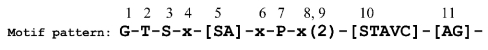Abstract
Sequence signature databases such as PROSITE, which include amino acid segments that are indicative of a protein's function, are useful for protein annotation. Lamentably, the annotation is not always accurate. A signature may be falsely detected in a protein that does not carry out the associated function (false positive prediction, FP) or may be overlooked in a protein that does carry out the function (false negative prediction, FN). A new approach has emerged in which a signature is replaced with a sequence profile, calculated based on multiple sequence alignment (MSA) of homologous proteins that share the same function. This approach, which is superior to the simple pattern search, essentially searches with the sequence of the query protein against an MSA library. We suggest here an alternative approach, implemented in the QuasiMotiFinder web server (http://quasimotifinder.tau.ac.il/), which is based on a search with an MSA of homologous query proteins against the original PROSITE signatures. The explicit use of the average evolutionary conservation of the signature in the query proteins significantly reduces the rate of FP prediction compared with the simple pattern search. QuasiMotiFinder also has a reduced rate of FN prediction compared with simple pattern searches, since the traditional search for precise signatures has been replaced by a permissive search for signature-like patterns that are physicochemically similar to known signatures. Overall, QuasiMotiFinder and the profile search are comparable to each other in terms of performance. They are also complementary to each other in that signatures that are falsely detected in (or overlooked by) one may be correctly detected by the other.
INTRODUCTION
Functionality assignment to proteins is one of the main goals in molecular biology. The classical way to accomplish this involves expansive and time-consuming mutagenesis studies in order to determine the residues comprising the functional site(s). Cumulative experimental data have been documented in databases such as PROSITE (1) and ELM (2), which are commonly used to suggest the function of unannotated proteins [reviewed, e.g. in Ref. (3)]. These databases contain stretches of amino acids, referred to as signatures or motifs, which mark function in proteins. Signatures were derived based on common amino acids in a multiple sequence alignment (MSA) of homologous proteins that share a similar function. However, signature derivation is error prone. For example, the signature of a particular functional site reflects the proteins currently documented as having this site, and a search using the signature might miss a true functional site, even if it is only marginally different from the documented signature. Furthermore, simple scans treat all deviations from the patterns equally; a substitution of leucine with isoleucine is considered equal to a substitution of leucine with aspartate (when both isoleucine and aspartate are not part of the signature). Indeed, stringent searches using the PROSITE signatures often fail to identify the functional sites in proteins, and the PROSITE documentation provides many well-documented cases of such false negative predictions (1).
As sequence databases grow, the simple sequence signatures are being replaced with sequence profiles, i.e. position-specific scoring matrices (PSSMs) and hidden Markov models that are calculated based on MSA of homologous proteins that share similar functions. The approach involves screening of profile databases, such as eMOTIF (4) and eBLOCKs (5), using the sequence of the query protein. The introduction of sequence profiles has led to significant improvements in accuracy and sensitivity compared with the simple search for sequence signatures.
We suggest here a complementary approach that relies on a search against the original signature databases. However, unlike the simple search, the sequence of the query protein is replaced with a search using a family of (multiply aligned) homologous proteins. Our working hypothesis is that highly conserved signatures are more likely to indicate the correct protein function. Thus, the degree of evolutionary conservation of the signature within the protein family is estimated, and this estimate is used as a measure that the likelihood of the signature is indicative of the protein's function. Our results show that the rate of false positive (FP) predictions can be significantly reduced (compared with a simple search) by a search for evolutionarily conserved signatures.
We also show that the rate of false negative (FN) prediction may be reduced by replacing the traditional search for exact signatures with a more permissive search for signature-like segments which are evolutionarily conserved within the protein family.
METHODS
The following is a brief description of the methods. A more detailed description is provided in the ‘Overview’ section at http://quasimotifinder.tau.ac.il/.
Evolutionary conservation
The preferable input for the QuasiMotiFinder web server is an MSA of homologous query proteins. Alternatively, the user can provide the sequence of a single query protein. In such a case, a PSI-BLAST (6) search for homologous sequences in the SWISS-PROT database (7) is carried out. An MSA of the homologous proteins is then built using the CLUSTALW program (8). The latter procedure may involve perturbation of the local BLAST alignment of homologous proteins in favor of CLUSTALW's global alignment. On average, the resultant MSA better reflects the evolutionary history of the homologous proteins than the BLAST alignment, which increases the accuracy of the estimated evolutionary conservation at each amino acid position. However, one may suspect that important information on local sequence motifs is lost this way. The results below demonstrate that, on average, QuasiMotiFinder performs well, even with CLUSTALW-based MSAs.
