Abstract
Respiratory syncytial virus (RSV) is a major cause of severe respiratory disease in infants and older people. Current RSV subunit vaccines are based on a fusion protein that is stabilized in the prefusion conformation and linked to a heterologous foldon trimerization domain to obtain a prefusion F (preF) trimer. Here we show that current RSV vaccines induce undesirable anti-foldon antibodies in non-human primates, mice and humans. To overcome this, we designed a foldon-free RSV preF trimer by elucidating the structural basis of trimerization-induced preF destabilization through molecular dynamics simulations and by introducing amino acid substitutions that negate hotspots of charge repulsion. The highly stable prefusion conformation was validated using antigenic and cryo-electron microscopy analysis. The preF is immunogenic and protective in naive mouse models and boosts neutralizing antibody titres in RSV-pre-exposed mice and non-human primates, while achieving similar titres to approved RSV vaccines in mice. This stable preF design is a promising option as a foldon-independent candidate for a next-generation RSV vaccine immunogen.
Subject terms: Protein vaccines, Viral membrane fusion, Preclinical research, Protein structure predictions
A targeted design to stabilize the respiratory syncytial virus prefusion F trimer enables the generation of a foldon-free immunogen that elicits similar humoral responses to approved vaccines.
Main
Respiratory syncytial virus (RSV) can induce severe respiratory disease in infants and the elderly1–5. RSV F, a class I fusion protein, facilitates the merging of viral and host cell membranes through irreversible protein refolding from the labile, high-energy prefusion conformation to the stable postfusion conformation6,7. The prefusion F (preF) protein comprises a globular head region connected to a helical stem formed by heptad repeat 2 (HR2), followed by a membrane-anchoring transmembrane region6,8. Like other class I fusion proteins, RSV F is synthesized as an inactive precursor (F0) that requires proteolytic cleavage during intracellular maturation to attain a fusion-competent state7,9. This activation step is unusual in the case of RSV as it involves two separate cleavage events by furin-like proteases, resulting in F2, p27 and the membrane-anchored F1 fusion domain10,11. The transition to the postfusion F (postF) protein conformation irreversibly alters the protein antigenic structure. Consequently, the preF protein is the primary target of RSV-neutralizing antibodies in human sera12,13. Most vaccines in clinical development therefore focus on RSV preF as the antigen of choice. Although all class I fusion proteins are metastable, an unusual aspect of RSV F is its high trimer instability that results in low expression levels and failure to form correctly folded trimers when the protein is expressed as a soluble ectodomain14,15. This inherent instability of the prefusion conformation has long impeded vaccine development until the recent approval of the ABRYSVO and AREXVY vaccines16–20. A heterologous foldon trimerization domain, derived from T4 fibritin21,22, has been used in recent vaccines to obtain a trimer, in conjunction with the introduction of prefusion-stabilizing amino acid substitutions14,15,23–26.
In this study, we show that current commercial preF proteins lead to induction and boosting of anti-foldon responses. Moreover, we demonstrate that the p27-mediated delay of preF trimerization is linked to the high trimer instability of RSV and that the use of foldon for RSV F actually leads to rapid refolding to the postF conformation. Using molecular dynamics (MD) simulations, we were able to pinpoint, and counteract, regions of trimerization-induced instability. This approach yielded highly stable trimeric RSV preF proteins that were no longer contingent on the use of a heterologous trimerization domain. This next-generation RSV vaccine candidate was successful at inducing and boosting neutralizing antibody responses and protecting against RSV challenge.
Results
Undesired anti-foldon responses after preF vaccination
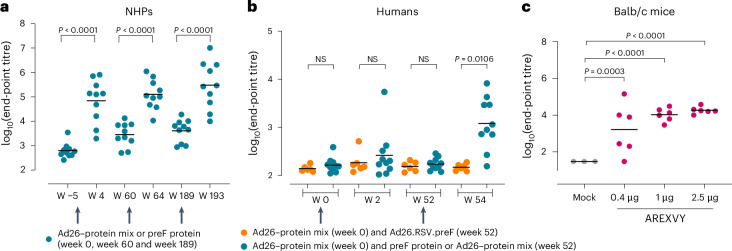
A disadvantage of the use of a heterologous foldon trimerization domain is the introduction of a neo-epitope that can potentially induce undesired off-target immune responses. These may be boosted upon repeated vaccination or when multiple foldon-containing vaccines are used and may interfere with the induction of the desired immune response. African green monkeys that were first intranasally pre-exposed with RSV-A and later immunized with RSV-A preF-based vaccine components (preF protein or preF protein combined with replication-incompetent adenovirus, Ad26.RSV.preF)27 indeed showed foldon-binding antibody titres that increased after every immunization and declined over time, but remained above background level (Fig. 1a).
