Abstract
One of the key challenges in the analysis of gene expression data is how to relate the expression level of individual genes to the underlying transcriptional programs and cellular state. Here we describe T-profiler, a tool that uses the t-test to score changes in the average activity of predefined groups of genes. The gene groups are defined based on Gene Ontology categorization, ChIP-chip experiments, upstream matches to a consensus transcription factor binding motif or location on the same chromosome. If desired, an iterative procedure can be used to select a single, optimal representative from sets of overlapping gene groups. T-profiler makes it possible to interpret microarray data in a way that is both intuitive and statistically rigorous, without the need to combine experiments or choose parameters. Currently, gene expression data from Saccharomyces cerevisiae and Candida albicans are supported. Users can upload their microarray data for analysis on the web at http://www.t-profiler.org.
INTRODUCTION
An important technique in the post-genomic era is the simultaneous measurement of the transcript levels of all genes from a genome by microarray experiments (1,2). In recent years, the amount of data from such experiments has rapidly increased (3,4). Furthermore, the combination of chromatin-immunoprecipitation and microarray technology (‘ChIP-chip’) has made it possible to globally measure the binding of transcription factors to gene promoters (5,6).
There has also been an explosion in the number of computational methods for analyzing microarray data. Among the most popular are algorithms such as hierarchical clustering (7), K-means clustering (8) and self-organizing maps (9). A limitation of these clustering methods is the need to have gene expression profiles across multiple hybridizations. Alternative methods have been developed that can take a single genome-wide expression pattern as input, such as motif-based correlation or regression (10–12).
To obtain easily interpretable information on changes in the cellular state in terms of functional annotation, methods such as Funspec (13), GO term finder (14), GOAL (15) and GeneXpress (http://genexpress.stanford.edu) score the significance of overlap between predefined gene groups [from Gene Ontology (GO) (16) or the MIPS database (17)] and the subset of induced or repressed genes. These methods are based on the cumulative hypergeometric distribution (also referred to as Fisher's exact test). A disadvantage of these methods is that they require individual genes to be significantly up- or down-regulated in order to contribute to the score.
We previously developed a method that can score GO categories without the need to apply cut-offs to the expression level of individual genes (18). This algorithm, now named T-profiler, uses the t-test to score the difference between the mean expression level of predefined groups of genes and that of all other genes on the microarray (see Methods). A similar approach was independently pioneered by Pavlidis et al. (19). T-profiler is currently suitable for the analysis of Saccharomyces cerevisiae and Candida albicans gene expression profiles, and in the near future will be extended to other organisms.
METHODS
For a given gene group G, the t-value is given by the following formula:
where
Here μG is the mean expression log-ratio of the NG genes in gene group G, μG′ is the mean expression log-ratio of the remaining NG′ genes and s is the pooled standard deviation, as obtained from the estimated variances for groups G and G′. The associated two-tailed P-value can be calculated from t using the t-distribution with NG − 2 degrees of freedom and is corrected for multiple testing by multiplying it by the number of gene groups that are being tested in parallel (Bonferroni correction). All groups with a corrected E-value of ≤0.05 are considered to be significantly regulated. To reduce the influence of outliers, which may result in false positives or false negatives, we discard the highest and lowest expression value in each gene group. This method is similar to the jack-knife procedure (20).
Gene groups sharing a common motif in their upstream region
Motif groups are defined as genes with a match to a particular consensus motif within 600 base pairs upstream of the open reading frame (ORF) (21), allowing no overlap with neighboring ORFs. The consensus motifs used in T-profiler are derived from three different sources. First, motifs were extracted from the SCPD database (http://cgsigma.cshl.org/jian/). Next, motifs were found by comparing the genome sequences of highly related yeast species (22,23). Finally, motifs discovered from various microarray experiments using the REDUCE algorithm (11,24) were added. Most of these motifs are similar or identical to motifs described in the literature. In total, 153 motif groups are included in the T-profiler calculation. Far less information is available about regulatory sequences of C.albicans. It was recently reported that about one-third of S.cerevisiae regulatory elements are conserved in C.albicans (25). T-profiler therefore uses the list of S.cerevisiae motifs, supplemented with newly discovered C.albicans regulatory motifs, to score C.albicans expression data.
Gene groups bound by a common transcription factor based on ChIP-chip data
The binding of transcription factors to their global DNA targets can be measured by ChIP-chip experiments. In S.cerevisiae this technique has been explored on a large scale by Lee et al. (5) and Harbison et al. (6). We used the transcription factor binding (TFB) data for 203 transcription factors from Harbison et al. (6) as input into T-profiler; the binding of 84 of these regulators was measured under various environmental conditions. A gene was considered to be part of a TFB group if the P-value reported by the authors was <0.001. In addition, TFB groups were required to have at least seven gene members. This resulted in 252 TFB groups that were used for T-profiler analysis.
