Summary
Glycemic traits are critical indicators of maternal and fetal health during pregnancy. We performed genetic analysis for five glycemic traits in 14,744 Chinese pregnant women. Our genome-wide association study identified 25 locus-trait associations, including established links between gestational diabetes mellitus (GDM) and the genes CDKAL1 and MTNR1B. Notably, we discovered a novel association between fasting glucose during pregnancy and the ESR1 gene (estrogen receptor), which was validated by an independent study in pregnant women. The ESR1-GDM link was recently reported by the FinnGen project. Our work enhances the findings in East Asian populations and highlights the need for independent studies. Further analyses, including genetic correlation, Mendelian randomization, and transcriptome-wide association studies, provided genetic insights into the relationship between pregnancy glycemic traits and hypertension. Overall, our findings advance the understanding of genetic architecture of pregnancy glycemic traits, especially in East Asian populations.
Keywords: pregnancy glycemic traits, gestational diabetes mellitus, genome-wide association study, ESR1, Mendelian randomization
Graphical abstract
Highlights
-
•
Glycemic traits GWAS involving 14,744 Chinese pregnant women identify 25 genetic loci
-
•
Newly discovered ESR1 association provides insights into maternal glucose levels
-
•
Highlighted a potential causal relationship between hypertension and glycemic traits
Zhu et al. analyzed 14,744 Chinese pregnant women, identifying 25 genetic associations with glycemic traits via GWAS. They discovered a novel link between fasting glucose and the ESR1 gene and validated this by an independent study. Further analyses provided a genetic insight into the relationship between pregnancy glycemic traits and hypertension.
Introduction
Pregnancy glycemic traits (e.g., fasting glucose) play a critical role in assessing and managing maternal health during pregnancy and serve as indicators of glucose metabolism and gestational diabetes mellitus (GDM).1 Understanding the genetic basis of pregnancy glycemic traits may provide insights into the underlying genetic factors influencing maternal glucose regulation during pregnancy. To date, numerous genome-wide association studies (GWASs) have identified candidate genetic variants associated with glycemic traits in normal populations.2,3,4 However, there has been limited research on glycemic traits during pregnancy, with only a few studies exploring this area, especially in the Chinese population.5,6,7
Clinical studies have focused on potential risk factors and adverse outcomes associated with abnormal glucose levels during pregnancy. In general, maternal age, obesity, polycystic ovary syndrome, and previous history of hyperglycemia increase a woman’s susceptibility to developing GDM.8,9,10 Elevated maternal blood glucose levels lead to increased fetal growth, causing macrosomia, infants born large for their gestational age, and a higher likelihood of cesarean section.11,12,13 Moreover, hyperglycemia during pregnancy is often associated with other complications during and after pregnancy, notably hypertension.11,14 However, the genetic architecture underlying the relationship between pregnancy glycemic traits and hypertension has not been fully explored.
Non-invasive prenatal testing (NIPT) has emerged as an effective approach to reduce the birth rate of children with fetal autosomal trisomies.15 NIPT involves sequencing cell-free DNA fragments, including fetal cell-free DNA, obtained from maternal peripheral plasma utilizing next-generation sequencing technology. Compared to the traditional genetic research pipeline of “sampling, sequencing, and analyzing,” using NIPT data significantly reduces both sequencing time and research costs, as genotype data are generated alongside clinical prenatal tests. Additionally, with the growing popularity of NIPT technology worldwide, it has generated an immense amount of genotype data, nearly ten times that of whole-genome sequencing samples.16 Previously, we demonstrated the validity of using large-scale NIPT sequencing data for population genetics and GWASs.17,18
In this study, we investigate the genetic basis of five pregnancy glycemic traits using NIPT sequence data, which include three glucose levels based on the oral glucose tolerance test (OGTT), the area under the curve of these three measurements, and GDM. In total, we identified 25 locus-trait associations, including several well-known examples such as the association of GDM with the genes CDKAL1 and MTNR1B, and fasting glucose levels with the gene PCSK1. A significant new association was discovered between fasting glucose levels during pregnancy and the ESR1 gene (estrogen receptor). This finding was subsequently confirmed by an independent study in pregnant women. Notably, the ESR1-GDM association was recently reported by the FinnGen project.19 Our work not only enriches the findings specific to East Asian populations but also underscores the necessity for independent studies. Additional analyses, including genetic correlation, Mendelian randomization (MR), transcriptome-wide association study (TWAS), and drug target enrichment analysis (DTEA), have provided deeper genetic insights into the relationship between glycemic traits during pregnancy and hypertension. These findings offer insights into the genetic landscape of pregnancy glycemic traits, particularly within East Asian populations.
Results
Participants and data types
In this study, we used two data types: whole-genome sequence (WGS) data from the NIPT test and pregnancy glycemic traits from the OGTT test. For participants with NIPT WGS data, we included those with a sequencing depth greater than 0.05 and a mapping rate above 90%, resulting in a cohort of 38,668 individuals. All were used in genotype imputation to ensure the highest possible accuracy. Additionally, there were 21,813 subjects who underwent the OGTT and had glycemic measurements. We employed all 21,813 subjects for subsequent regression analysis involving other clinical phenotypes. For the GWAS analysis, we used participants who possessed both NIPT sequence data and glycemic data, totaling 14,889 individuals (Figure S1A).
For the 38,668 individuals with NIPT sequence data, we assessed gestational age at the time of the NIPT test and fetal fraction in maternal plasma (Figure S1B), as provided in the NIPT reports. Specifically, gestational age information was available for 28,037 participants, with an average of 16.16 weeks (113.13 days). Additionally, fetal fraction data were obtained for 15,837 participants, with an average of 9.76%, aligning with our literature review (STAR Methods). For the 21,813 subjects with pregnancy glycemic traits, we provided distributions of maternal information and birth outcomes (Figures S1C–S1D). Maternal characteristics included age (29.58 ± 4.03 years), BMI (21.39 ± 3.08 kg/m2), first-time pregnancy status, and hypertension. Birth outcomes included gestational age (38.74 ± 1.58 weeks), birth weight (3286.43 ± 473.71 g), delivery type, and newborn gender.
Pregnancy glycemic traits
Applying the criteria of 0hGlu ≥ 5.1 mmol/L, 1hGlu ≥ 10.0 mmol/L, and 2hGlu ≥ 8.5 mmol/L, 2,502 out of 21,813 pregnant women were diagnosed with GDM, resulting in a disease rate of 11.47% (Figure S2A). This rate is slightly lower than the 14.8% reported in a 2019 meta-analysis conducted in mainland China.20 After integrating NIPT sequence data, we retained 14,744 subjects, among which 1,619 were GDM cases, and 13,125 were controls. This subset was used for the GWAS analysis. For the 0hGlu, 1hGlu, and 2hGlu measurements, the available sample sizes for the GWAS were 14,889, 14,790, and 14,760, respectively. We provided the distributions of the three glucose levels among the GDM case and control groups (Figure S2A).
Additionally, we computed the area under the curve (AUC) for the three OGTT glucose levels, which served as a metric for glucose tolerance during pregnancy and was denoted as GDMAUC (Figure S2B). The sample size for the GWAS of GDMAUC was 14,726.
Regression analysis between GDM and multi-phenotypes
Using a significance threshold of 5.15E−4, univariate regression analysis identified 26 risk factors for GDM and 29 for GDMAUC (Tables 1 and S1 and Figure S2C). Among the maternal information, age was most significant (for GDM: odds ratio [OR] = 1.112 and p value = 4.19E−98, for GDMAUC: beta = 0.135 and p value = 5.20E−283), followed by BMI, multiparity, and hypertension. Among the 93 early-stage laboratory tests, the most significant features were blood glucose (GLU), prealbumin (PA), γ-glutamyl transferase (GGT), urine glucose (GLU_U), and uric acid, being correlated with GDM in previous studies.21,22,23,24,25,26,27,28,29 In the multivariate regression analysis, to ensure an effective sample size of 3,000 for each variable, we tested 20 out of 26 factors for GDM and 24 out of 29 for GDMAUC, identifying five and nine significant risk factors, respectively (Tables S2 and S3). Notably, age was the only significant maternal factor for GDM multivariately (p value = 2.78E−10).
Table 1.
Regression results for GDM/GDMAUC and risk factors and outcomes
| Traits info |
Mean ± std./n (%) |
Regression results of GDM |
Regression results of GDMAUC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | Traits | All samples | In GDM control | In GDM case | n | OR | Std. | p value | n | Beta | Std. | p value |
| Risk factors | age (years) | 29.58 ± 4.03 | 29.37 ± 3.94 | 31.20 ± 4.32 | 21,808 | 1.112 | 0.005 | 4.19E−98 | 21,786 | 0.135 | 0.004 | 5.20E−283 |
| BMI (kg/m2) | 21.39 ± 3.08 | 21.22 ± 2.98 | 22.67 ± 3.45 | 19,161 | 1.143 | 0.007 | 3.42E−87 | 19,140 | 0.159 | 0.005 | 3.04E−199 | |
| first-time pregnancy, n (%) | 7,769 (55.73%) | 7,176 (57.09%) | 593 (43.25%) | 13,941 | 0.573 | 0.057 | 2.98E−22 | 13,931 | −0.539 | 0.038 | 3.36E−46 | |
| hypertension, n (%) | 308 (1.41%) | 233 (1.21%) | 75 (3.00%) | 21,813 | 2.530 | 0.134 | 5.12E−12 | 21,791 | 1.143 | 0.129 | 1.06E−18 | |
| Outcomes | gestational weeks (weeks) | 38.74 ± 1.58 | 38.78 ± 1.55 | 38.37 ± 1.74 | 21,808 | 0.662 | 0.033 | 8.84E−35 | 21,786 | −0.077 | 0.005 | 1.88E−60 |
| birth weight (g) | 3,286.43 ± 473.71 | 3,287.08 ± 464.74 | 3,281.42 ± 537.96 | 21,672 | 0.003 | 10.096 | 5.75E−01 | 21,650 | 4.475 | 1.424 | 1.68E−03 | |
| low birthweight, n (%) | 979 (4.52%) | 821 (4.28%) | 158 (6.35%) | 21,672 | 1.517 | 0.090 | 3.26E−06 | 21,650 | 0.069 | 0.013 | 1.48E−07 | |
| giant baby, n (%) | 1,054 (4.86%) | 889 (4.63%) | 165 (6.63%) | 21,672 | 1.462 | 0.088 | 1.43E−05 | 21,650 | 0.100 | 0.012 | 1.37E−16 | |
| c-section, n (%) | 11,789 (54.10%) | 10,225 (53.01%) | 1,564 (62.56%) | 21,790 | 1.481 | 0.044 | 2.76E−19 | 21,768 | 0.096 | 0.006 | 4.11E−52 | |
| newborn gender, n (%) | 10,418 (48.07%) | 9,218 (48.05%) | 1,200 (48.25%) | 21,673 | 1.008 | 0.043 | 8.47E−01 | 21,651 | 0.005 | 0.006 | 3.89E−01 | |
Notes: in the columns of “All sample,” “In GDM control” and “In GDM case,” for quantitative traits, we provide the mean and standard deviation of each variable; for dichotomous traits, we provide the number of positive samples for the corresponding trait in each group and the percentage of positive samples to the total sample size in that group.