Fig. 1. Foldon-containing preF protein induces off-target responses in mice, NHPs and humans.
a, RSV-pre-exposed NHPs (n = 10) were immunized at week 0, week 60 and week 189 (arrows) with preF protein or preF protein–Ad26-preF. Foldon-binding antibody titres were measured using ELISA in serum isolated before immunization and 4 weeks after every immunization. b, Human participants were immunized with preF protein combined with Ad26-preF at week 0 and with preF protein or preF protein–Ad26-preF (blue symbols, n = 10) or Ad26-preF alone (orange symbols, n = 6) at week 52 (arrows). Just before and 2 weeks after immunization, foldon-binding antibody titres were measured using ELISA. c, Balb/c mice were immunized twice with a 4 week interval with AREXVY, an AS01E-adjuvanted preF-protein-based commercially available RSV vaccine (n = 6 per group) or buffer (n = 3). Foldon-binding antibody titres were measured using ELISA 2 weeks after the second immunization. End-point titres were calculated and expressed as log10. The bars represent mean titres per group. The NHP and human ELISAs have no lower limit of detection (LLOD) defined while the mouse ELISA has an LLOD of 1.47 (log10). NS, not statistically different. Statistical testing (two sided) was performed using paired t-test with Bonferroni correction (a), ANOVA with Bonferroni correction (b) or a Tobit model with Bonferroni correction (c).
Next, foldon-binding antibody responses were determined in a subset of participants from clinical trial NCT03982199 that were immunized and boosted with foldon-containing preF protein in combination with Ad26.RSV.preF. No clear foldon-binding antibody titres were induced 2 weeks after a first immunization with Ad26.RSV.preF–preF protein. However, 2 weeks after the second immunization, robust foldon-binding antibody titres were induced in eight of ten participants boosted with preF protein or Ad26.RSV.preF–preF protein, while participants that were boosted with Ad26.RSV.preF alone did not develop foldon-binding antibody titres (Fig. 1b).
Mice were immunized twice with AREXVY, a licensed foldon-containing preF-protein-based vaccine, and a dose-dependent induction of foldon-binding antibody titres was detected 2 weeks after the second immunization (Fig. 1c).
These data show that an RSV vaccine that contains a preF protein with a foldon induces off-target foldon responses in animals as well as humans, which increase after repeated immunizations.
RSV preF destabilizes upon trimerization
To prevent the induction of off-target anti-foldon responses, we set out to design a foldon-free RSV preF protein. First, we studied the impact of foldon-driven trimerization on the stability of the preF protein. We reasoned that the unique processing of RSV F0, which involves the release of the bulky, trimerization-impeding p27 domain14, together with the highly unstable nature of the mature RSV preF trimer, hints at a destabilizing effect upon trimerization.