Evolutionary conservation scores are calculated for each position in the MSA using the maximum likelihood-based algorithm Rate4Site (9). A phylogenetic tree is built from the MSA using the neighbor-joining method (10). The most likely branch lengths are calculated and subsequently used to estimate the evolutionary rate of each amino acid site. These rates are standardized and used as conservation scores. A negative score indicates high conservation, and a positive score indicates a variable residue. The conservation score for a sequence signature or a quasi-signature is an arithmetic mean (average) of the conservation scores of all the residues in the signature. The numerical experiments described below suggest that a sequence motif that is assigned with an average conservation score of ≤−0.445 is likely to be indicative of the protein's function.
Physicochemical similarity
The query sequence is scanned for patterns that resemble PROSITE signatures using a physicochemical amino acid replacement matrix (11). A score is assigned to each pattern in the query sequence, based on its average physicochemical distance from the corresponding signature.
Total score
For each signature at every putative location in the query sequence, the conservation and physicochemical scores are standardized and a total score that amalgamates both is calculated. The total score is calculated as the Pythagorean distance, in the physicochemical-similarity versus evolutionary-conservation plane, between the assigned score and a hypothetical point that represents the ‘ideal’ signature. The ‘ideal’ signature has the highest possible physicochemical resemblance and evolutionary conservation scores.
Statistical analysis
The statistical significance of a putative pseudo-signature is estimated based on its pre-calculated distribution in a population of ∼3000 proteins and domains taken from the Pfam database (12). It is reported in terms of a P-value that takes into account a Bonferroni correction for multiple comparisons (13).
RESULTS AND DISCUSSION
An example: the bovine furin
Subtilases are an extensive family of serine proteases whose catalytic activity is provided by a charge relay system. They appear to have independently and convergently evolved an aspartate–serine–histidine catalytic triad, like that found in the trypsin serine proteases (14). The sequence around the residues involved in the catalytic triad is completely different from that of the analogous residues in the trypsin serine proteases and can be used as specific to that category of proteases.
In the PROSITE database there are three different signatures (motifs) that represent the active site of the subtilase family. Each motif indicates the sequence around one residue of the catalytic triad: the PS00136 entry relates to the sequence around the aspartate, the PS00137 entry relates to the sequence around the histidine and the PS00138 entry relates to the sequence around the serine.
We exemplified a QuasiMotiFinder calculation using the latter motif, which contains 11 residues, the third of which is the catalytic serine (Figure 1). We selected the sequence of bovine furin (SWISS-PROT accession code: FURIN_BOVIN), which appears in the FN list of the PS00138 motif in PROSITE. The protein, which releases mature proteins from their pro-proteins (e.g. albumin and Von Willebrand factor), is known to be part of the subtilase family. However, a normal PROSITE scan detects only two out of the three representative signatures of this family (the PS00136 and PS00137 motifs), overlooking the PS00138 motif.
Figure 1.
PROSITE motif PS00138. The first, second, third and seventh amino acid positions can accommodate only glycine, threonine, serine and proline, respectively. The fifth position can accommodate serine or alanine. The eleventh position can accommodate alanine or glycine. The tenth position can accommodate serine, threonine, alanine, valine or cysteine, and the fourth, sixth, eighth and ninth positions are wildcard residues (x). The serine residue in the third position is part of the catalytic triad.