Women with GDM tend to have shorter gestational lengths (OR = 0.662, p value = 8.84E−35), undergo cesarean sections at higher rates (OR = 1.481, p value = 2.76E−19), and deliver a low-birth-weight (OR = 1.517, p value = 3.26E−06) or macrosomic infant (OR = 1.462, p value = 1.43E−05) (Table 1). The gender of newborn is not a significant birth outcome with GDM (OR = 1.008, p value = 0.847). At a 3.33E−03 (= 0.05/15) threshold, four diagnoses were significantly associated with GDM and seven with GDMAUC, including pregnancy with uterine scar and intrauterine distress (Table S4). Additional diagnoses significantly associated with GDMAUC were eclampsia (t = 5.272, p value = 1.37E−07), pregnancy with vomiting (t = −3.924, p value = 8.75E−05), and gestational hypertension (t = 3.795, p value = 1.48E−04) (Figure S2D). Numerous studies have reported positive associations between preeclampsia/eclampsia and GDM.14,30,31
Genotype imputation performance
Note that, from December 2018, the sequencing depth of the NIPT product was increased. We excluded 1,719 samples with unknown test dates, resulting in 10,795 pregnant women with an average depth of 0.1049× before December 2018 and 26,154 participants with an average depth of 0.1644× afterward, with an overall average depth of 0.1525× (Figure S3A).
After STITCH imputation (STAR Methods), we obtained a total of 8,134,302 SNPs. Filtering out SNPs with a Hardy-Weinberg equilibrium (HWE) p value < 1E−6 and minor allele frequency (MAF) < 0.05 resulted in 2,818,480 high-quality SNPs. We assessed imputation accuracy of these high-quality SNPs using Pearson’s correlation between the imputed genotype dosage and the original high-depth genotype of 30 Chinese samples from the 1000 Genomes Project (1KGP). On average, the imputation accuracy R was 87.61%, and R2 was 79.34% (Figure S3B). More imputation evaluation results, including additional datasets and results at the specific SNP level, are described in our companion work.32 Additionally, we computed the Pearson’s correlation (R2) for the frequency of non-reference alleles of these high-quality SNPs in our dataset and the East Asian samples from the 1KGP (Figure S3C). The R2 of 0.97 indicates a high consistency of our imputed variants with those in the 1KGP.
We utilized BaseVar for population variation detection and conducted principal component analysis (PCA) on the identified variants to reveal population structure (STAR Methods). Additionally, we acquired population structure information derived from the PCA plot of 585 samples representing 30 administrative divisions in China. These divisions were further categorized into seven main geographical regions spanning from north to south China. These samples were independently recruited in a study approved by the institutional review board at BGI (BGI-IRB 21163) and authorized by the Human Genetic Resources Administration of China ([2022] CJ0197). We projected the principal components plot of the main data onto the reference data and presented the plot in Figure S3D. Notably, the main samples exhibited a significant overlap with samples from the Huazhong area, which corresponds to central China and aligns with the location of Wuhan.
GWAS analysis of glycemic traits
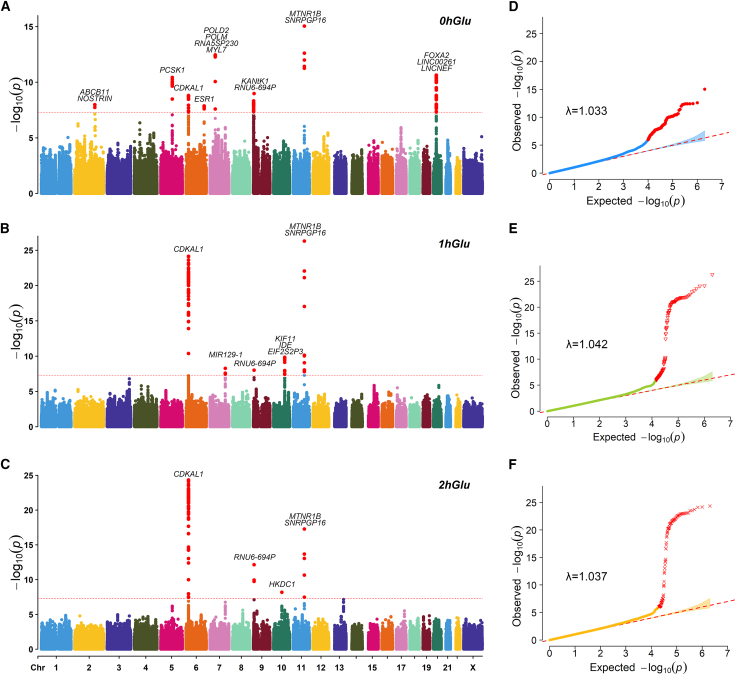
We performed GWAS analysis for three OGTT traits, GDM, and GDMAUC (Figures 1 and 2 and Table S5). The full GWAS summary statistics for these traits are available on our MANE (Maternal and Neonatal) PheWeb website.33 In total, we identified 25 trait-locus associations, with many previously associated either with pregnancy glycemic traits (e.g., CDKAL1, MTNR1B, and PCSK1)6 or with glucose levels in the general population (e.g., POLD2).34,35,36
Figure 1.
The GWAS results of three OGTT measurements
(A–C) Manhattan plots of three OGTT measurements; SNPs that pass the genome-wide threshold (5E−8) were colored in red.
(D–F) QQ-plots of three OGTT measurements with genomic inflation factor λ.
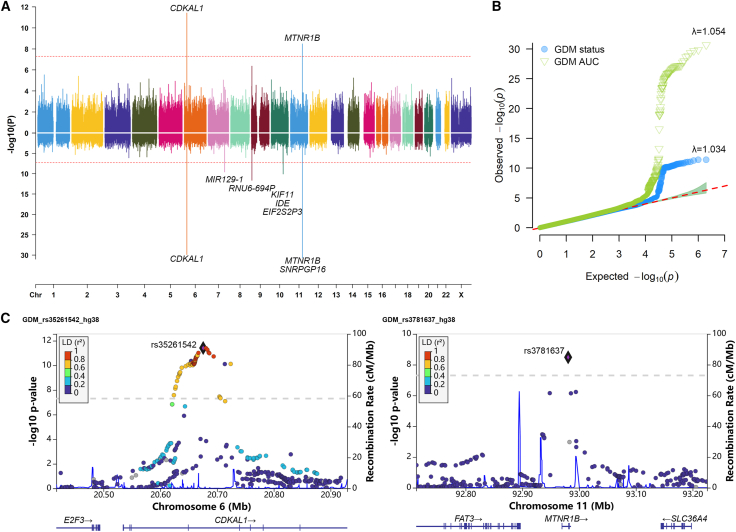
Figure 2.
The GWAS results of GDM status and GDMAUC
(A) Mirrored Manhattan plot indicates the GWAS results of GDM status and GDMAUC, located on top and bottom, respectively; the red dashed line represents the genome-wide significance threshold (5E−8).
(B) QQ plot for GDM status and GDMAUC with genomic inflation factor λ.
(C) Regional plots of two loci CDKAL1 and MTNR1B for testing GDM.
Two genes, CDKAL1 and MTNR1B, were consistently associated with all five traits, as previously reported to be associated with GDM.5,7,19,37,38 CDKAL1 has been associated with proinsulin conversion defect and glucose-stimulated insulin response defect.39,40 MTNR1B (melatonin receptor 1B) encodes one of the two high-affinity forms of a receptor for the hormone melatonin, playing a role in glucose homeostasis.41 For GDM status, the lead SNPs mapped on the two genes were rs35261542 (CDKAL1, p value = 3.88E−12) and rs3781637 (MTNR1B, p value = 3.29E−09), respectively. In Table S6, we provided details on the two SNPs and known GDM-associated SNPs. Notably, rs35261542 was significant in the mixed population study,37 while rs3781637 has not previously been reported in association with GDM. Additionally, allele frequency of rs3781637 significantly differed between East Asian and European populations.
The heritabilities (standard error) for 0hGlu, 1hGlu, 2hGlu, GDM, and GDMAUC were 0.1308 (0.0372), 0.1395 (0.04), 0.0611 (0.04), 0.0817(0.0977), and 0.1429 (0.0421), respectively.
Validation study
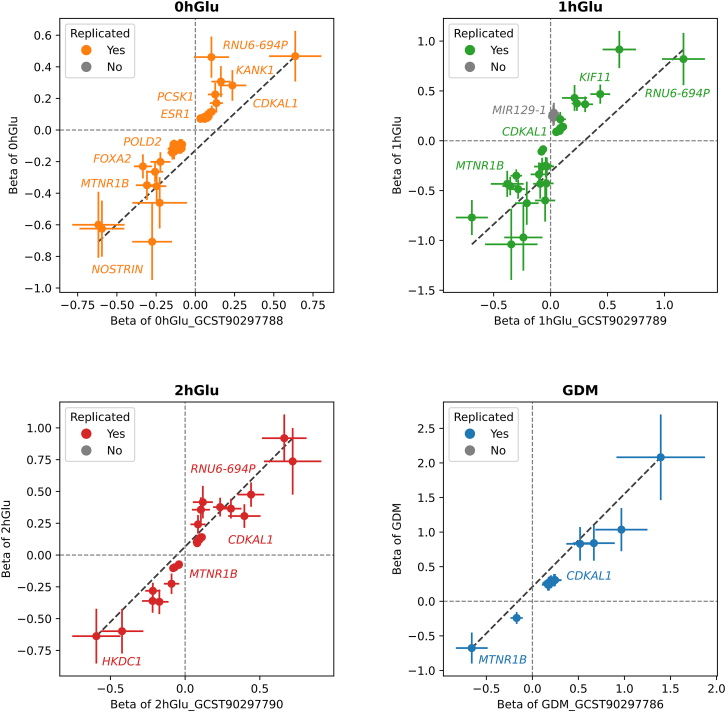
Here, we selected a recently published study involving 22,882 Chinese pregnant women from Shenzhen Bao’an Women’s and Children’s Hospital,7 referred to as the Shenzhen cohort. This cohort underwent the same OGTT protocol as our study participants. We downloaded their GWAS results of fasting glucose, 1-h and 2-h glucose levels after consuming 75 g of sugar, and GDM status from the GWAS catalog.
