Abstract
POWER, the PhylOgenetic WEb Repeater, is a web-based service designed to perform user-friendly pipeline phylogenetic analysis. POWER uses an open-source LAMP structure and infers genetic distances and phylogenetic relationships using well-established algorithms (ClustalW and PHYLIP). POWER incorporates a novel tree builder based on the GD library to generate a high-quality tree topology according to the calculated result. POWER accepts either raw sequences in FASTA format or user-uploaded alignment output files. Through a user-friendly web interface, users can sketch a tree effortlessly in multiple steps. After a tree has been generated, users can freely set and modify parameters, select tree building algorithms, refine sequence alignments or edit the tree topology. All the information related to input sequences and the processing history is logged and downloadable for the user's reference. Furthermore, iterative tree construction can be performed by adding sequences to, or removing them from, a previously submitted job. POWER is accessible at http://power.nhri.org.tw.
INTRODUCTION
Phylogenetic analysis in biological macromolecule sequences is an important and common strategy for research into evolution and taxonomy. One major advantage of the proposed approach is that it estimates evolutionary distance using genetic information instead of traditional, standard morphological measurements (1,2). Therefore, biologists frequently utilize phylogenetic analysis to present or interpret sequence data. The significant quantity and constant multiplication of macromolecular sequences in public databanks can be a huge biological resource if researchers can efficiently extract information from a wide range of species to analyze and develop a sophisticated theory to determine phylogeny (2,3).
A phylogenetic analysis process for a collection of nucleic acid sequences or protein sequences typically requires successive steps of multiple sequence alignment (MSA), a phylogenetic analysis and a tree builder for final presentation. Several standalone software and web-based servers have been developed for these steps. First, an MSA is conducted using ClustalW in the command mode (4,5), GUI ClustalX (6) or Internet-based bioinformatics services, such as those of the Centre for Molecular and Biomolecular Informatics (SWISS EMBnet node server, EBI), the Bioinformatics Institute (Institute for Chemical Research, Kyoto University) and the Baylor College of Medicine. Next, the phylogenetic relationships among target sequences can be analyzed using software packages such as PHYLIP (7) and WebPHYLIP (8) based on prealigned sequences from the MSA results. The tree file resulting from this step is in text format and is displayed using tree plotting programs such as DRAWTREE, WebPHYLIP, TREEVIEW, NJPLOT (9), ATV (10), PhyloDraw (11) and DRAWGRAM in the PHYLIP package. A standalone software package, MEGA2 (12), can also be employed to create tree topologies from the MSA results. Other commercial or free packages and utilities supporting phylogenetic analysis of macromolecular data are also available (13). A comprehensive list of these phylogeny programs can be found on the PHYLIP website (http://evolution.genetics.washington.edu/phylip.html).
Since each step is normally conducted separately, effort cannot be avoided in data formatting and transferring. Consequently, the process is tedious and time consuming. Some web servers, such as PhyloBLAST (14), provide a chained phylogenetic analysis process. However, these programs are not designed for general-purpose phylogenetic analysis processes. For example, PhyloBLAST is designed specifically for analyzing protein sequences. The phylogenetic analysis function of PhyloBLAST initially performs an MSA on sequences obtained from a BLAST search. Users cannot define their own parameters in these chained processes.
Tree plotting also causes problems during the final step. Some tree building utilities produce branch-crossing trees or label-overlapping topologies when managing a large number of input sequences. Such incorrectly displayed results are difficult to interpret and confusing to users (11). Web applications cannot always successfully visualize graphical trees because of problems with postscript conversion, incompatibility of different Java virtual machines and server errors.
This article presents a web-based application, PhylOgenetic WEb Repeater (POWER), to conduct phylogenetic analysis of protein and nucleic acid sequences efficiently. POWER seamlessly integrates the three steps of phylogenetic analysis. MSA and phylogenetic analysis are executed using the well-established methods ClustalW and PHYLIP. With the novel tree builder presented here, a graphical tree can be constructed without problems of branch-crossing and label-overlapping. Through a simple interface, users can analyze molecules effortlessly using default settings and can also modify parameters. Starting from a sequence input in FASTA format, an MSA report and a final tree image, as a PNG file, are automatically generated. Instead of real-time manipulation on the web, POWER provides a link or delivers an optional email message to inform the user that the job has been completed. The link in the result page allows users to manipulate their primary input items, retrieve all the files and repeatedly optimize the resulting trees.