GO categories
The third type of gene group is based on membership of a specific GO category (16). In GO, each gene is classified according to biological process, molecular function and cellular component. The GO gene group contains the genes associated with a specific GO category as well as all of its child categories. Only GO groups with more than six members were used for calculation. This resulted in 1389 GO-derived gene groups that were used for T-profiler analysis. Significant scores of GO groups give direct information about which functions or cellular processes are expected to have changed as a result of the altered gene expression. It should be kept in mind, however, that, unlike in the case of motif and ChIP-chip based gene groups, the t-values for GO categories are not directly related to a molecular mechanism.
Iterative removal of redundant gene groups
Several of the predefined gene groups scored by T-profiler show strong mutual overlap: the GO categories used by T-profiler are hierarchically organized; consensus motifs can match similar sequences; and ChIP-chip experiments can reveal that similar sets of genes are bound by different transcription factors and/or under different conditions. The t-values for overlapping gene groups are strongly correlated and therefore mutually redundant. Following the idea of forward selection of non-redundant motifs in REDUCE (11), we implemented an iterative procedure to select a non-redundant set of gene groups among those that have t-values significantly different from zero. At each step, we subtract the mean expression level of the genes in the gene group with the highest absolute t-value from all genes in that gene group. The t-values are then recalculated for all other gene groups, and the procedure is repeated until even the most significantly regulated gene group has a P-value > 0.05. In the case of nested GO categories at different levels in the hierarchy, this procedure will naturally select the most appropriate level for a given branch of annotation.
Aneuploidy test
Hughes et al. (26) described the discovery of chromosomal aberrations in yeast deletion mutants based on gene expression profiles. These are often duplications or deletions of an entire chromosome. By applying T-profiler at the level of whole chromosomes, where gene groups are defined as the set of all genes on a specific chromosome, it is possible to detect such aneuploidy. A statistically significant chromosomal t-value does not necessarily point, however, to aneuploidy, as it may also be caused by normal differential regulation by a transcription factor whose targets are preferentially located on the same chromosome. In the aneuploid dataset from Hughes et al. (26) we observed an absolute t-value > 10 for almost all deleted or duplicated chromosomes; such extreme t-values are therefore a good indicator of aneuploidy.
AN EXAMPLE
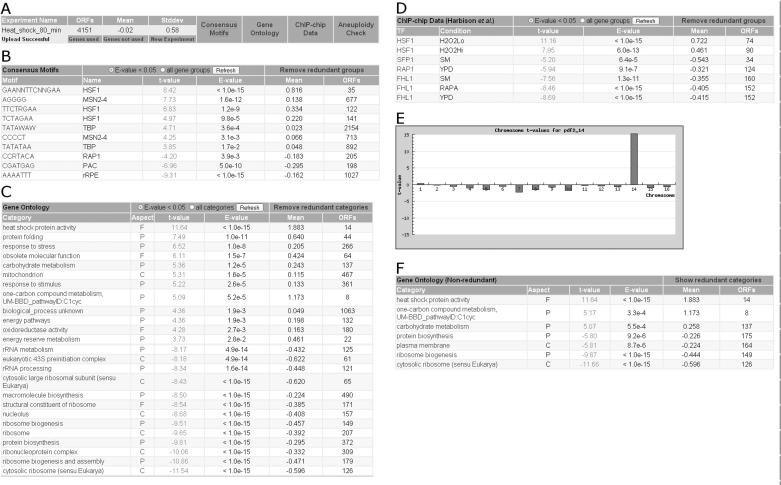
Gene expression datasets can be uploaded as a tab-delimited text file with the systematic ORF name in the first column and the log-transformed expression data in the second column. The upload of an expression profile comparing cells 80 min after a heat shift from 30 to 37°C from the Environmental Stress Response data set of Gasch et al. (23) will serve as an example. After uploading, the user is presented with some basic information about the dataset, including the number of genes, the average and the standard deviation (Figure 1A). Importantly, no cut-offs are applied; all values are used for calculation.
Figure 1.
Screenshots of the various T-profiler analysis results. (A) Statistics of the uploaded gene expression dataset for cells assayed 80 min after the temperature shift from 30 to 37°C (23). The type of analysis can be selected from the panels to the right. (B) Scoring consensus motifs. Only significantly scoring motifs are shown (E-value < 0.05). By selecting the motifs in the left column, information about the genes containing this motif and their expression levels can be obtained. (C) Scoring GO categories. Only a subset of the 50 significantly changed categories is shown. (D) Scoring ChIP-chip based gene groups. (E) Graph showing the t-value for each chromosome, obtained from the gene expression profile of the mutant pfd2Δ, in which chromosome 14 is duplicated. (F) The same result as in (C), but now with redundant gene groups removed by our iterative procedure.
Next, the user can follow links to results for four different types of predefined gene groups: genes whose promoter region matches a specific consensus motif (Figure 1B), genes that belong to a specific GO category (Figure 1C), genes whose promoter is significantly bound by a specific transcription factor according to a ChIP-chip experiment (Figure 1D) and genes that reside on a specific chromosome (Figure 1E). The statistical parameters that are output by T-profiler for any group scored are (i) a t-value measuring the up-regulation (t > 0) or down-regulation (t < 0) in units of the standard error of the difference and (ii) an E-value that is Bonferroni corrected for the parallel testing of the large number of categories, which represents the number of groups with the same t-value or higher that would be observed by chance. Typically, only a small subset of the gene groups considered will score as differentially expressed (Figure 1).