In detail, 20 out of the 25 trait-locus associations were subject to replication, as 5 GDMAUC-associated loci were removed because the Shenzhen cohort did not test for GDMAUC. For each of the 20 trait-locus associations, we examined a flanking window of 500 kb in the Shenzhen cohort. An association was considered replicated if at least one SNP within this window had a p value less than 1E−4 and a linkage disequilibrium (LD) r2 greater than 0.1. Among the 20 associations, 19 were successfully replicated, with the sole exception being 1hGLU-MIR129-1 (Figure 3). The gene MIR129-1 has previously been reported to be associated with type 2 diabetes in East Asian populations.42,43 The replication study underscores the robustness of our GWAS findings on the pregnancy glycemic traits.
Figure 3.
The comparison between the GWAS results in this study and the replication study
The forest plots between this study and the replication study, for 0hGlu, 1hGlu, 2hGlu, and GDM, respectively; the position of the dots represents the beta values of the two studies, and the lines represent the range of one standard deviation above and below; the successfully replicated variants in both studies are marked in color, or otherwise are marked in gray.
A novel gene ESR1
We identified ESR1 as a candidate gene associated with fasting glucose (0hGlu), with lead SNP rs3020430 (p value = 1.36E−08). Recently, the FinnGen project, with 12,332 GDM cases and 131,109 parous female controls, identified ESR1 as a novel gene associated with GDM through GWAS, with rs537224022 as the lead SNP.19 The MAF of this SNP is 0.006 in European populations and undetectable (0.00) in East Asian populations, which explains its absence in our dataset.
The gene ESR1 encodes an estrogen receptor and ligand-activated transcription factor, which regulates estrogen-inducible genes that play a role in growth, metabolism, sexual development, gestation, and other reproductive system.44 To validate our finding of the effects of ESR1 on fasting glucose, we performed a GWAS-GWAS colocalization analysis45 with the Shenzhen cohort7 using the coloc::coloc.abf46 in R. In the Shenzhen cohort’s GWAS summary of fasting glucose, the lead SNP in the ESR1 gene is rs9322351, with a p value of 6.91E−06 (in LD with rs3020430, r2 = 0.4008, in the 1KGP EAS samples, GRCh38). The posterior probability of H4 (indicating one common causal variant) was 0.84, surpassing the threshold of 0.75, signifying a shared causal signal. We employed the R function locuscompare47 to generate regional plots (Figure S4). Additional literature has reported associations between ESR1 and pregnancy, such as its link to susceptibility to recurrent pregnancy loss in a Tunisian study,48 as well as estrogen dysregulation contributing to preeclampsia occurrence.49,50
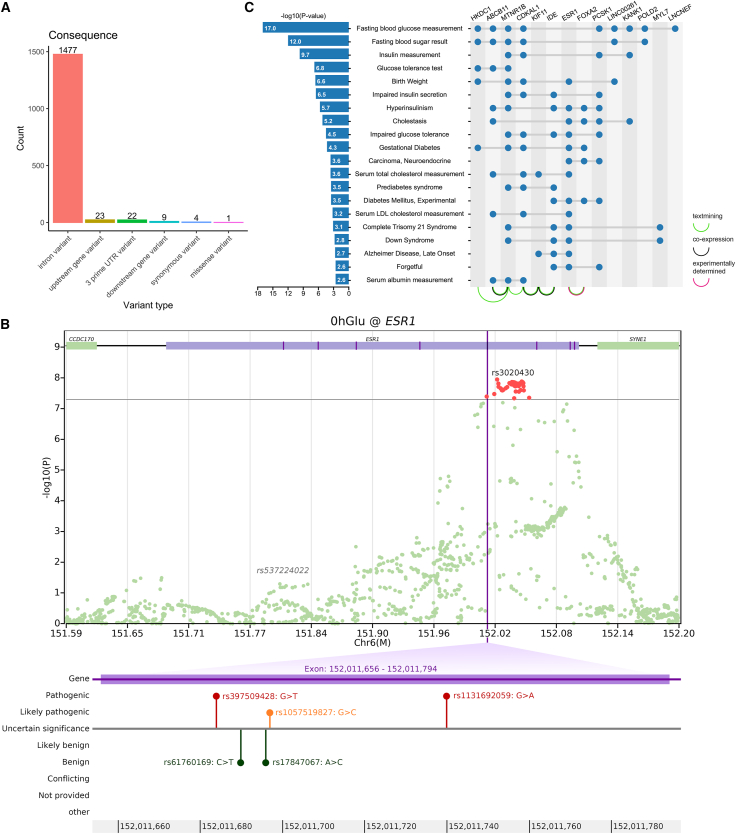
Upon analyzing all 1,536 SNPs within the ESR1 gene from our dataset, we conducted annotation analysis to glean functional insights for each mutation site (Figure 4A). Notably, 96.2% of the SNP sites are situated within intron regions, lacking direct involvement in protein encoding. Upon further investigation beyond our dataset to encompass all SNPs in ESR1, we observed a region within the ESR1 association peak that falls within exon 5 (Figure 4B). This region harbors three pathogenic/likely pathogenic variants (rs397509428, rs1131692059, and rs1057519827) according to the ClinVar database,51 with linked diseases encompassing estrogen resistance syndrome and breast neoplasms.52,53 We hypothesize that these SNPs may exert influence on glycemic levels during pregnancy by modulating estrogen resistance and interacting with insulin levels.54,55,56
Figure 4.
The GWAS result and annotation information on ESR1 and the results of gene set enrichment analysis
(A) The bar plot showing the variant count of each category for functional annotation on ESR1.
(B) The regional plot for 0hGlu on ESR1, with the fifth exon and potential pathogenic variants indicated below.
(C) The results of the gene set enrichment analysis for all genes correlated to glycemic traits, the −log10 p values are displayed on the left side using a bar plot, the genes corresponding to each pathway are marked with blue dots on the right side, and the interactions between each gene are indicated with curves below.
Gene set enrichment analysis
A total of 21 mapped genes associated with pregnancy glycemic traits were identified, including 14 protein-coding genes, 4 pseudogenes, and 3 non-coding RNAs (Table S5). Since PPI (protein-protein interactions) analysis excludes pseudogenes and non-coding RNAs, the 14 protein-coding genes were included in the STRING PPI analysis (Figure 4C). The majority of PPIs were derived from text mining, with a minority being co-expressed and one interaction experimentally determined. Among the gene pairs potentially influencing pregnancy glycemic levels through co-expression are MTNR1B and ABCB11, CDKAL1 and KIF11, as well as KIF11 and IDE. Notably, an experimentally determined interaction was observed between ESR1 and FOXA2. Studies using a Foxa2 knockout mouse model have demonstrated that Foxa2 regulates the estrogen responsiveness and influences ESR1 transcriptional activity in the uterus.57
At a significance threshold of p value <0.01, requiring a minimum gene count of 3, and an enrichment factor >1.5, the gene set enrichment analysis analysis identified enrichment in 20 functional pathways (Figure 4C). The top five pathways included fasting blood glucose measurement, fasting blood sugar result, insulin measurement, glucose tolerance test, and birth weight. Notably, genes such as HKDC1, MTNR1B, CDKAL1, ESR1, and LINC00261 were found in the birth weight pathway. Additionally, the pathway associated with gestational diabetes featured genes including HKDC1, MTNR1B, CDKAL1, ESR1, and FOXA2. Both the birth weight and the gestational diabetes pathway involved gene ESR1, suggesting its essential role in maternal and fetal conditions during pregnancy.
Genetic correlation analysis
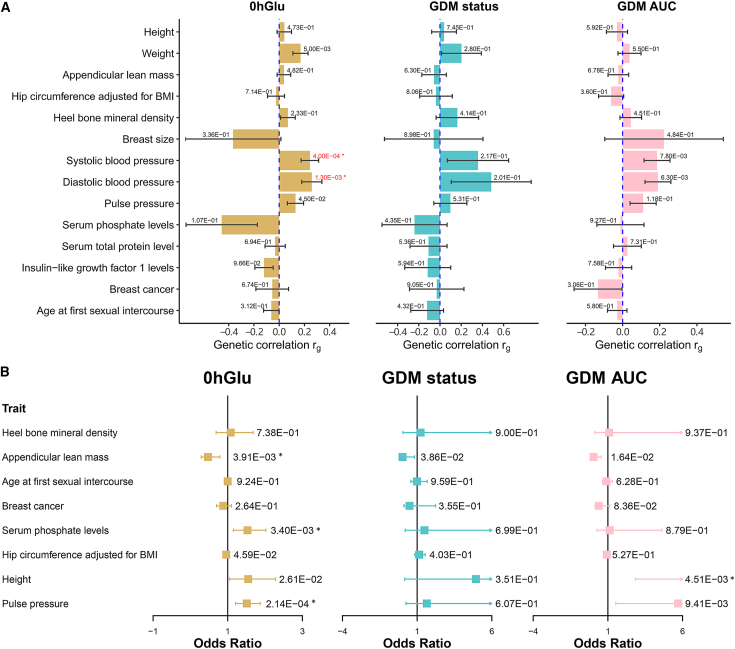
Building upon our earlier discovery that ESR1 is a gene associated with pregnancy glycemic traits and considering the prominence of hormone signaling revealed in the pathway-based analysis, we delved deeper into the association between ESR1 and pregnancy glycemic traits (Figure 5A). First, we evaluated the genetic correlation between established ESR1-associated phenotypes and pregnancy glycemic traits, including 0hGlu, GDM, and GDMAUC. The ESR1-associated phenotypes were acquired by searching “ESR1” in the GWAS catalog,58 and we downloaded the available full summary statistics. In total, GWAS summaries for 15 phenotypes were downloaded, encompassing traits such as height (GCST90018959), systolic blood pressure (GCST90018972), and pulse pressure (GCST90018970), among others (Table S7).
Figure 5.
Results of genetic correlation and Mendelian randomization between glycemic traits and ESR1-associated phenotypes from GWAS catalog
(A) The bar plots indicate genetic correlation results. The value marked on each bar indicates the p value of the genetic correlation test.
(B) The forest plots indicate MR results. The value marked on each bar indicates the p value of the MR test, and the asterisks (∗) means that the p value has reached the Bonferroni corrected threshold 6.25E−3 (= 0.05/8).