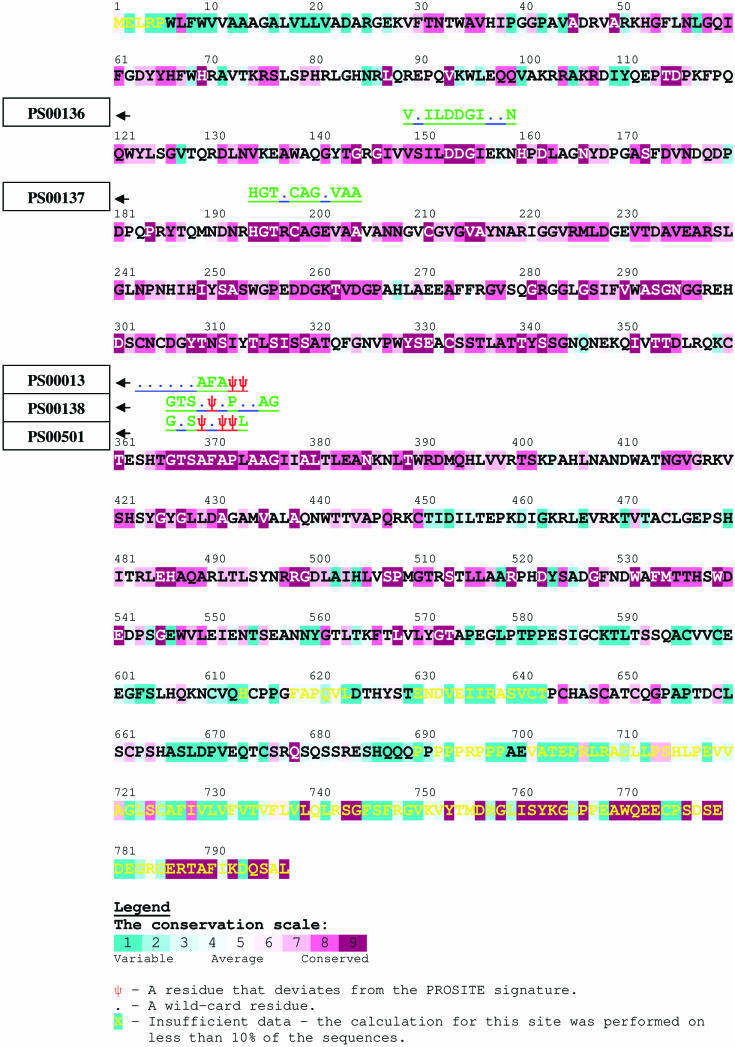
We uploaded the sequence into the QuasiMotiFinder web server and carried out a run with default parameters and all the 99 homologous sequences that were automatically collected. The results are presented in Figure 2. As anticipated, the PROSITE entries PS00136 and PS00137 were detected as strict PROSITE motifs, with average conservation scores of −0.740 and −0.735, respectively. Such low scores are well below the −0.445 cutoff (see below), which is indicative of their significance.
Figure 2.
A QuasiMotiFinder analysis of the bovine furin protein. A part of the output is presented here, and the full output is available as supplementary material at http://quasimotifinder.tau.ac.il/sm_QMF.htm. The query sequence is color-coded by evolutionary conservation (see the color bar), with burgundy-through-turquoise indicating conserved-through-variable residues. Amino acid positions that were occupied with 9 or fewer residues (the rest of the homologous proteins included gaps) are marked in yellow. The inferred evolutionary conservation grade of these positions is unreliable. Residues in the query sequence that are identical to the ones of the PROSITE signature are marked in green above the sequence according to the single-letter code; residues that deviate from the PROSITE signature are marked as ‘Ψ’, and wildcard residues are marked with dots. Three strict PROSITE motifs were detected. Two of them, the PS00136 and PS00137 signatures, are related to the aspartic and histidine residues of the catalytic triad, which is consistent with the biological function of the protein. In addition, the server detected three pseudo-motifs: PS00013, PS00501 and PS00138. The first two differ from their original motifs in two and three positions, respectively, thus suggesting that they are FP hits. The third (residues 366–376) involves a single change compared with the PS00138 motif, which is typical for the furin protein family.
Three statistically significant pseudo-PROSITE-motifs were detected (Figure 2): PS00138, PS00013 and PS00501, all in the vicinity of the same (evolutionarily conserved) sequence segment. The PS00013 and PS00501 motifs differ from their original pattern in two and three positions, respectively. This suggests that they are FP hits, in spite of the fact that they were assigned sufficiently low average conservation scores and P-values.
Of these three quasi-motifs, PS00138, which starts at position 366, was assigned the lowest average conservation score (−0.937; well below the −0.445 cutoff) and the smallest P-value (3.42524 × 10−5). The fifth residue of the pattern, which is listed as serine or alanine according to the original motif, was substituted by a phenylalanine in the bovine furin protein (Figure 2). This is considered to be a radical substitution because of the bulkiness of the phenyl. However, it was found in the bovine furin alone; all the closely related furin orthologs, e.g. from human, mouse and rat, contain a serine residue in the equivalent position.
The outcome of the analysis may be used to revise the definition of the PS00138 motif, such that phenylalanine will be allowed in the fifth amino acid position in addition to the original alanine and serine. The suggested change will eliminate FN sequences, such as the bovine furin, from the PROSITE list. However, it may also increase the rate of FP predictions.