Figure 1B shows consensus motifs associated with differential regulation. The heat shock response motif (HSF1) and the general stress response motif (MSN2/4) score positively, whereas the PAC and rRPE motifs, both over-represented in genes involved in rRNA biosynthesis (27), score negatively. The up-regulation of genes under the control of the HSF1 motif is specific to heat-shocked cells, whereas the down-regulation of genes involved in rRNA biosynthesis and genes containing MSN2/4 motifs is typical of the environmental stress response (23). Figure 1D shows which transcription factors and corresponding ChIP-chip conditions are associated with differential regulation. The fact that genes bound by the transcription factor Hsf1p score positively whereas the genes bound by the ribosome-regulating transcription factors Rap1p, Sfp1p and Fhl1p score negatively is consistent with the motif-based results. Figure 1C shows the results of T-profiler analysis based on GO; in total, 50 categories have a significant t-value. Most of the positively scoring categories are involved in heat shock and stress response, whereas most of the negatively scoring categories are comprised mainly of ribosomal genes. Again, the results compare well with the results obtained by T-profiler using motif and ChIP-chip based gene groups. However, the large number of similar GO categories reported makes it harder to interpret the results. Figure 1F shows how this problem is resolved by the iterative removal of redundant categories. Finally, in Figure 1E, the high t-value of chromosome 14 points to a duplication of chromosome 14 in the deletion mutant pfd2Δ.
CONCLUSION
T-profiler analyzes genome-wide expression patterns one experiment at a time, without the need to tune any parameters. Our use of the t-test to score gene groups eliminates the need to impose a threshold on the expression level of individual genes. A group can be scored as significantly induced or repressed even if the expression of none of its individual member genes changes significantly. This feature greatly increases the sensitivity to small-amplitude coordinate changes in the expression of groups of genes. Representing a transcriptome by a relatively small set of statistically robust and easily interpretable t-values allows for seamless comparison between hybridizations, even across different platforms and laboratories. We plan to extend the functionality of T-profiler to multiple experiments in the near future.
Acknowledgments
We would like to thank Merijn Schuurmans and Ania Zakrzewska for helpful discussions and for testing T-profiler, Reka Letso for a critical reading of the manuscript, and Xiang-Jun Lu for assistance in setting up the webserver. This work was supported by grants from the Netherlands Foundation for Technical Research (STW) to F.K. (APB.5504) and from the National Institutes of Health to H.J.B. (R01HG003008). Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 3.Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 6.Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharan R., Shamir R. CLICK: a clustering algorithm with applications to gene expression analysis. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:307–316. [PubMed] [Google Scholar]
- 9.Tamayo P., Slonim D., Mesirov J., Zhu Q., Kitareewan S., Dmitrovsky E., Lander E.S., Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen L.J., Knudsen S. Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics. 2000;16:326–333. doi: 10.1093/bioinformatics/16.4.326. [DOI] [PubMed] [Google Scholar]
- 11.Bussemaker H.J., Li H., Siggia E.D. Regulatory element detection using correlation with expression. Nature Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 12.Conlon E.M., Liu X.S., Lieb J.D., Liu J.S. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson M.D., Grigull J., Mohammad N., Hughes T.R. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volinia S., Evangelisti R., Francioso F., Arcelli D., Carella M., Gasparini P. GOAL: automated Gene Ontology analysis of expression profiles. Nucleic Acids Res. 2004;32:W492–W499. doi: 10.1093/nar/gkh443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mewes H.W., Heumann K., Kaps A., Mayer K., Pfeiffer F., Stocker S., Frishman D. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 1999;27:44–48. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lascaris R., Bussemaker H.J., Boorsma A., Piper M., van der Spek H., Grivell L., Blom J. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 2003;4:R3. doi: 10.1186/gb-2002-4-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlidis P., Lewis D.P., Noble W.S. Exploring gene expression data with class scores. Pac. Symp. Biocomput. 2002:474–485. [PubMed] [Google Scholar]
- 20.Heyer L.J., Kruglyak S., Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Helden J., Andre B., Collado-Vides J. A web site for the computational analysis of yeast regulatory sequences. Yeast. 2000;16:177–187. doi: 10.1002/(SICI)1097-0061(20000130)16:2<177::AID-YEA516>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Kellis M., Patterson N., Birren B., Berger B., Lander E.S. Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. J. Comput. Biol. 2004;11:319–355. doi: 10.1089/1066527041410319. [DOI] [PubMed] [Google Scholar]
- 23.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roven C., Bussemaker H.J. REDUCE: an online tool for inferring cis-regulatory elements and transcriptional module activities from microarray data. Nucleic Acids Res. 2003;31:3487–3490. doi: 10.1093/nar/gkg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasch A.P., Moses A.M., Chiang D.Y., Fraser H.B., Berardini M., Eisen M.B. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PloS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes T.R., Roberts C.J., Dai H., Jones A.R., Meyer M.R., Slade D., Burchard J., Dow S., Ward T.R., Kidd M.J., et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nature Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 27.Hughes J.D., Estep P.W., Tavazoie S., Church G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]