At a significance threshold of 3.33E−3 (= 0.05/15), we observed two significant associations for 0hGlu, including systolic blood pressure (SBP, rg = 0.2436 ± 0.0694, p value = 4.00E−04) and diastolic blood pressure (DBP, rg = 0.2586 ± 0.0803, p value = 1.30E−03). Notably, for GDMAUC, the two most associated phenotypes were also SBP (rg = 0.185 ± 0.070, p value = 7.80E−03) and DBP (rg = 0.190 ± 0.070, p value = 6.30E−03) (Table S8). While the phenotypic correlation between GDM and hypertension has been confirmed,59,60 only limited studies have explored this correlation from a genetic perspective.
We also conducted a literature review to explore the potential influence of hypertension during pregnancy on blood glucose levels, summarizing our findings into three pathways. Firstly, it may worsen insulin resistance, impeding insulin’s capacity to transport glucose into cells effectively.61 Secondly, hypertension may impact maternal vascular function and hemodynamics, thereby impairing the blood supply to the placenta and uterus, disrupting insulin metabolism.62 Additionally, hypertension may lead to increased inflammation and oxidative stress, negatively affecting insulin sensitivity and secretion.63 These factors collectively may result in elevated glycemic levels during pregnancy, increasing the risk of gestational diabetes.
Mendelian randomization analysis
In this section, we investigated the causal effects of the associated GWAS phenotypes on pregnancy glycemic traits with variants associated with ESR1. Specifically, for each exposure phenotype, we identified all independently significant (ind.sig) variants (r2 < 0.2, p value < 5E−8) within 50 kb on both sides of ESR1 (chr6:151606672–152179619, GRCh38). This identified eight phenotypes with more than three ind.sig SNPs. Inference of phenotypes with fewer than three SNPs may lack power; therefore, we excluded the corresponding phenotypes. At the significant threshold of 6.25E−3 (= 0.05/8), we observed three, zero, and one causal relationships for 0hGlu, GDM, and GDMAUC, respectively (Figure 5B and Table S9). For 0hGlu, the most significant causality was observed in pulse pressure (p value = 2.14E−04), followed by serum phosphate density (p value = 3.40E−03) and appendicular lean mass (p value = 3.91E−03). For GDMAUC, the top two potentially causal phenotypes were height (p value = 4.51E−03) and pulse pressure (p value = 9.41E−03). The MR analysis highlighted the relationship between blood pressure and pregnancy glycemic traits in a causal manner, supposed by observations that women with chronic hypertension are more prone to developing GDM.64
To ensure the validity of our MR analysis, we examined the pleiotropic effects of instrumental variables and also performed a reverse MR analysis. We combined all instrumental variables across the exposure phenotypes and checked their GWAS p values relative to glycemic traits. We found no genome-wide significant signals with the smallest p value for rs488133-GDMAUC (p value = 0.050). For the reverse MR analysis, we employed 0hGlu as the exposure variable and pulse pressure as the outcome variable and conducted a two-sample MR analysis using the R function TwoSampleMR.65 The p value from the inverse variance weighted method was 0.250 (>0.05), signifying the absence of reverse causality.
Transcriptome-wide association study
TWASs can identify significant associations underlying genetically regulated genes and traits of interest.66,67 We thus performed a TWAS analysis to investigate whether ESR1 affects GDM by modulating its expressions (Figure S5 and Table S10). To predict the genetically regulated gene expression (GReX), we multiplied the independent significant cis-eQTL (expression quantitative trait loci) effects of ESR1 by the corresponding genotype dosages in our dataset.
At the significance threshold of 1.0E−03 (= 0.05/49), we identified 17, 0, and 2 tissues with potentially causal GReX on 0hGlu, GDM, and GDMAUC, respectively. The top three tissues for 0hGlu were brain basal ganglia (p value = 1.69E−07), artery tibial (p value = 3.61E−07), and cell-transformed fibroblasts (p value = 5.00E−07). For GDMAUC, the two significant tissues were artery tibial (p value = 1.81E−04) and cell-transformed fibroblasts (p value = 6.92E−04). Here, we wish to underscore the hypothesis that ESR1 plays a crucial role in influencing pregnancy glycemic traits through the regulation of gene expressions in the tibial artery. This is expected given the well-established association between diabetes and arterial disease.68,69,70 Our study offers a potential pathway through which these two diseases may be interconnected.
Significant overlap of GWAS loci and drug targets
The DTEA translates GWAS results into potential applications. Analysis of 19,365 genes and 10,722 drug gene sets, followed by filtering (STAR Methods), retained 1,072 drug gene sets from 66 classifications. At a significance threshold of 7.58E−04 and AUC >0.5, we noted significant overlaps of 3, 0, and 6 Anatomical Therapeutic Chemical classifications for 0hGlu, GDM, and GDMAUC, respectively. Notably, for GDMAUC, significant associations were found with hypertension (C07A, p value = 1.21E−07; C08C, p value = 7.01E−04) and hypotension (C01C, p value = 5.93E−05) (Figure S6). This observation aligns with the genetic correlation and MR analysis results, indicating a putative causal relationship between blood pressure and pregnancy glycemic traits. The potential utility of drug targets for hypertension in addressing hyperglycemia necessitates comprehensive evaluation through rigorous experimental and clinical trials.
Discussion
Glycemic traits during pregnancy play a crucial role in maternal and fetal health. Abnormal glycemic traits during pregnancy may indicate a risk of developing gestational diabetes, which can have adverse outcomes for both the mother and the baby. Understanding the genetic architecture of pregnancy glycemic traits is essential for facilitating the effective management of associated complications. In our study, we uncovered an associated signal, ESR1, which encodes an estrogen receptor known to play a significant role in metabolism and gestation. Our investigation revealed potential mechanisms through which ESR1 was linked to pregnancy glycemic traits. One pathway involved its impact on blood pressure, while another pathway involved the regulation of gene expressions in the tibial artery. Further investigation is required to elucidate the underlying biological mechanisms involved in these associations.
In recent years, the rapid advancement of high-throughput multi-omics technologies has facilitated the acquisition of transcriptomics, proteomics, and metabolomics data from study participants. Numerous placental multi-omics studies have been conducted to identify biomarkers derived from diverse omics datasets and subsequently construct predictive models to distinguish women at high risk of pregnancy hyperglycemia.71,72,73,74,75,76 While these analyses enable the identification of associated biomarkers from each omics data, the explanations of the underlying biological mechanisms and the genetic factors involved are still obscure. The integration of genomic data with other multi-omics data is essential for improving our understanding of disease etiology comprehensively. A TWAS offers an approach to integrate genomic and transcriptomic data, revealing disease-associated genes through the regulation of expressions.66 Additionally, proteome-wide association study represents a novel method for detecting gene-phenotype associations mediated by alterations in protein function.77 Metabolome-wide association study has unveiled associations between metabolic phenotypes and disease risk, taking into account metabolic quantitative trait loci.78 We believe that the integrative analysis of genomic and other multi-omics data holds significant promise for providing a comprehensive exploration of the disease etiology of pregnancy hyperglycemia.
Limitations of the study
While our study offered valuable insights into the genetic landscape of pregnancy glycemic traits, it is essential to acknowledge its limitations. First, despite the recruitment of nearly 40,000 participants, the valid sample size for the GWAS was limited to 14,000 due to the absence of laboratory measurements in 65% of the participants. Conducting a GWAS analysis requires larger sample sizes to achieve sufficient statistical power. Additionally, it’s important to acknowledge that our sample collection was confined to Wuhan, central China, potentially limiting the generalizability of our findings to the entire Chinese population. To address this, we are currently working on recruiting additional samples with both genotype and laboratory data from various regions across China. Simultaneously, we are planning a large-scale meta-analysis within the East Asian population. Second, while our analytical approaches have provided insights into how ESR1 may influence glycemic traits during pregnancy, biological experiments remain the gold standard for validation. These experiments typically include mouse and cell models. However, our literature review revealed that knocking out the estrogen receptor (ESR1) can result in infertility in female mice,79,80 which poses significant challenges for designing models to study glycemic traits in pregnant mice. We are actively pursuing ongoing efforts for future biological replication. Additionally, most of the GWAS hits are non-coding intronic variants, which complicates the direct inference of their functional consequences on glycemic traits. This adds another layer of complexity to experimental replication, as it is challenging to pinpoint the precise regulatory mechanisms affected by these non-coding variants in ESR1. Third, we acknowledge that the ESR1-associated phenotypes retrieved from the GWAS catalog predominantly originated from European populations. Despite our efforts to harmonize data on allele type and frequency, minimizing disparities in variants spectrum between European and East Asian populations, there may persist some bias in our results. Furthermore, we downloaded some ESR1 associations from East Asian populations; however, they either did not provide full summary statistics81,82,83,84 or had a limited number of signals that were subsequently excluded during harmonization.85,86 We strive for undertaking robust replication studies involving larger East Asian populations to enhance the reliability of our findings. Fourth, we note that studying environmental factors alongside genetic analysis in GDM research is crucial. Environmental influences such as diet, lifestyle, and pollutants play a significant role in GDM development. Understanding these factors enhances our ability to predict, prevent, and manage GDM effectively, improving maternal and fetal health outcomes. Unfortunately, we did not include a questionnaire to gather data on environmental factors in this study, but we plan to collect these variables in future investigations.
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact Xin Jin (jinxin@genomics.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA)87 of China National GeneBank DataBase (CNGBdb)88 with accession number CNP0003674 and can also be downloaded directly from https://db.cngb.org/MANE.PheWeb/. A summary of analysis software and tools were provided in key resources table. All original code has been deposited at Zenodo and is publicly available through https://doi.org/10.5281/zenodo.11614736. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This study was supported by the Central Guidance on Local Science and Technology Development Fund of Hubei Province (2022BGE261), National Natural Science Foundation of China (32171441 and 32000398), Guangdong-Hong Kong Joint Laboratory on Immunological and Genetic Kidney Diseases (2019B121205005), Top Medical Young Talents (2019) of Hubei Province, Guangdong Provincial Key Laboratory of Genome Read and Write (2017B030301011), open project of BGI-Shenzhen, Shenzhen 518000 China (BGIRSZ20200008), and the China National GeneBank. We are grateful to Dr. Likuan Xiong for helpful discussions and assistance with external replication studies.
