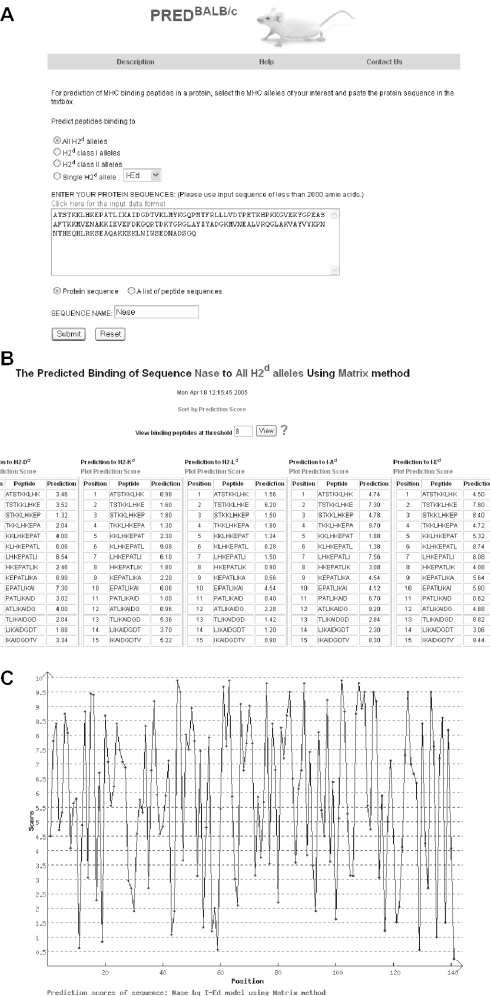
Users have the option to predict peptides binding to all H2d molecules, H2d class I molecules, H2d class II molecules or a single H2d molecule. The default selection on the webpage is ‘all H2d’ molecules. To perform predictions using PREDBALB/c, the user must paste a protein sequence into a textbox and assign a name to the sequence. The sequence must contain between 9 and 2000 amino acids. If the prediction is run with an input sequence containing symbols other than the 20 amino acid codes (spaces and carriage returns are allowed) or the total sequence length is outside the 9–2000 amino acids range, an error message will be displayed. The input can be either a contiguous protein sequence or a list of peptides, one per line. The default selection on the webpage is ‘protein sequence’ (Figure 1A), which means the input sequence is treated as a contiguous protein sequence (carriage returns and line breaks will be ignored). The PREDBALB/c input processing program decomposes protein sequence (or the list of peptides) into a series of 9mer peptides overlapping by eight amino acids. Individual 9mer peptides are then submitted for prediction. Predicted binding scores for all 9mers are displayed in the result tables (Figure 1B). The 9mer binding scores are within the range 0–10; the higher the score, the higher the probability of the peptide being a binder. PREDBALB/c has the option to plot the binding scores of all the overlapping 9mer peptides as a graph, in which the x-axis represents the start position of a 9mer peptide and the y-axis represents the binding score of the 9mer peptide (Figure 1C). The user can sort the peptides by their binding scores and choose to view only predicted binders with binding scores above a certain threshold. To assess prediction accuracy, we used measures of sensitivity SE = TP/(TP + FN) and specificity SP = TN/(TN + FP) (TP: true positives; TN: true negatives; FP: false positives; FN: false negatives). The higher the value of SP, the lower is the value of SE, which results in lower number of both TPs and FPs. The lower the value of SP, the higher is the value of SE, which results in higher number of both TPs and FPs. Raw binding scores are mapped to a linear scale that corresponds to SP values, and therefore the prediction thresholds across different models have similar meaning. For example, when a user sets the threshold to 8, the specificity of the predictions to all five alleles is 0.8. The corresponding sensitivities of each model can be viewed at http://antigen.i2r.a-star.edu.sg/predBalbc/HTML/specificity.html.
Figure 1.
An example of the output pages of PREDBALB/c when the input is a single protein. The input protein sequence is Staphylococcal nuclease (Nase), the sequence type chosen is ‘protein sequence’ and the H2d alleles of interest are all H2d alleles. (A) The input page. (B) The main result page. The input sequence is decomposed into overlapping 9mers for the prediction of binding scores to each allele. (C) Graphical view of the predicted binding scores to the I-Ed molecule.