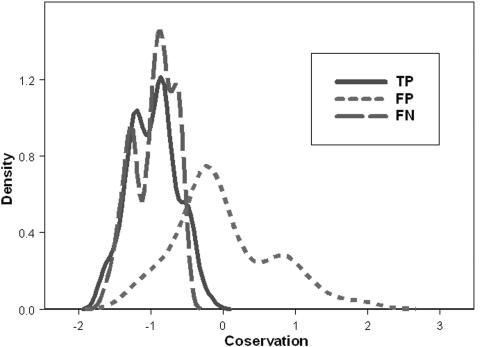
The distribution of evolutionary conservation scores in TP, FN and FP signature predictions
Each sequence signature in the PROSITE database includes a list of proteins with FP, FN and TP predictions (1). We randomly picked 22 of these signatures that have 6 characters or more (Table 1) (as described in Appendix A, Supplementary Material; http://quasimotifinder.tau.ac.il/sm_QMF.htm). These signatures were selected such that each of them includes proteins from each of the three subgroups TP, FP and FN, and such that an MSA for each of the proteins exists in the Pfam database (12). Overall, our test set included 181 proteins: 65 in the TP subgroup, 65 in the FP subgroup and 51 in the FN subgroup. The QuasiMotiFinder web server was used to search for both strict and quasi-PROSITE signatures, and the results are summarized in the density plots of Figure 3. The distributions that were obtained for signatures of proteins in the TP and FN subgroups were very similar, in both their average values and ‘widths’, as they should be. Average evolutionary conservation scores of ∼−1 were obtained for both distributions, and these indicate that, on average, the signatures in these two subgroups are composed of highly conserved residues. This is expected since, in general, the residues in the signatures are important for maintaining the protein's structure and function. In summary, these distributions provide support for our working hypothesis that a true signature known to be indicative of protein function is evolutionarily conserved.
Table 1.
The set of 22 sequence signatures used in the statistical analysis
| PROSITE identifier | Signature description | γi |
|---|---|---|
| PS00485 | Adenosine and AMP deaminase signature | −1.4940 |
| PS00197 | 2Fe-2S ferredoxins, iron–sulfur-binding region signature | 2.7733 |
| PS00636 | dnaJ domains signatures and profile | −2.4318 |
| PS00693 | Riboflavin synthase alpha chain family Lum-binding site signature | −1.3764 |
| PS00147 | Arginase family signatures | 1.8978 |
| PS00152 | ATP synthase alpha- and beta-subunits signature | −1.5161 |
| PS00043 | Bacterial regulatory proteins, gntR family signature | −0.9389 |
| PS00104 | EPSP synthase signatures | −0.6271 |
| PS00453 | FKBP-type peptidyl–prolyl cis–trans isomerase signatures/profile | −1.7875 |
| PS00178 | Aminoacyl-transfer RNA synthetases class-I signature | −1.0039 |
| PS00227 | Tubulin subunits alpha, beta and gamma signature | 4.5902 |
| PS00296 | Chaperonins cpn60 signature | −1.3363 |
| PS00061 | Short-chain dehydrogenases/reductases family signature | 0.7331 |
| PS00036 | Basic-leucine zipper (bZIP) domain signature and profile | 0.2328 |
| PS00559 | Eukaryotic molybdopterin oxidoreductases signature | −1.5359 |
| PS00287 | Cysteine protease inhibitors signature | −2.3378 |
| PS00107 | Protein kinases ATP-binding region signature | −0.0815 |
| PS00118 | Phospholipase A2 histidine active site | 2.3733 |
| PS00283 | Soybean trypsin inhibitor (Kunitz) protease inhibitors family signature | −0.7184 |
| PS00606 | Beta-ketoacyl synthases active site | −0.0164 |
| PS00697 | ATP-dependent DNA ligase AMP-binding site | 2.0708 |
| PS01228 | Hypothetical cof family signatures | 0 |
γi is the value of the coefficient associated with the signature in the logistic model of Equation 1; PS01228 was selected as a reference and its coefficient was set to zero.
Figure 3.
The distribution of the conservation scores within the three subgroups: true positive (TP, solid line), false negative (FN, dashed line) and false positive (FP, dot–dashed line).
In contrast, the distribution that was obtained for falsely predicted signatures from proteins in the FP subgroup is significantly different from those of the TP and FN subgroups. The mean conservation score obtained for the FP distribution was close to zero, which indicates that, on average, the ‘signatures’ in this subgroup are composed of residues of moderate conservation. The FP distribution is also much broader than the TP and FN distributions.