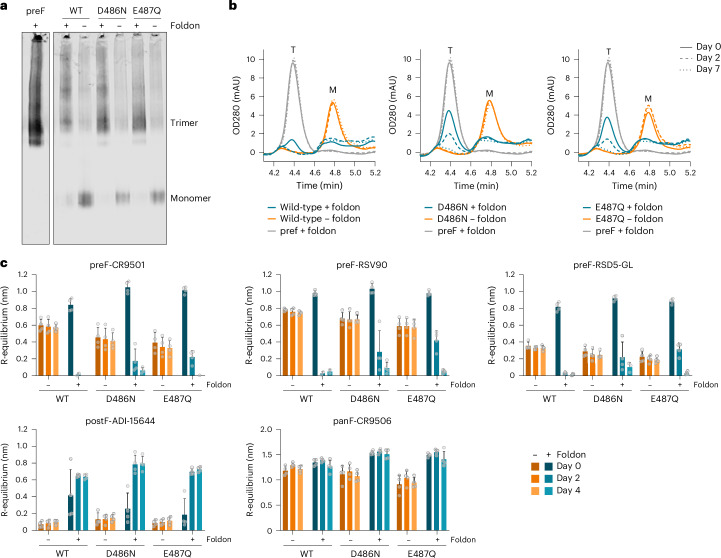
Soluble, non-stabilized RSV-A F protein ectodomain expressed as a monomer and was efficiently trimerized when C-terminally fused to a foldon, as shown by native polyacrylamide gel electrophoresis (PAGE) followed by western blot analysis of supernatants of transfected cells (Fig. 2a). Introduction of single stabilizing substitutions D486N or E487Q (ref. 14) in wild-type RSV-A F similarly resulted in monomeric F expression and also trimerized upon addition of a foldon (Fig. 2a). The elution pattern on analytical size-exclusion chromatography (SEC) confirmed the quaternary structure of these variants, which were compared with a previously described stabilized trimeric variant fused to a foldon as a positive control (Fig. 2b and Supplementary Fig. 1)14. Interestingly, while the wild-type RSV F monomer and the positive control stabilized preF trimer remained stable after storage for 7 days, the wild-type, foldon-fused RSV F trimer was no longer detectable in analytical SEC after 2 days. Semi-stabilized foldon variants with either D486N or E487Q were more stable, but still lost all trimers after 7 days of storage (Fig. 2b and Supplementary Fig. 1).
Fig. 2. Trimerization of RSV F destabilizes the prefusion conformation.
a, Multimerization profile of RSV-A F protein in the supernatant of Expi293F cells, as determined by native PAGE followed by western blot and probed with anti-RSV F antibody CR9506. RSV F proteins either were wild-type consensus sequences (‘WT’) or contained stabilizing substitutions D486N or E487Q. The presence or absence of a C-terminal foldon trimerization domain is indicated with a plus or a minus sign, respectively. The monomer and trimer bands are indicated. A stabilized (N67I, S215P, D486N) RSV preF trimer (‘preF’) with foldon was taken along as positive control for trimer formation. Native PAGE was performed three times with similar results. b, Analytical SEC analysis of RSV-A F protein expressed in the supernatant of Expi293F cells. RSV F proteins are as described in a, with the presence or absence of a C-terminal foldon trimerization domain indicated. Analytical SEC was performed 3 days after transfection at the day of collection (day 0) and after 2 and 7 days of storage at 4 °C (day 2 and day 7). As positive control for the preF trimer, a stabilized (N67I, S215P, D486N) construct was taken along with a foldon. The approximate retention time of trimeric (T) or monomeric (M) F is indicated. OD280, optical density at 280 nm. c, Antigenicity profile of wild-type, D486N or E487Q RSV-A F protein with (plus sign) or without (minus sign) a C-terminal foldon trimerization domain, as expressed in the supernatant of Expi293F cells. Measurements were performed with biolayer interferometry using a panel of monoclonal antibodies on the day of collection (day 0) and after 2 and 4 days of storage at 4 °C. The specificity of the anti-RSV F antibodies is indicated as binding to the preF or postF conformation, or as pan-specific (panF), indicating binding to both prefusion and postfusion conformations. R-equilibrium binding is reported as the average of n = 4 biological replicates + s.d., and individual data points are represented by open circles.
To investigate this process, binding studies were performed using biolayer interferometry (BLI) with a panel of monoclonal antibodies (mAbs) to probe F conformation in solution (see Supplementary Fig. 2 for epitope specificities). Antigenic analysis in supernatant at the time of collection and after 2 and 4 days of storage confirmed striking differences between monomeric RSV F and RSV F trimerized with a foldon (Fig. 2c). For monomeric wild-type RSV F, stable binding of prefusion-specific RSV F antibodies CR9501 (ref. 28), RSV90 (ref. 29) and RSD5-GL (ref. 30) and pan-specific RSV F antibody CR9506 and no binding to postF-specific mAb ADI-15664 (ref. 31) was observed. By contrast, binding of preF-specific antibodies to foldon-containing wild-type RSV F trimers was lost after storage at 4 °C for 2 days and binding to foldon-containing D486N and E487Q variants showed only residual prefusion-specific mAb binding while binding to the postF-specific antibody increased (Fig. 2c). Thus, although initially the foldon-containing variants were expressed as a preF trimer, this conformation proved unstable, switching to a postfusion conformation.