Author contributions
A.Z. and X.J. conceived the study, designed the research program, and managed the project. H.X., M.Y., J. Zhou, Y. Zhou, J.L., Y. Zhong, Z.C., H.M., X.C., L.H., and R.Z. collected the data. L.L., X.L., and P.L. preprocessed the data and finished the quality control. H.Z., H.X., L.L., M.Y., Y.L., X.Z., X.Q., and J. Zeng performed the statistical analysis and results visualization. H.Z., H.X., L.L., M.Y., and Y.L. wrote the manuscript. All authors participated in revising the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| GWAS summary statistics | This paper; China National GeneBank DataBase | CNSA: CNP0003674 |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| 1000 Genomes Project reference panel EAS | The International Genome Sample Resource | https://www.internationalgenome.org/data |
| Summary statistics from GWAS Catalog (See Table S7) | Sollis E. et al., 202358 | https://www.ebi.ac.uk/gwas/ |
| The reference transcriptome data form GTEx | Gamazon E.R. et al., 201566 | https://www.gtexportal.org/home/ |
| The effect sizes for all 49 tissues | Barbeira A.N. et al., 202189 | Zenodo: 5709385 |
| Phase 3 downloaded from the MAGMA website | de Leeuw C.A. et al., 201590 | https://ctg.cncr.nl/software/magma |
| The drug-gene-set built from DGIdb v5 | Freshour S.L. et al., 202191 | https://www.dgidb.org/ |
| Anatomical Therapeutic Chemical (ATC) classification of the drug data | The WHO Collaborating Center for Drug Statistics Methodology | https://www.whocc.no/ |
| DisGeNET | Piñero J. et al., 201792 | https://www.disgenet.org |
| Software and algorithms | ||
| Original Code for analyses | This paper | Zenodo: 11614736 |
| BWA-MEM 0.7.16a-r1181 | Li H. and Durbin R., 200993 | https://bio-bwa.sourceforge.net/ |
| SAMtools 1.3 | Danecek P. et al., 202194 | https://www.htslib.org/ |
| BQSR of the GATK 4.0.4.0 toolset | McKenna A. et al., 201095 | https://gatk.broadinstitute.org/hc/en-us/articles/360035890531-Base-Quality-Score-Recalibration-BQSR- |
| STITCH 1.6.6 | Davies R.W. et al., 201696 | https://github.com/rwdavies/STITCH |
| PLINK2 | Chang C.C. et al., 201597 | https://www.cog-genomics.org/plink/2.0/ |
| LDSC 1.0.1 | Bulik-Sullivan B.K. et al., 201598 | https://github.com/bulik/ldsc |
| Metascape | Zhou Y. et al., 201999 | https://metascape.org/ |
| STRING | Szklarczyk D. et al., 2023100 | https://string-db.org/cgi/ |
| R software version 4.0.2 | R project | https://www.r-project.org/ |
| BaseVar | Liu S. et al., 201817 | https://pypi.org/project/basevar/ |
| LiftOver | Hinrichs A.S. et al., 2006101 | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| R package TwoSampleMR: clump_data | Hemani G. et al., 201865 | https://mrcieu.github.io/TwoSampleMR/reference/clump_data.html |
| MAGMA v1.10 | de Leeuw C.A. et al., 201590 | https://ctg.cncr.nl/software/magma |
| R package coloc | Wallace C. et al., 202046 | https://cran.r-project.org/web/packages/coloc/ |
| R package locuscomparer | Liu B. et al., 201947 | https://github.com/boxiangliu/locuscomparer |
| Other | ||
| MANE (Maternal and Neonatal) PheWeb websiteThis paper | This paper | https://db.cngb.org/MANE.PheWeb/ |
Experimental model and subject details
Subjects
All pregnant women were recruited from the Wuhan Children’s Hospital in Wuhan, Hubei Province, China, where they underwent routine prenatal tests—including blood tests, urinary tests, oral glucose tolerance tests, ultrasound screenings, and NIPT test—from 2017 to 2020. All the participants have signed the informed consent forms. This study was approved by the IRB at Wuhan Children’s Hospital (2021R062) and BGI Research (BGI-IRB 21088) and also authorized by the Human Genetic Resources Administration of China ([2021] CJ2002). We used NIPT sequencing data as the sole inclusion criterion and then performed filtering based on sequencing depth, mapping rate, and outlier values.
Phenotype
We collected four sets of phenotypic data: (1) maternal information from the NIPT questionnaire, (2) laboratory tests from prenatal exams, (3) neonatal information from childbirth outcomes, and (4) clinical diagnoses from doctor’s notes. Maternal data included age, height, weight, systolic blood pressure (SBP), diastolic blood pressure (DBP), and obstetric history, collected pre-NIPT screening. BMI was calculated as weight (kg) divided by height (m2). Hypertension was defined as SBP/DBP over 140/90 mmHg. As a result, four maternal factors—age, BMI, obstetric history (first-time pregnancy status), and hypertension—were used for regression analysis.
The laboratory tests were primarily blood and urinary biochemical measurements. To detect early risk indicators of GDM, we focused on laboratory features in the first trimester. A total of 93 available phenotypes were measured and classified into 9 categories, including hematological (n = 24), urinalysis (n = 24), infection (n = 14), liver-related (n = 8), metabolism (n = 6), protein (n = 5), electrolyte (n = 4), hormone (n = 4), and kidney-related (n = 4). For certain phenotypes, some women had multiple records due to more than one examination or test. We defined an overall observation by taking average or ascertained incidence for quantitative and dichotomous traits, respectively.
The neonatal information included gestational week, birth weight, newborn gender, and delivery options. Low birth weight is defined as a birth weight under 2500g, while fetal macrosomia is diagnosed when a baby weighs over 4000g. The delivery options included natural birth and cesarean section. The clinical diagnosis was the doctor’s descriptions of pregnancy status which spanned a wide spectrum of information, for example, pregnancy complications, how much amniotic fluid, what fetal positions for birth, etc.
Maternal glycemic traits
The oral glucose tolerance test (OGTT) was usually taken during the 24th to 28th week of pregnancy. Specifically, the pregnant women were asked to drink about 237mL liquid that contained 75g glucose. Before drinking the liquid, the pregnancy women would have blood drawn that for measuring the fasting glucose (0hGlu). After drinking the solution, blood was drawn two more times at 1-h (1hGlu) and 2-h (2hGlu) post consumption. By following the recommendations of international association of diabetes and pregnancy study groups,102 we defined a GDM case if at least one of the following factors were true: 0hGlu >5.1 mmol/L, 1hGlu >10.0 mmol/L, or 2hGlu >8.5 mmol/L. To consider the contributions of the three OGTT values simultaneously, we further computed the area under the curve (AUC) as 1hGlu + (0hGlu + 2hGlu)/2.103,104 We used GDMAUC as a complement to the GDM status and performed same statistical analysis.
Method details
Whole-genome sequencing
During gestational weeks 12–22, pregnant women who consented to the NIPT test had 5 mL of peripheral blood drawn. The blood samples were centrifuged to separate the plasma, which contained both maternal and fetal cell-free DNA (cfDNA). We then employed single-end whole genome sequencing using the BGISEQ-500 platform, with a read length of 35 base pairs (bp). On average, the sequencing depth ranged between 0.1× and 0.2×. Notably, at the time of NIPT testing, the fetal DNA fraction in maternal plasma is approximately 10%,105,106 with half of it being identical to the maternal DNA. For subsequent analyses, we did not distinguish between maternal and fetal DNA; instead, we treated all cfDNA as maternal.
Variants calling
The sequencing reads were stored in fastq (.fq) format. First, we used BWA-MEM algorithm93 to align input reads to the human reference genome assembly GRCh38/hg38 and obtained the bam alignment files. Then, we used SAMtools94 to sort sequences and remove duplicate sequences, thus to keep those sequences with highest mapping quality. The following step was to check the base quality in the aligned files and correct the quality value of the original bases to ensure the accuracy of base detection, which was achieved by Base Quality Score Recalibration (BQSR) of the GATK toolset.95 Finally, to obtain the population-based variants, we used BaseVar that was designed to call variants especially for ultra-low-pass WGS data.107 The detected variants were stored in VCF format.
Genotype imputation
Since the NIPT sequencing data was ultra-low depth and only about 10% of the genome were measured, resulting in a high rate of genotype missingness. To make the NIPT sequence data qualified for genetic analysis, we performed genotype imputation for the aligned bam files in STITCH,96 which produced allele dosages of the imputed genotype. To evaluate the imputation performance, we used 30 randomly selected Chinese samples from the high-coverage 1000 genomes project (1KGP)108 as the true set, down-sampled them to about 0.1×, and performed imputation for the down-sample data together with the original NIPT data. Then, we calculated the Pearson’s correlation R2 between the imputed dosages (test) and high coverage genotypes (truth) as the imputation accuracy.
Quantification and statistical analysis
Regression analysis
For the four types of phenotypes, we performed regression analysis to examine their relationship with GDM and GDMAUC. In general, for potential risk factors (e.g., maternal age, early-stage laboratory tests), we fitted logistic and linear model by regressing GDM and GDMAUC as dependent variable, respectively. For potential outcomes (e.g., birthweight, delivery type), we fitted either linear or logistic regression for quantitative or binary outcomes while treating GDM and GDMAUC as predictive variable. For the clinical diagnosis that were all binary variables, it was uncertain whether they were potential risks or outcomes, we used chi-square and t testes for GDM and GDMAUC, respectively.
For significant risk factors with p-value less than 5.15E−4 (=0.05/97), we further performed multivariate logistic and linear regression analysis for GDM and GDMAUC, respectively. It is important to acknowledge that not all phenotypes had measurements available for every participant, which could reduce the effective sample size in a multivariate model. To ensure robustness, we selected a subset of significant factors, ensuring that the sample size in the multivariate model remained at least 3,000.
Genome-wide association study
We executed GWAS analysis for the three glycemic traits (0hGlu, 1hGlu, and 2hGlu), GDM status and GDMAUC. This was achieved in PLINK 2.097 with argument --glm --maf 0.05 --hwe 1e−06 --geno 0.1 by testing SNPs with minor allele frequency (MAF) > 0.05, Hardy-Weinberg equilibrium (HWE) > 1E−6, and genotype missing rate (geno) < 0.1. The logistic regression model was employed to assess GDM status, whereas linear regression models were utilized for evaluating OGTT traits and GDMAUC. The covariates comprised maternal age, gestational week when taking OGTT screening, and the five principal components (PCs) of genotype variants. All covariates were standardized to have a mean of zero and a standard deviation of one by using the argument --covar-variance-standardize and the quantitative phenotype were normalized by --pheno-quantile-normalize. We set the genome-wide significance threshold as 5E−08.