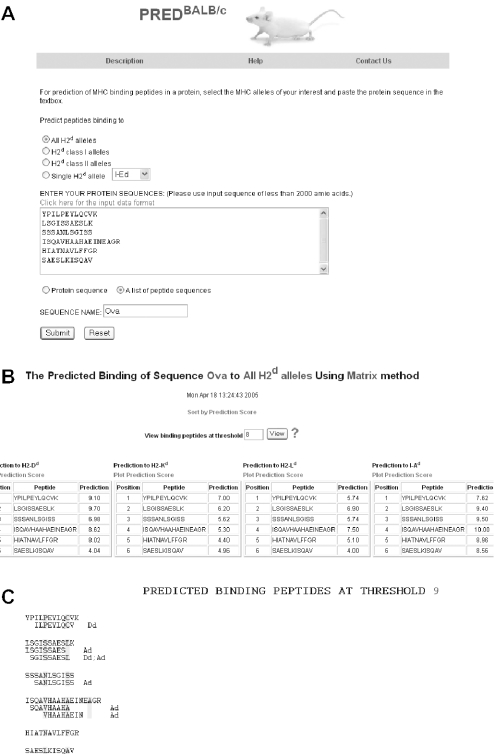
When users select the input sequence type to be ‘a list of peptide sequences’, the input sequences separated by carriage returns or line breaks are treated as different peptides (Figure 2A). All overlapping 9mers in each peptide are submitted for prediction. In the result tables, predicted binding scores are represented by the highest individual 9mer binding score within the input peptide. The predicted binding scores of individual 9mers in each peptide in the list are not shown (Figure 2B). To display the top-scoring 9mer peptides from each input peptide, the user can use the function ‘View binding peptides at threshold 9’ (Figure 2B). In the result page (Figure 2C), the 9mers with binding scores equal to or above the threshold of 9 are aligned with the input peptides. The predicted 9mers are displayed with the names of the H2d alleles to which the 9mer binding scores are above the threshold. For example, the first input peptide, YPILPEYLQCVK, has binding scores 9.10, 7.00, 5.74, 7.62 and 8.62 to H2-Dd, H2-Kd, H2-Ld, I-Ad and I-Ed, respectively (Figure 2B). At threshold 9 (SP level 0.9), there are no 9mer binders to H2d class II alleles and the 9mer ILPEYLQCV has the highest binding score to H2-Dd, 9.10. Thus, in Figure 2C, this 9mer is aligned with the input peptide and followed by ‘Dd’.
Figure 2.
An example of the output pages of PREDBALB/c when the input is a list of peptides. The input is a list of peptides from chicken ovalbumin and the H2d alleles of interest are all H2d alleles. (A) The input page. (B) The main result page. All 9mers in each peptide are submitted for prediction. The predicted binding scores are represented by the highest individual 9mer binding score of each input peptide. (C) The alignment view of the predicted binding peptides at threshold 9, which indicates that the specificity of the prediction is 0.9.
CONCLUSION
PREDBALB/c marks a new direction in predictive modeling of MHC-binding peptides and T-cell epitopes. The main advantage is that PREDBALB/c focuses on a complete organism and its predictions represent a complete set of predicted targets of T-cell immune responses. The focus on the complete set of MHC alleles is closer to studies involving laboratory animals. This approach provides a more complete view of the immune responses of an organism. The BALB/c mouse is an important laboratory model and PREDBALB/c is, therefore, useful for the analysis of immunization regimens and deciphering responses to infections. Further development of PREDBALB/c will include addition of matrices for prediction of 8mer and 10mer binders to H2d class I molecules and further improvement of prediction matrices by cyclical refinement—using newly defined binders and non-binders from experiments.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This project has been funded in part by US Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Grant No. 5 U19 AI56541 and Contract No. HHSN266200400085C. Funding to pay the Open Access publication charges for this article was provided by the Institute for Infocomm Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pamer E., Cresswell P. Mechanisms of MHC class I–restricted antigen processing. Annu. Rev. Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 2.Villadangos J.A., Bryant R.A., Deussing J., Driessen C. Proteases involved in MHC class II antigen presentation. Immunol. Rev. 1999;172:109–120. doi: 10.1111/j.1600-065x.1999.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 3.Yewdell J.W., Bennink J.R. Cut and trim: generating MHC class I peptide ligands. Curr. Opin. Immunol. 2001;13:13–18. doi: 10.1016/s0952-7915(00)00175-8. [DOI] [PubMed] [Google Scholar]
- 4.Bryant P., Ploegh H. Class II MHC peptide loading by the professionals. Curr. Opin. Immunol. 2004;16:96–102. doi: 10.1016/j.coi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Zhong W., Reche P.A., Lai C.C., Reinhold B., Reinherz E.L. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J. Biol. Chem. 2003;278:45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 6.Brusic V., Zeleznikow J. Computational binding assays of antigenic peptides. Lett. Pept. Sci. 1999;6:313–324. [Google Scholar]