These results show that the addition of a foldon destabilizes the prefusion conformation of the soluble RSV F protein and that this effect can be reduced by the introduction of stabilizing substitutions.
Preventing RSV preF destabilization upon trimerization
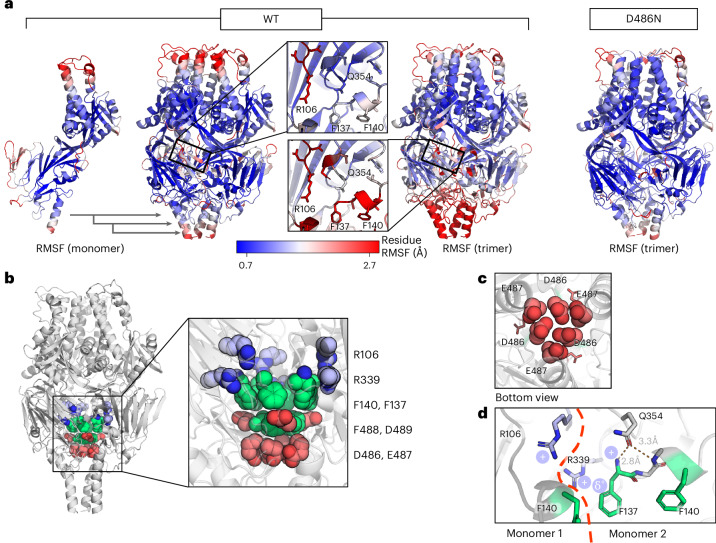
The trimer interface was investigated for possible regions of instability to inform design efforts to stabilize the preF trimer without the requirement for foldon. To systematically uncover hotspots of instability in preF upon trimerization, MD simulations for both the wild-type trimer and monomer were performed32 (Fig. 3a and Supplementary Fig. 3a). Notably, whereas the monomer showed fluctuations mainly at the apex, the trimer showed pronounced destabilization in the lower region. This observed destabilization focused around the ring of negative charge repulsion at the head–stem nexus near the three-fold axis (Fig. 3b,c), implicating residues D486 and E487 and corroborating previous findings14. However, the instability in MD analysis was more extensive, affecting the entire lower head and stem regions of RSV preF. To validate the approach, another set of MD simulations were performed on a trimer with the stabilizing D486N substitution that neutralizes the electrostatic repulsion in the repulsive ring. The mutation substantially decreases the root mean square fluctuation (RMSF) in the trimer, alleviating the wild-type instability (Fig. 3a (right panel) and Supplementary Fig 3b). Additional instability hotspots in the trimer were identified within the fusion peptide and the 353–357 loop (Fig. 3a,d). The destabilization of the fusion peptide and adjacent loops aligns with their functional necessity to detach during the conformational transformation to the postfusion state. Instability in the F2 C-terminus was evident in both monomer and trimer simulations, adding to the fusion peptide instability. This MD analysis informed our design strategy for preF trimer stabilization, focusing on these identified instability regions and the HR2 triple-helix interface that forms the stem.
Fig. 3. Regions of instability in RSV F form upon trimerization.
a, Left panels: RMSFs from MD simulations of wild-type RSV F monomer and trimer, projected on the PDB 4MMS trimer structure. The monomer fluctuations are projected on a trimer for easier comparison with the trimer simulation (central zoom on the fusion peptide region). Right panel: RMSF of the D486N mutant trimer. Also see Supplementary Fig. 3. The proteins are coloured according to the residue RMSF, with the colour white corresponding to the average fluctuations, blue to low fluctuations (less than mean − σ) and red to high fluctuations (more than mean + σ). b, Side view of the RSV F trimer. The architecture of the core is shown consisting of the negatively charged layer (red), aromatic layer (green) and positively charged layer (blue). c, Bottom view of the region of instability composed of the ring of six negative charges encircling the three-fold axis, depicting the concentrated area of electrostatic repulsion. D489 is shown as sticks. d, Positively charged region of instability at the interface of the protomers. Details of the interprotomeric repulsion between R106 and R339, the contribution of the F1 N-terminus positive charge and the positive charge at the edge of the aromatic ring of F137 are shown. Stabilizing interactions between Q354 and the F1 N-terminus are shown as dotted orange lines. Modelling of the wild-type RSV F trimer is as described in Methods.