Heritability is a metric indicating the proportion of phenotypic variation in a trait that can be ascribed to genetic factors.109 We used the LDSC (Linkage Disequilibrium Score Regression) method98 to estimate the heritability of glycemic traits, GDM, and GDMAUC. We note that, given the binary nature of GDM status, calculating heritability should be conducted using the liability scale rather than the observed scale. In particular, we specified the population prevalence of GDM as 0.1 and the sample prevalence as 0.113 by using arguments --pop-prev and --samp-prev, respectively.
Protein-protein interaction and gene set enrichment analysis
Protein-protein interaction (PPI) analysis aims to identify and characterize the network of interactions between two or more proteins, which is crucial for deciphering cellular processes and functions.110 Gene set enrichment analysis (GSEA) focuses on identifying biological pathways that are significantly enriched within a list of genes.111 We conducted the PPI and GSEA analysis by combining all mapped genes associated with the five pregnancy glycemic traits. For PPI analysis, we utilized STRING to visualize the network.100 This included known interactions (e.g., experimentally determined), predicted interactions (e.g., gene neighborhood), and other interactions (e.g., text-mining). GSEA analysis was performed using Metascape,99 leveraging the DisGeNET database, which catalogs gene-disease associations.92
Genetic correlation analysis
Genetic correlation refers to the correlation established at the genetic level between two phenotypes.112 Understanding the genetic correlation between traits is helpful to understand the shared genetic etiology and reveal the genetic pleiotropy. We employed LDSC98 to estimate the genetic correlations between 0hGlu, GDM, GDMAUC, and potentially correlated traits. Specifically, we acquired full summary statistics for these traits from the GWAS catalog.58 To ensure compatibility, we utilized LiftOver101 to convert the reference genome of the downloaded summary statistics to GRCh38. LD calculations were performed using the East Asian population data from the 1000 Genomes Project as a reference.
Mendelian randomization analysis
Mendelian randomization (MR) is a statistical technique to assess causal effects of a modifiable exposure on an outcome by using genetic variants as instrumental variables (IVs).113 In this study, we would like to explore the causal relationships between certain potentially correlated phenotypes and glycemic traits. For each examined exposure phenotype, we chose highly significant SNPs with p-value less than 5E−08 and conducted LD clumping to ensure the retained SNPs were independent. Specifically, we used R function TwoSampleMR: clump_data65 by setting clumping r2 as 0.2 and other parameters in default values. The remaining SNPs were utilized to compute the polygenic score (PGS) for our participants, achieved by multiplying the exposure’s genetic effects with individual-level genotype dosages. To assess the causality of the PGS on glycemic traits, we constructed appropriate regression models with the PGS as the predictor.
Transcriptome-wide association study
TWAS is a method that integrates gene expression data and genotype information to identify genes whose expression levels are associated with complex traits or diseases.66,67 It involves two datasets: reference transcriptome data and the main dataset. The reference data includes samples with genotype and gene expression information, used to study genetic impacts on gene expression. The main dataset consists of samples with genotype and complex traits. By integrating genetic effects from the reference data with the main dataset, we derived genetically regulated expression (GReX). Finally, we tested the association between GReX and the trait of interest.
In this study, we used the GTEx database (v8) as our reference transcriptome data to estimate GReX levels for our participants.66 Specifically, we chose to use the cross-tissue gene expression imputation framework (CTIMP)114 and downloaded the effect sizes from Zenodo89 for all 49 tissues. The cis-eQTL effects were obtained from all variants located within 500kb upstream and downstream of a target gene. These variants were clumped using an r2 threshold of 0.2 to retain independent effects using R:clump_data.65 GReX was calculated by multiplying eQTL effects with genotype dosages. Logistic regression was then used to analyze GDM status, and linear regression was applied to 0hGlu and GDMAUC, with a significance threshold adjusted to 0.001 using Bonferroni correction.
Drug target enrichment analysis
In this section, we conducted drug target enrichment analysis (DTEA) to determine if a group of candidate genes was significantly enriched for known drug targets. This analysis can provide insights into drug repurposing opportunities, uncover novel therapeutic targets, and aid in understanding the molecular mechanisms underlying diseases.115 Using MAGMA v1.10,90 we performed drug-gene set analysis on glycemia-associated genes. The drug-gene set was curated from the Drug-Gene Interaction Database (DGIdb) version 5 interactions file.91 Gene-level analyses were performed using the default competitive model.116
The follow-up analysis involved drug classification. For this, we needed data on the Anatomical Therapeutic Chemical (ATC) classification of drugs.117,118 Using previously described enrichment curves, we annotated the drug-gene set analysis results according to ATC classifications, excluding those with fewer than 10 valid drug-gene sets.116,119 The sets were ranked by association (-log10(p)) and divided into groups based on ATC classification. We then calculated the area under the curve (AUC) and p-value using the Wilcoxon Mann-Whitney test, with a Bonferroni-corrected significance threshold set at p < 7.58E−04 for 66 drug classifications.
Published: October 9, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2024.100631.
Contributor Information
Xin Jin, Email: jinxin@genomics.cn.
Aifen Zhou, Email: april1972@163.com.
Supplemental information
References
- 1.Sweeting A., Wong J., Murphy H.R., Ross G.P. A Clinical Update on Gestational Diabetes Mellitus. Endocr. Rev. 2022;43:763–793. doi: 10.1210/endrev/bnac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung R.H., Chiu Y.F., Wang W.C., Hwu C.M., Hung Y.J., Lee I.T., Chuang L.M., Quertermous T., Rotter J.I., Chen Y.D.I., et al. Multi-omics analysis identifies CpGs near G6PC2 mediating the effects of genetic variants on fasting glucose. Diabetologia. 2021;64:1613–1625. doi: 10.1007/s00125-021-05449-9. [DOI] [PubMed] [Google Scholar]
- 5.Kwak S.H., Kim S.H., Cho Y.M., Go M.J., Cho Y.S., Choi S.H., Moon M.K., Jung H.S., Shin H.D., Kang H.M., et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes. 2012;61:531–541. doi: 10.2337/db11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes M.G., Urbanek M., Hivert M.F., Armstrong L.L., Morrison J., Guo C., Lowe L.P., Scheftner D.A., Pluzhnikov A., Levine D.M., et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes. 2013;62:3282–3291. doi: 10.2337/db12-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhen J., Gu Y., Wang P., Wang W., Bian S., Huang S., Liang H., Huang M., Yu Y., Chen Q., et al. Genome-wide association and Mendelian randomisation analysis among 30,699 Chinese pregnant women identifies novel genetic and molecular risk factors for gestational diabetes and glycaemic traits. Diabetologia. 2024;67:703–713. doi: 10.1007/s00125-023-06065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morisset A.S., St-Yves A., Veillette J., Weisnagel S.J., Tchernof A., Robitaille J. Prevention of gestational diabetes mellitus: a review of studies on weight management. Diabetes. Metab. Res. Rev. 2010;26:17–25. doi: 10.1002/dmrr.1053. [DOI] [PubMed] [Google Scholar]
- 9.Reece E.A. The fetal and maternal consequences of gestational diabetes mellitus. The journal of maternal-fetal & neonatal medicine : the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies. J. Matern. Fetal Neonatal Med. 2010;23:199–203. doi: 10.3109/14767050903550659. [DOI] [PubMed] [Google Scholar]
- 10.Petry C.J. Gestational diabetes: risk factors and recent advances in its genetics and treatment. Br. J. Nutr. 2010;104:775–787. doi: 10.1017/s0007114510001741. [DOI] [PubMed] [Google Scholar]
- 11.Buchanan T.A., Xiang A.H., Page K.A. Gestational diabetes mellitus: risks and management during and after pregnancy. Nat. Rev. Endocrinol. 2012;8:639–649. doi: 10.1038/nrendo.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kc K., Shakya S., Zhang H. Gestational diabetes mellitus and macrosomia: a literature review. Ann. Nutr. Metab. 2015;66:14–20. doi: 10.1159/000371628. [DOI] [PubMed] [Google Scholar]
- 13.Moses R.G., Knights S.J., Lucas E.M., Moses M., Russell K.G., Coleman K.J., Davis W.S. Gestational diabetes: is a higher cesarean section rate inevitable? Diabetes Care. 2000;23:15–17. doi: 10.2337/diacare.23.1.15. [DOI] [PubMed] [Google Scholar]
- 14.Jiang L., Tang K., Magee L.A., von Dadelszen P., Ekeroma A., Li X., Zhang E., Bhutta Z.A. A global view of hypertensive disorders and diabetes mellitus during pregnancy. Nat. Rev. Endocrinol. 2022;18:760–775. doi: 10.1038/s41574-022-00734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benn P., Borell A., Chiu R., Cuckle H., Dugoff L., Faas B., Gross S., Johnson J., Maymon R., Norton M., et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat. Diagn. 2013;33:622–629. doi: 10.1002/pd.4139. [DOI] [PubMed] [Google Scholar]
- 16.Shendure J., Findlay G.M., Snyder M.W. Genomic Medicine-Progress, Pitfalls, and Promise. Cell. 2019;177:45–57. doi: 10.1016/j.cell.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Huang S., Chen F., Zhao L., Yuan Y., Francis S.S., Fang L., Li Z., Lin L., Liu R., et al. Genomic Analyses from Non-invasive Prenatal Testing Reveal Genetic Associations, Patterns of Viral Infections, and Chinese Population History. Cell. 2018;175:347–359.e14. doi: 10.1016/j.cell.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Liu S., Huang S., Liu Y., Gu Y., Lin X., Zhu H., Liu H., Xu Z., Cheng S., Lan X., et al. Utilizing Non-Invasive Prenatal Test Sequencing Data Resource for Human Genetic Investigation. bioRxiv. 2023;56:377. doi: 10.1101/2023.12.11.570976. Preprint at. [DOI] [PubMed] [Google Scholar]