SYSTEM IMPLEMENTATION
Parameter collection, job processing and result display
The POWER system is built with an open-source LAMP structure: Linux (operating system), Apache (web server), MySQL (relational database) and PHP (html-embedded scripting language).
POWER conducts MSA and phylogenetic analysis using algorithms from ClustalW v1.83 (5) and the PHYLIP package v3.5 (7). Users can adjust most ClustalW and PHYLIP parameters. The ClustalW and PHYLIP source code was modified for system integration, and all output file names were normalized to prevent name conflicts among different jobs.
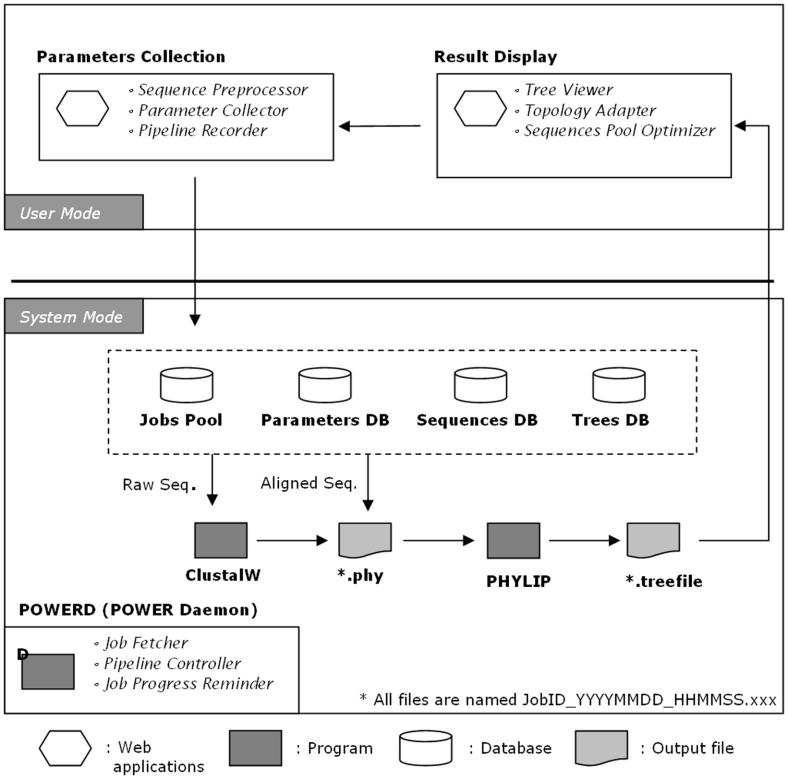
The POWER system separates a job into two modes, ‘user mode’ and ‘system mode’ (Figure 1). In ‘user mode’, POWER users can manipulate all the parameters for MSA and phylogenetic analysis and view the results on a web browser. Initially, users should define sequence type (nucleotides or amino acids) and input data (by pasting sequences directly into a form or uploading a pre-edited file). Both raw sequence data in FASTA format and aligned data in PHYLIP-compatible format are permissible. POWER guides the user step by step through the whole analysis process. Although POWER runs well with the default parameter settings, the parameters for MSA and phylogenetic analysis can be adjusted for advanced manipulation. After users verify all the parameters through the guiding steps and submit their jobs, the process enters ‘system mode’, during which the POWER daemon, called POWERD, takes over the process.
Figure 1.
Inside POWER. In ‘user mode’ (top), the user inputs parameters on the parameter input page and views the result on the result display page. In ‘system mode’ (bottom), POWERD controls the entire process and calls programs to process input data and create output files.
All input sequences in a query are parsed and stored in a temporary database as the source of entries for the following analysis. For system security, reliability and automatic bounce handling, POWER is equipped with the Qmail (http://cr.yp.to/qmail.html) SMTP server to send users email notification messages when requested. To optimize system performance, the POWER daemon can process up to four jobs simultaneously. POWERD calls the appropriate analysis programs depending on user-defined parameters. Eventually, a phylogenetic tree file (*.treefile) is generated. The system then returns to ‘user mode’. PHYLIP output files are processed by a tree image maker written in PHP, which calls on the GD library modules for high-quality graphical display in PNG format. By optimizing the relative distance between nodes, the tree builder creates an optimized tree topology which avoids improper crossing of branches and overlapping of sequence identifiers.