Currently available preF-stabilized proteins of RSV type A and B were investigated to determine whether they remained trimeric without the support of a foldon trimerization domain. Stabilization of the first-generation type A RSV preF was achieved by substitutions N67I, S215P and D486N (ref. 14). Stabilization of type B RSV preF was achieved by substitutions P101Q, I152M, L203I, S215P, D486N and D489Y (ref. 33). The three stabilizing substitutions in type A (Fig. 4a, top panel) or the six stabilizing substitutions in type B (Fig. 4a, bottom panel) were insufficient to maintain a trimeric structure when the foldon was removed.
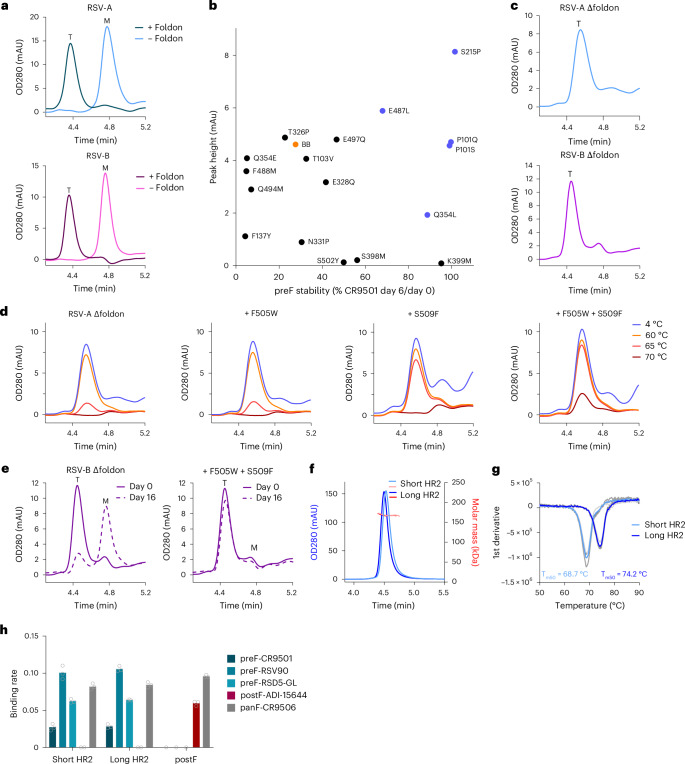
Fig. 4. Stable prefusion RSV F without a foldon domain through stabilization of the fusion peptide cavity and HR2 region.
a, Analytical SEC analysis of RSV-A and RSV-B F protein with or without foldon, as determined in cell supernatant. RSV-A F carries substitutions N67I, S215P and D486N. RSV-B F carries substitutions P101Q, I152M, L203I, S215P, D486N and D489Y. Trimeric (T) or monomeric (M) F is indicated. b, PreF expression and stability depicted as trimer peaks in analytical SEC at day 0 versus the CR9501 binding ratio at day 0 and 6. Stabilizing substitutions were screened in a semi-stable consensus backbone (‘BB’; RSV-A F with foldon and D486N). Substitutions in blue were selected for follow-up. c, Analytical SEC on RSV-A and RSV-B F without a foldon (Δfoldon) in cell supernatant. RSV-A Δfoldon contains P101Q, S215P, Q354L, D486N, E487L and D489Y. RSV-B Δfoldon contains P101Q, I152M, L203I, S215P, Q354L, D486N, E487L and D489Y. T is indicated. d, Stability of indicated variants as determined by analytical SEC on supernatants after 15 min of incubation at indicated temperatures. The backbone is RSV-A Δfoldon described in c. e, Stability of indicated variants as determined by analytical SEC on supernatants stored for 0 or 16 days at 4 °C. The backbone is RSV-B Δfoldon described in c. f, SEC-MALS (~162 kDa) of RSV-A F Δfoldon trimers with stabilizing substitutions P101Q, S215P, Q354L, D486N, E487L, D489Y, F505W and S509F. Variants contain either a short HR2 (residues 1–513) or a long HR2 (residues 1–524). g, Melting temperature (Tm50) of RSV-A F Δfoldon variants with a short or long HR2 sequence, as described in f, determined by differential scanning fluorimetry (DSF). Data reported as the average (blue line) of n = 3 technical replicates (grey lines). h, Antigenicity profile of RSV-A F Δfoldon variants with a short or long HR2, as described in f, measured by BLI. RSV postF protein is included as a control52. Antibody specificity is indicated. The binding rate is reported as the average of three technical replicates, and individual data points are represented by open circles.