- 19.Elliott A., Walters R.K., Pirinen M., Kurki M., Junna N., Goldstein J.I., Reeve M.P., Siirtola H., Lemmelä S.M., Turley P., et al. Distinct and shared genetic architectures of gestational diabetes mellitus and type 2 diabetes. Nat. Genet. 2024;56:377–382. doi: 10.1038/s41588-023-01607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao C., Sun X., Lu L., Liu F., Yuan J. Prevalence of gestational diabetes mellitus in mainland China: A systematic review and meta-analysis. J. Diabetes Investig. 2019;10:154–162. doi: 10.1111/jdi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powe C.E. Early Pregnancy Biochemical Predictors of Gestational Diabetes Mellitus. Curr. Diabetes Rep. 2017;17:12. doi: 10.1007/s11892-017-0834-y. [DOI] [PubMed] [Google Scholar]
- 22.Falcone V., Kotzaeridi G., Breil M.H., Rosicky I., Stopp T., Yerlikaya-Schatten G., Feichtinger M., Eppel W., Husslein P., Tura A., Göbl C.S. Early Assessment of the Risk for Gestational Diabetes Mellitus: Can Fasting Parameters of Glucose Metabolism Contribute to Risk Prediction? Diabetes Metab. J. 2019;43:785–793. doi: 10.4093/dmj.2018.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omazić J., Viljetić B., Ivić V., Kadivnik M., Zibar L., Müller A., Wagner J. Early markers of gestational diabetes mellitus: what we know and which way forward? Biochem. Med. 2021;31 doi: 10.11613/bm.2021.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M., Chen Y., Chen D. Association between transthyretin concentrations and gestational diabetes mellitus in Chinese women. Arch. Gynecol. Obstet. 2020;302:329–335. doi: 10.1007/s00404-020-05599-y. [DOI] [PubMed] [Google Scholar]
- 25.Luo C., Li Z., Lu Y., Wei F., Suo D., Lan S., Ren Z., Jiang R., Huang F., Chen A., et al. Association of serum vitamin D status with gestational diabetes mellitus and other laboratory parameters in early pregnant women. BMC Pregnancy Childbirth. 2022;22:400. doi: 10.1186/s12884-022-04725-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alanbay I., Coksuer H., Ercan M., Keskin U., Karasahin E., Ozturk M., Tapan S., Ozturk O., Kurt I., Ergun A. Can serum gamma-glutamyltransferase levels be useful at diagnosing gestational diabetes mellitus? Gynecol. Endocrinol. 2012;28:208–211. doi: 10.3109/09513590.2011.588756. [DOI] [PubMed] [Google Scholar]
- 27.Kong M., Liu C., Guo Y., Gao Q., Zhong C., Zhou X., Chen R., Xiong G., Yang X., Hao L., Yang N. Higher level of GGT during mid-pregnancy is associated with increased risk of gestational diabetes mellitus. Clin. Endocrinol. 2018;88:700–705. doi: 10.1111/cen.13558. [DOI] [PubMed] [Google Scholar]
- 28.Khan F.Y., Kauser H., Palakeel J.J., Ali M., Chhabra S., Lamsal Lamichhane S., Opara C.O., Hanif A. Role of Uric Acid Levels in the Development of Gestational Diabetes Mellitus: A Review. Cureus. 2022;14 doi: 10.7759/cureus.31057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue C., Ying C., Li X. Elevated Serum Uric Acid Is Associated With Gestational Diabetes Mellitus: An Observational Cohort Study. J. Clin. Endocrinol. Metab. 2023;108:e480–e486. doi: 10.1210/clinem/dgac760. [DOI] [PubMed] [Google Scholar]
- 30.Schneider S., Freerksen N., Röhrig S., Hoeft B., Maul H. Gestational diabetes and preeclampsia--similar risk factor profiles? Early Hum. Dev. 2012;88:179–184. doi: 10.1016/j.earlhumdev.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Wu N. Gestational Diabetes Mellitus and Preeclampsia: Correlation and Influencing Factors. Front. Cardiovasc. Med. 2022;9 doi: 10.3389/fcvm.2022.831297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H., Li L., Yang M., Zeng J., Zhou J., Tao Y., Zhou Y., Cai M., Liu J., Huang Y., et al. Genetic analysis of 104 pregnancy phenotypes in 39,194 Chinese women. medRxiv. 2023;56:377. doi: 10.1101/2023.11.23.23298979. Preprint at. [DOI] [Google Scholar]
- 33.2023. MANE pheweb. https://db.cngb.org/MANE.PheWeb/ [Google Scholar]
- 34.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 35.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., Venkataraman G.R., Wainberg M., Ollila H.M., Kiiskinen T., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson T.G., Leyden G.M., Wang Q., Bell J.A., Elsworth B., Davey Smith G., Holmes M.V. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pervjakova N., Moen G.H., Borges M.C., Ferreira T., Cook J.P., Allard C., Beaumont R.N., Canouil M., Hatem G., Heiskala A., et al. Multi-ancestry genome-wide association study of gestational diabetes mellitus highlights genetic links with type 2 diabetes. Hum. Mol. Genet. 2022;31:3377–3391. doi: 10.1093/hmg/ddac050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Changalidis A.I., Maksiutenko E.M., Barbitoff Y.A., Tkachenko A.A., Vashukova E.S., Pachuliia O.V., Nasykhova Y.A., Glotov A.S. Aggregation of Genome-Wide Association Data from FinnGen and UK Biobank Replicates Multiple Risk Loci for Pregnancy Complications. Genes. 2022;13 doi: 10.3390/genes13122255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascoe L., Tura A., Patel S.K., Ibrahim I.M., Ferrannini E., Zeggini E., Weedon M.N., Mari A., Hattersley A.T., McCarthy M.I., et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 40.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 41.Rubio-Sastre P., Scheer F.A.J.L., Gómez-Abellán P., Madrid J.A., Garaulet M. Acute melatonin administration in humans impairs glucose tolerance in both the morning and evening. Sleep. 2014;37:1715–1719. doi: 10.5665/sleep.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spracklen C.N., Horikoshi M., Kim Y.J., Lin K., Bragg F., Moon S., Suzuki K., Tam C.H.T., Tabara Y., Kwak S.H., et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582:240–245. doi: 10.1038/s41586-020-2263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki K., Akiyama M., Ishigaki K., Kanai M., Hosoe J., Shojima N., Hozawa A., Kadota A., Kuriki K., Naito M., et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat. Genet. 2019;51:379–386. doi: 10.1038/s41588-018-0332-4. [DOI] [PubMed] [Google Scholar]
- 44.Sun J.W., Collins J.M., Ling D., Wang D. Highly Variable Expression of ESR1 Splice Variants in Human Liver: Implication in the Liver Gene Expression Regulation and Inter-Person Variability in Drug Metabolism and Liver Related Diseases. J. Mol. Genet. Med. 2019;13 [PMC free article] [PubMed] [Google Scholar]
- 45.Giambartolomei C., Zhenli Liu J., Zhang W., Hauberg M., Shi H., Boocock J., Pickrell J., Jaffe A.E., CommonMind Consortium. Pasaniuc B., Roussos P. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34:2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., Gloudemans M.J., Rao A.S., Ingelsson E., Montgomery S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahia W., Soltani I., Haddad A., Soua A., Radhouani A., Mahdhi A., Ferchichi S. Association of genetic variants in Estrogen receptor (ESR)1 and ESR2 with susceptibility to recurrent pregnancy loss in Tunisian women: A case control study. Gene. 2020;736 doi: 10.1016/j.gene.2020.144406. [DOI] [PubMed] [Google Scholar]
- 49.El-Beshbishy H.A., Tawfeek M.A., Al-Azhary N.M., Mariah R.A., Habib F.A., Aljayar L., Alahmadi A.F. Estrogen Receptor Alpha (ESR1) Gene Polymorphisms in Pre-eclamptic Saudi Patients. Pakistan J. Med. Sci. 2015;31:880–885. doi: 10.12669/pjms.314.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berkane N., Liere P., Oudinet J.P., Hertig A., Lefèvre G., Pluchino N., Schumacher M., Chabbert-Buffet N. From Pregnancy to Preeclampsia: A Key Role for Estrogens. Endocr. Rev. 2017;38:123–144. doi: 10.1210/er.2016-1065. [DOI] [PubMed] [Google Scholar]
- 51.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernard V., Kherra S., Francou B., Fagart J., Viengchareun S., Guéchot J., Ladjouze A., Guiochon-Mantel A., Korach K.S., Binart N., et al. Familial Multiplicity of Estrogen Insensitivity Associated With a Loss-of-Function ESR1 Mutation. J. Clin. Endocrinol. Metab. 2017;102:93–99. doi: 10.1210/jc.2016-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toy W., Shen Y., Won H., Green B., Sakr R.A., Will M., Li Z., Gala K., Fanning S., King T.A., et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan H., Yang W., Zhou F., Li X., Pan Q., Shen Z., Han G., Newell-Fugate A., Tian Y., Majeti R., et al. Estrogen Improves Insulin Sensitivity and Suppresses Gluconeogenesis via the Transcription Factor Foxo1. Diabetes. 2019;68:291–304. doi: 10.2337/db18-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoh K., Ikeda K., Horie K., Inoue S. Roles of Estrogen, Estrogen Receptors, and Estrogen-Related Receptors in Skeletal Muscle: Regulation of Mitochondrial Function. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24031853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuasa T., Takata Y., Aki N., Kunimi K., Satoh M., Nii M., Izumi Y., Otoda T., Hashida S., Osawa H., Aihara K.I. Insulin receptor cleavage induced by estrogen impairs insulin signaling. BMJ Open Diabetes Res. Care. 2021;9 doi: 10.1136/bmjdrc-2021-002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelleher A.M., DeMayo F.J., Spencer T.E. Uterine Glands: Developmental Biology and Functional Roles in Pregnancy. Endocr. Rev. 2019;40:1424–1445. doi: 10.1210/er.2018-00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Birukov A., Glintborg D., Schulze M.B., Jensen T.K., Kuxhaus O., Andersen L.B., Kräker K., Polemiti E., Jensen B.L., Jørgensen J.S., et al. Elevated blood pressure in pregnant women with gestational diabetes according to the WHO criteria: importance of overweight. J. Hypertens. 2022;40:1614–1623. doi: 10.1097/hjh.0000000000003196. [DOI] [PubMed] [Google Scholar]
- 60.Hedderson M.M., Ferrara A. High blood pressure before and during early pregnancy is associated with an increased risk of gestational diabetes mellitus. Diabetes Care. 2008;31:2362–2367. doi: 10.2337/dc08-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salvetti A., Brogi G., Di Legge V., Bernini G.P. The inter-relationship between insulin resistance and hypertension. Drugs. 1993;46:149–159. doi: 10.2165/00003495-199300462-00024. [DOI] [PubMed] [Google Scholar]
- 62.Scioscia M., Gumaa K., Kunjara S., Paine M.A., Selvaggi L.E., Rodeck C.H., Rademacher T.W. Insulin resistance in human preeclamptic placenta is mediated by serine phosphorylation of insulin receptor substrate-1 and -2. J. Clin. Endocrinol. Metab. 2006;91:709–717. doi: 10.1210/jc.2005-1965. [DOI] [PubMed] [Google Scholar]