We carried out statistical analysis in order to further characterize the differences between the three distributions of Figure 3. A two-sided comparison of the conservation score between the proteins of the TP and FP subgroups, using the Wilcoxon signed rank test (13), indicated that the distributions are significantly different from each other in their mean conservation scores, with P-value < 0.0001. Further examination showed that the subgroups also differ from each other in the variance among the conservation scores; P-value < 0.001 was obtained using both a two-sided Wilcoxon signed rank test and a Levene test for equality of variance (13). Similar tests showed that the FN and FP subgroups are significantly different from each other in the average and variance of the conservation scores (P-value < 0.001, P-value = 0.0005 and P-value < 0.001, respectively).
The same tests were applied for comparison of the TP and FN subgroups. Both the conservation score and the variance among the proteins in each subgroup were not significantly different (P-value = 0.8391, P-value = 0.7136 and P-value = 0.31, respectively).
Overall, Figure 3 and our statistical analysis demonstrate that the conservation score assigned to a signature can be used to estimate the likelihood that it is indicative of the protein's function.
Signatures with average conservation of ≤−0.445 are likely to be indicative of the protein's function
The CART (Classification And Regression Tree) program using the GINI index in combination with cross-validation techniques (15) was used to build the optimal decision tree for discrimination between the TP and FP subgroups. An optimal tree of one node and two leaves was obtained. The decision rule in the node was as follows: if the signature's average conservation score is smaller than −0.445, decide TP; otherwise, decide FP. The misclassification error rate of the tree was about 0.14 (18 of 130 proteins) and the deviance was 0.508.
Logistic models can be used to distinguish between true and false signature assignments
A logistic model was created to distinguish between the TP and the FP subgroups. After examining all the recorded parameters and by the use of analysis of deviance while comparing only hierarchal models of all the logistic models that were tested, the model that appeared to be most appropriate included two main parameters: the conservation score and the pattern. According to the model, the probability (q) of a pattern to belong to the FP subgroup is given by
| 1 |
where C is the pattern's average conservation score and Mi (i = 1,2,…,21) is an index that is associated with the signature's identity. Mi equals 1 when the pattern type is i; it equals −1 when the pattern is PS01228 (as this pattern was arbitrarily chosen to be the reference) and equals zero otherwise. The coefficients α = 2.819, β = 7.058 and γi (Table 1) were derived to optimize the discriminating power of the model; that is, they maximize the distinction between entries in the TP and FP subgroups. An analysis of the relative contributions of the different descriptors, based on deviants (13), showed that the average evolutionary conservation score (C) is the most important factor in the model and that the set of signature-specific coefficients (Mi) was secondary.
After optimizing the model on the discrimination between the TP and the FP subgroups, we tested its performance on entries from the FN subgroup. Based on the similarity between the FN and TP subgroups, we expect that most of the former entries will be assigned Q-value of ≤0.5. Indeed, our examination showed that 45 of the 51 entries in this subgroup (∼88%) had q < 0.5; that is, they were more likely to be true rather than false signatures. Moreover, 37 (∼73%) of these entries were likely to be true signatures within a 95% confidence interval.
QuasiMotiFinder is similar in performance and complementary to eMOTIF
We compared the performance of QuasiMotiFinder with that of the eMOTIF server (4). A new set of proteins from the TP, FP and FN lists in PROSITE was compiled for this purpose. From each subgroup, 30 sequence signatures were selected at random, and a protein that was not used in the previous analysis was chosen by chance from the corresponding list. Each of these 90 proteins was analyzed using the eMOTIF and QuasiMotiFinder web servers. The outcome of the analysis included an indication of whether the protein contains the right sequence motif. Suitable thresholds were automatically used to segregate between proteins that did and did not contain the motif. In the case of QuasiMotiFinder, the average evolutionary conservation of the motif among the protein and its homologs was calculated, and the −0.445 cutoff value was used as a threshold. The eMOTIF server assigns a probability to each detected motif of appearing in the protein. This probability was used to segregate the motifs; a value of ≥50% was taken to indicate that the protein contains the motif. The results are summarized in Table 2.
Table 2.
Comparison of the performance of the QuasiMotiFinder (QMF) and the eMOTIF web servers
| TP | FP | FN | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Present | Absent | Total | Present | Absent | Total | Present | Absent | Total | |
| QMF | 29 | 1 | 30 | 6 | 24 | 30 | 23 | 7 | 30 |
| eMOTIF | 25 | 5 | 30 | 1 | 29 | 30 | 24 | 6 | 30 |
The three subgroups are marked as TP, FP and FN. The number of proteins with correctly detected motifs is listed in the ‘Present’ column in each subgroup. The number of proteins whose motifs were overlooked is listed in the ‘Absent’ column. The total number of proteins in each subgroup is listed in the ‘Total’ column.