- 7.Brusic V., Bajic V.B., Petrovsky N. Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods. 2004;34:436–443. doi: 10.1016/j.ymeth.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.De Groot A.S., Saint-Aubin C., Bosma A., Sbai H., Rayner J., Martin W. Rapid determination of HLA B*07 ligands from the West Nile virus NY99 genome. Emerg. Infect. Dis. 2001;7:706–713. doi: 10.3201/eid0704.010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchwald U.K., Lees A., Steinitz M., Pirofski L.A. A peptide mimotope of type 8 pneumococcal capsular polysaccharide induces a protective immune response in mice. Infect. Immun. 2005;73:325–33. doi: 10.1128/IAI.73.1.325-333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakai Y., Morrison B.J., Burke J.D., Park J.M., Terabe M., Janik J.E., Forni G., Berzofsky J.A., Morris J.C. Vaccination by genetically modified dendritic cells expressing a truncated neu oncogene prevents development of breast cancer in transgenic mice. Cancer Res. 2004;64:8022–8028. doi: 10.1158/0008-5472.CAN-03-3442. [DOI] [PubMed] [Google Scholar]
- 11.Rammensee H.G., Bachmann J., Emmerich N.P., Bachor O.A., Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 12.Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 13.Reche P.A., Glutting J.P., Reinherz E.L. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 2002;63:701–709. doi: 10.1016/s0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
- 14.Udaka K., Wiesmuller K.H., Kienle S., Jung G., Tamamura H., Yamagishi H., Okumura K., Walden P., Suto T., Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- 15.Hattotuwagama C.K., Guan P., Doytchinova I.A., Flower D.R. New horizons in mouse immunoinformatics: reliable in silico prediction of mouse class I histocompatibility major complex peptide binding affinity. Org. Biomol. Chem. 2004;2:3274–3283. doi: 10.1039/B409656H. [DOI] [PubMed] [Google Scholar]
- 16.Yu K., Petrovsky N., Schönbach C., Koh J.Y.L., Brusic V. Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol. Med. 2002;8:137–148. [PMC free article] [PubMed] [Google Scholar]
- 17.Quesnel A., Hsu S.C., Delmas A., Steward M.W., Trudelle Y., Abastado J.P. Efficient binding to the MHC class I K(d) molecule of synthetic peptides in which the anchoring position 2 does not fit the consensus motif. FEBS Lett. 1996;387:42–46. doi: 10.1016/0014-5793(96)00446-2. [DOI] [PubMed] [Google Scholar]
- 18.Brusic V., Rudy G., Harrison L.C. MHCPEP, a database of MHC-binding peptides. Nucleic Acids Res. 1994;22:3663–3665. doi: 10.1093/nar/22.17.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhasin M., Singh H., Raghava G.P. MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 2003;19:665–666. doi: 10.1093/bioinformatics/btg055. [DOI] [PubMed] [Google Scholar]
- 20.McSparron H., Blythe M.J., Zygouri C., Doytchinova I.A., Flower D.R. JenPep: a novel computational information resource for immunobiology and vaccinology. J. Chem. Inf. Comput. Sci. 2003;43:1276–1287. doi: 10.1021/ci030461e. [DOI] [PubMed] [Google Scholar]
- 21.Rammensee H.G., Falk K., Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu. Rev. Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 22.Rammensee H.G. Chemistry of peptides associated with MHC class I and class II molecules. Curr. Opin. Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 23.Scott C.A., Peterson P.A., Teyton L., Wilson I.A. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 1998;8:319–329. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 24.Khan A.M., Srinivasan K.N., August J.T., Brusic V. Neural Models for predicting viral vaccine targets. J. Bioinform. Comput. Biol. 2005 doi: 10.1142/s0219720005001466. in press. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G.L., Khan A.M., Srinivasan K.N., August J.T., Brusic V. MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki452. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sette A., Buus S., Appella E., Smith J.A., Chesnut R., Miles C., Colon S.M., Grey H.M. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl Acad. Sci. USA. 1989;86:3296–3300. doi: 10.1073/pnas.86.9.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mata M., Travers P.J., Liu Q., Frankel F.R., Paterson Y. The MHC class I-restricted immune response to HIV-gag in BALB/c mice selects a single epitope that does not have a predictable MHC-binding motif and binds to Kd through interactions between a glutamine at P3 and pocket D1. J. Immunol. 1998;161:2985–2993. [PubMed] [Google Scholar]
- 28.Saikh K.U., Martin J.D., Nishikawa A.H., Dillon S.B. Influenza A virus-specific H-2d restricted cross-reactive cytotoxic T lymphocyte epitope(s) detected in the hemagglutinin HA2 subunit of A/Udorn/72. Virology. 1995;214:445–452. doi: 10.1006/viro.1995.0055. [DOI] [PubMed] [Google Scholar]
- 29.Zatechka D.S., Jr, Hegde N.R., Hariharan K., Srikumaran S. Identification of murine cytotoxic T-lymphocyte epitopes of bovine herpesvirus 1. Vaccine. 1999;17:686–69. doi: 10.1016/s0264-410x(98)00251-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.