- 63.Zhou M.S., Wang A., Yu H. Link between insulin resistance and hypertension: What is the evidence from evolutionary biology? Diabetol. Metab. Syndrome. 2014;6:12. doi: 10.1186/1758-5996-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leon M.G., Moussa H.N., Longo M., Pedroza C., Haidar Z.A., Mendez-Figueroa H., Blackwell S.C., Sibai B.M. Rate of Gestational Diabetes Mellitus and Pregnancy Outcomes in Patients with Chronic Hypertension. Am. J. Perinatol. 2016;33:745–750. doi: 10.1055/s-0036-1571318. [DOI] [PubMed] [Google Scholar]
- 65.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., GTEx Consortium. Nicolae D.L., et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.American Diabetes Association Peripheral arterial disease in people with diabetes. Diabetes Care. 2003;26:3333–3341. doi: 10.2337/diacare.26.12.3333. [DOI] [PubMed] [Google Scholar]
- 69.Forsythe R.O., Apelqvist J., Boyko E.J., Fitridge R., Hong J.P., Katsanos K., Mills J.L., Nikol S., Reekers J., Venermo M., et al. Effectiveness of bedside investigations to diagnose peripheral artery disease among people with diabetes mellitus: A systematic review. Diabetes. Metab. Res. Rev. 2020;36 doi: 10.1002/dmrr.3277. [DOI] [PubMed] [Google Scholar]
- 70.Tsuchiya M., Suzuki E., Egawa K., Nishio Y., Maegawa H., Inoue S., Mitsunami K., Morikawa S., Inubushi T., Kashiwagi A. Stiffness and impaired blood flow in lower-leg arteries are associated with severity of coronary artery calcification among asymptomatic type 2 diabetic patients. Diabetes Care. 2004;27:2409–2415. doi: 10.2337/diacare.27.10.2409. [DOI] [PubMed] [Google Scholar]
- 71.Tang L., Li P., Li L. Whole transcriptome expression profiles in placenta samples from women with gestational diabetes mellitus. J. Diabetes Investig. 2020;11:1307–1317. doi: 10.1111/jdi.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Assi E., D'Addio F., Mandò C., Maestroni A., Loretelli C., Ben Nasr M., Usuelli V., Abdelsalam A., Seelam A.J., Pastore I., et al. Placental proteome abnormalities in women with gestational diabetes and large-for-gestational-age newborns. BMJ Open Diabetes Res. Care. 2020;8 doi: 10.1136/bmjdrc-2020-001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q.Y., You L.H., Xiang L.L., Zhu Y.T., Zeng Y. Current progress in metabolomics of gestational diabetes mellitus. World J. Diabetes. 2021;12:1164–1186. doi: 10.4239/wjd.v12.i8.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pineda-Cortel M.R.B., Bunag J.A.A., Mamerto T.P., Abulencia M.F.B. Differential gene expression and network-based analyses of the placental transcriptome reveal distinct potential biomarkers for gestationaldiabetes mellitus. Diabetes Res. Clin. Pract. 2021;180 doi: 10.1016/j.diabres.2021.109046. [DOI] [PubMed] [Google Scholar]
- 75.Sriboonvorakul N., Hu J., Boriboonhirunsarn D., Ng L.L., Tan B.K. Proteomics Studies in Gestational Diabetes Mellitus: A Systematic Review and Meta-Analysis. J. Clin. Med. 2022;11 doi: 10.3390/jcm11102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Razo-Azamar M., Nambo-Venegas R., Meraz-Cruz N., Guevara-Cruz M., Ibarra-González I., Vela-Amieva M., Delgadillo-Velázquez J., Santiago X.C., Escobar R.F., Vadillo-Ortega F., Palacios-González B. An early prediction model for gestational diabetes mellitus based on metabolomic biomarkers. Diabetol. Metab. Syndrome. 2023;15:116. doi: 10.1186/s13098-023-01098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brandes N., Linial N., Linial M. PWAS: proteome-wide association study-linking genes and phenotypes by functional variation in proteins. Genome Biol. 2020;21:173. doi: 10.1186/s13059-020-02089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang C., Qin N., Zhu M., Chen M., Xie K., Cheng Y., Dai J., Liu J., Xia Y., Ma H., et al. Metabolome-wide association study identified the association between a circulating polyunsaturated fatty acids variant rs174548 and lung cancer. Carcinogenesis. 2017;38:1147–1154. doi: 10.1093/carcin/bgx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh S.P., Wolfe A., Ng Y., DiVall S.A., Buggs C., Levine J.E., Wondisford F.E., Radovick S. Impaired estrogen feedback and infertility in female mice with pituitary-specific deletion of estrogen receptor alpha (ESR1) Biol. Reprod. 2009;81:488–496. doi: 10.1095/biolreprod.108.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nalvarte I., Antonson P. Estrogen Receptor Knockout Mice and Their Effects on Fertility. Receptors. 2023;2:116–126. doi: 10.3390/receptors2010007. [DOI] [Google Scholar]
- 81.Han M.R., Long J., Choi J.Y., Low S.K., Kweon S.S., Zheng Y., Cai Q., Shi J., Guo X., Matsuo K., et al. Genome-wide association study in East Asians identifies two novel breast cancer susceptibility loci. Hum. Mol. Genet. 2016;25:3361–3371. doi: 10.1093/hmg/ddw164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishigaki K., Akiyama M., Kanai M., Takahashi A., Kawakami E., Sugishita H., Sakaue S., Matoba N., Low S.K., Okada Y., et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 2020;52:669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yengo L., Vedantam S., Marouli E., Sidorenko J., Bartell E., Sakaue S., Graff M., Eliasen A.U., Jiang Y., Raghavan S., et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J.Y., Kim J., Kim S.W., Park S.K., Ahn S.H., Lee M.H., Suh Y.J., Noh D.Y., Son B.H., Cho Y.U., et al. BRCA1/2-negative, high-risk breast cancers (BRCAX) for Asian women: genetic susceptibility loci and their potential impacts. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Yamamoto K., Akiyama M., et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PubMed] [Google Scholar]
- 87.Guo X., Chen F., Gao F., Li L., Liu K., You L., Hua C., Yang F., Liu W., Peng C., et al. CNSA: a data repository for archiving omics data. Database. 2020;2020:baaa055. doi: 10.1093/database/baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen F.Z., You L.J., Yang F., Wang L.N., Guo X.Q., Gao F., Hua C., Tan C., Fang L., Shan R.Q., et al. CNGBdb: China National GeneBank DataBase. Yi Chuan. 2020;42:799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
- 89.Barbeira A.N., Melia O., Liang Y., Bonazzola R., Wang G., Wheeler H., Aguet F., Ardlie K., Wen X., Im H.K. CTIMP models on GTEx v8[Data set] Zenodo. 2021 doi: 10.5281/zenodo.5709385. [DOI] [Google Scholar]
- 90.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freshour S.L., Kiwala S., Cotto K.C., Coffman A.C., McMichael J.F., Song J.J., Griffith M., Griffith O.L., Wagner A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., García-García J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davies R.W., Flint J., Myers S., Mott R. Rapid genotype imputation from sequence without reference panels. Nat. Genet. 2016;48:965–969. doi: 10.1038/ng.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.International Association of Diabetes and Pregnancy Study Groups Consensus Panel. Metzger B.E., Gabbe S.G., Persson B., Buchanan T.A., Catalano P.A., Damm P., Dyer A.R., Leiva A.d., Hod M., et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–682. doi: 10.2337/dc09-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang C., Wei Y., Sun W., Yang H. The Area under the Curve (AUC) of Oral Glucose Tolerance Test (OGTT) Could Be a Measure Method of Hyperglycemia in All Pregnant Women. Open J. Obstet. Gynecol. 2019;09:186–195. doi: 10.4236/ojog.2019.92019. [DOI] [Google Scholar]
- 104.Sakaguchi K., Takeda K., Maeda M., Ogawa W., Sato T., Okada S., Ohnishi Y., Nakajima H., Kashiwagi A. Glucose area under the curve during oral glucose tolerance test as an index of glucose intolerance. Diabetol. Int. 2016;7:53–58. doi: 10.1007/s13340-015-0212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deng C., Liu J., Liu S., Liu H., Bai T., Jing X., Xia T., Liu Y., Cheng J., Wei X., et al. Maternal and fetal factors influencing fetal fraction: A retrospective analysis of 153,306 pregnant women undergoing noninvasive prenatal screening. Front. Pediatr. 2023;11 doi: 10.3389/fped.2023.1066178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang E., Batey A., Struble C., Musci T., Song K., Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat. Diagn. 2013;33:662–666. doi: 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- 107.ShujiaHuang. basevar (GitHub). https://github.com/ShujiaHuang/basevar.
- 108.Fairley S., Lowy-Gallego E., Perry E., Flicek P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020;48:D941–D947. doi: 10.1093/nar/gkz836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wray N.R., Visscher P.M. Estimating Trait Heritability. Nat. Educ. 2008;1:29. [Google Scholar]
- 110.Grindrod P., Kibble M. Review of uses of network and graph theory concepts within proteomics. Expert Rev. Proteomics. 2004;1:229–238. doi: 10.1586/14789450.1.2.229. [DOI] [PubMed] [Google Scholar]
- 111.Wang L., Jia P., Wolfinger R.D., Chen X., Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L., et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu Y., Li M., Lu Q., Weng H., Wang J., Zekavat S.M., Yu Z., Li B., Gu J., Muchnik S., et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019;51:568–576. doi: 10.1038/s41588-019-0345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loscalzo J. Personalized cardiovascular medicine and drug development: time for a new paradigm. Circulation. 2012;125:638–645. doi: 10.1161/circulationaha.111.089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaspar H.A., Breen G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-12325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.WHO . 2023. Collaborating Centre for Drug Statistics Methodology.https://www.whocc.no/ [Google Scholar]
- 118.WHO . 2023. Anatomical Therapeutic Chemical (ATC) Classification.https://www.who.int/tools/atc-ddd-toolkit/atc-classification [Google Scholar]
- 119.Docherty A.R., Mullins N., Ashley-Koch A.E., Qin X., Coleman J.R.I., Shabalin A., Kang J., Murnyak B., Wendt F., Adams M., et al. GWAS Meta-Analysis of Suicide Attempt: Identification of 12 Genome-Wide Significant Loci and Implication of Genetic Risks for Specific Health Factors. Am. J. Psychiatr. 2023;180:723–738. doi: 10.1176/appi.ajp.21121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA)87 of China National GeneBank DataBase (CNGBdb)88 with accession number CNP0003674 and can also be downloaded directly from https://db.cngb.org/MANE.PheWeb/. A summary of analysis software and tools were provided in key resources table. All original code has been deposited at Zenodo and is publicly available through https://doi.org/10.5281/zenodo.11614736. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.