Table 2 demonstrates that the difference in performance between the two servers is insignificant (TP: P-value = 0.19, FP: P-value = 0.1028, FN: P-value = 0.8339, total number of errors versus the total number of correct results for all groups: P-value ≈ 1). A closer analysis of the FN subgroup revealed that, even though the lists of motifs detected by QuasiMotiFinder and the eMOTIF servers are similar to each other in their numbers of items, they differ in the identity of the motifs. For example, the interleukin-related motif PS00253 of the protein IL-1F7 (SWISS-PROT accession code: IL1F7_HUMAN) was overlooked by the eMOTIF search but was correctly found by QuasiMotiFinder. Thus, these two web servers appear to be complementary to each other.
CONCLUSIONS
Motif pattern searches are complementary to the more widely used profile/hidden Markov model methods. In the right context, they can be very effective, e.g. Bork et al. analysis of convergent evolution in the α/β-barrel subclass (16). A new motif pattern search method was presented here. The results (Figure 3) convincingly demonstrate the capacity of the method to reduce the rates of FP and FN predictions compared with a traditional search for strict signatures in the sequence of a single protein, without taking into account its homologs. The analysis also shows that the method is complementary to searches against databases of sequence profiles. The permissive search in QuasiMotiFinder is beneficial in that it allows the detection of signature-like patterns that are often indicative of the protein's function. However, at times, it may lead to errors. For example, in cases where the signature includes residues that are involved in catalysis, even minor alterations may not be allowed, and the detected quasi-signature may be meaningless. Thus, the quasi-signatures should best be regarded as suggestions for further analysis in view of our knowledge about the signature and the protein. The method, which was implemented in a web server (http://quasimotifinder.tau.ac.il/), can be used to guide experiments for the determination of protein function.
The current version of QuasiMotiFinder is based on sequence signatures that were taken from PROSITE. In the future, more motifs, e.g. from the recently established ELM database (2), will be added. We plan to further develop the server to include profiles, e.g. from the PROSITE and the eMOTIF databases. We will also add the option to graphically display the phylogenetic tree online and to repeat the calculations using clades (sub-trees). In addition, we plan to develop tools to aid in the detection of change and loss-of-function in protein families. Such cases are likely to be characterized by changes in amino acids of the sequence motif that is associated with the given function.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Allegra Via, Burkhard Rost and David Steinberg for helpful discussions, and to Eyal Privman and the Bioinformatics Service Unit at the George S. Wise Faculty of Life Sciences at Tel Aviv University for providing technical assistance and computational facilities. This work was supported by the European Commission FP6 Integrated Project EUROHEAR, LSHG-CT-20054-512063. Funding to pay the Open Access publication charges for this article was provided by N.B-T., Tel Aviv University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D.M.A., Ausiello G., Brannetti B., Costantini A., et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonassen I. Methods for discovering conserved patterns in protein sequences and structures. In: Higgins D., Taylor W., editors. Bioinformatics: Sequence, Structure and Databanks. NY: Oxford University Press; 2000. [Google Scholar]
- 4.Huang J.Y., Brutlag D.L. The EMOTIF database. Nucleic Acids Res. 2001;29:202–204. doi: 10.1093/nar/29.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Q.J., Lu L., Saxonov S., Brutlag D.L. eBLOCKs: enumerating conserved protein blocks to achieve maximal sensitivity and specificity. Nucleic Acids Res. 2005;33:D178–D182. doi: 10.1093/nar/gki060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., Pilbout S., Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 10.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 11.Miyata T., Miyazawa S., Yasunaga T. Two types of amino acid substitutions in protein evolution. J. Mol. Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- 12.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conover W.J. Practical Nonparametric Statistics. NY: Wiley; 1980. [Google Scholar]
- 14.Siezen R., Leunissen J. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L., Friedman J.H., Olshen R.A., Stone C.J. Classification and Regression Trees. NY: Chapman & Hall; 1984. [Google Scholar]
- 16.Bork P., Gellerich J., Groth H., Hooft R., Martin F. Divergent evolution of a beta/alpha-barrel subclass: detection of numerous phosphate-binding sites by motif search. Protein Sci. 1995;4:268–274. doi: 10.1002/pro.5560040213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.