Normally, a phylogenetic analysis job is completed within a matter of minutes and its result is displayed directly in the same web browser window after the job is done. If a job takes longer than expected, the user can save the link provided in the result page as a bookmark to check later or input an email address on the job submission page to be notified by the POWER daemon when the job is complete. By clicking on the hyperlink provided in the email, the user can retrieve the final output trees. The result page has three parts, Tree Image, Job Information and Download Area. The arrangement of branches in the tree image can be flipped and rotated directly on the result webpage by clicking on any branching point. Additionally, users can download all output files including tree images from the Download Area, and can remove or add some sequences and run the analysis again.
CONCLUSIONS
POWER integrates several novel features for parallel calculation and an optimized database structure for parameter collection, job processing and result visualization. Several programs were written specifically for the system, including the sequence preprocessor, file format converter, topology illustrator and job controller. Users can easily manipulate the parameters using online help and query logs for iterative jobs.
This work involved constructing a simple and friendly user interface with default parameters for quick analysis and the option to define specific parameter settings. The system covers almost all combinations of ClustalW and PHYLIPS parameter settings and provides online help for each step to satisfy most needs to analyze molecular evolution. Using the graphical tool, users can easily interpret a phylogenetic tree without improper crossing.
Tree building iteration is a common process among biologists trying to find a proper tree topology. Tracking changes in a tree resulting from repeated parameter tuning and addition or deletion of sequences is difficult. POWER overcomes this problem by providing detailed logs of user-defined parameters, all output files and process history.
In summary, this work presents a web service, POWER, that seamlessly and flexibly combines successive steps of phylogenetic analysis. The system, which is based on well-developed algorithms including ClustalW and PHYLIP, can conveniently and reliably align sequences and depict trees of many biological macromolecules. POWER is intended to assist biologists from a broad range of disciplines and is likely to be particularly helpful for non-experts.
Acknowledgments
The authors would like to thank the National Health Research Institutes, Taiwan (Bioinformatics Core Laboratory) and the National Science Council, Taiwan (National Science and Technology Program for Genomic Medicine) for financially supporting this research under Contract Nos BS-092-PP-05 and NSC 92-3112-B-400-007-Y. Funding to pay the Open Access publication charges for this article was provided by National Science Council, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Campbell N.A., Reece J.B. Biology. Menlo Park, CA: Benjamin Cummings; 2002. Phylogeny and systematics; pp. 484–507. [Google Scholar]
- 2.Nei M., Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. Molecular basis of evolution; pp. 3–16. [Google Scholar]
- 3.Gibas C., Jambeck P. Mutiple Sequence Alignments, Trees, and Profiles. Sebastopol, CA: O'Reilly; 2001. pp. 191–214. [Google Scholar]
- 4.Higgins D.G., Thompson J.D., Gibson T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 5.Thompson J.D., Higgins D.G., Gibson T.J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. WA: University of Washington; 1993. PHYLIP: Phylogeny Inference Package version 3.5. [Google Scholar]
- 8.Lim A., Zhang L. WebPHYLIP: a web interface to PHYLIP. Bioinformatics. 1999;15:1068–1069. doi: 10.1093/bioinformatics/15.12.1068. [DOI] [PubMed] [Google Scholar]
- 9.Perriere G., Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 10.Zmasek C.M., Eddy S.R. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 11.Choi J.H., Jung H.Y., Kim H.S., Cho H.G. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16:1056–1058. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 13.Misener S., Krawetz S.A. Bioinformatics Methods and Protocols. Totowa, NJ: Humana Press; 2000. [Google Scholar]
- 14.Brinkman F.S., Wan I., Hancock R.E., Rose A.M., Jones S.J. PhyloBLAST: facilitating phylogenetic analysis of BLAST results. Bioinformatics. 2001;17:385–387. doi: 10.1093/bioinformatics/17.4.385. [DOI] [PubMed] [Google Scholar]