Additional substitutions were screened for improved trimer expression and prefusion stability using an RSV-A F parental variant with intermediate expression and stability (Fig. 4b). This parental construct contained a foldon trimerization domain and one stabilizing substitution in the ring of negative charge repulsion (D486N). As positive controls, the previously described S215P substitution, stabilizing the hinge loop, and P101Q/S, substitutions at the protomer interface, were included14,33. Q354L successfully stabilized the prefusion conformation compared with the metastable backbone. E487L increased trimer expression and improved trimer stability more than twofold.
To evaluate the impact of additional stabilizing substitutions on RSV F trimers that were not fused to a foldon domain (Δfoldon), the selected amino acid substitutions were combined with substitutions that were previously used to stabilize RSV F of subgroup A (S215P and D486N) or subgroup B (S215P, P101Q, D486N, D489Y, I152M and L203I)33. While RSV-A F Δfoldon with S215P and D486N substitutions was expressed as a monomer (Fig. 4a), the addition of P101Q, Q354L, E487L and D489Y yielded a trimeric elution peak on analytical SEC, as shown by an elution shift towards a shorter retention time (Fig. 4c, top panel). As expected, this foldon-free trimer eluted slightly later than the foldon-containing RSV F trimer owing to the lower molecular weight (Fig. 4a,c). Similarly, introduction of substitutions Q354L and E487L additively improved the trimer-to-monomer ratio in an RSV-B F Δfoldon variant (Supplementary Fig. 4a), and the combined introduction of P101Q, I152M, L203I, S215P, Q354L, D486N, E487L and D489Y resulted in predominant trimeric expression without the need for foldon (Fig. 4c, bottom panel).
As the stability of RSV-A preF-Δfoldon was lost at temperatures over 60 °C (Fig. 4d), additional stabilizing substitutions were evaluated in the membrane-proximal HR2 stem region that forms a triple helix. Large hydrophobic residues were used for heptad positions F505 and S509 to fill the internal cavities and improve trimerization34,35. Substitution of S509 to F, I, L and M improved the trimer-to-monomer ratio in an RSV-A preF-Δfoldon backbone carrying D486N and S215P stabilizing mutations (Supplementary Fig. 4b). Importantly, introduction of S509F in the heat-sensitive RSV-A F Δfoldon protein significantly improved trimer stability after 65 °C heat stress (Fig. 4d). Next, F505W was introduced alongside S509F as a cavity-filling substitution to further increase stability. Introduction of F505W without S509F had no impact on trimer stability compared with the backbone, but the double substitution variant with F505W and S509F showed improved heat stress stability, reflected in minimal trimer loss after incubation at 65 °C and some trimer preservation at 70 °C (Fig. 4d). Synergistic effects of F505W and S509F were also observed in an RSV-B F Δfoldon variant with intermediate stability that was expressed as a monomer (Supplementary Fig. 4c). Whereas individual substitution of either F505W or S509F did not lead to trimer formation, the combination of both substitutions gave a detectable trimer peak in analytical SEC (Supplementary Fig. 4c) and improved the stability of RSV-B F Δfoldon by retaining the trimeric conformation during storage (Fig. 4e).
As the HR2 ‘inverted pyramid’ architecture of the upper stem probably continues into a more classical coiled-coil architecture in the lower stem, 11 additional C-terminal residues were added to extend the triple helix and potentially increase trimer stability. Stabilized RSV-A F Δfoldon variants with either a short stem (residues 1–513) or an extended stem (residues 1–524) were expressed and purified (Fig. 4f). The extended stem variant showed higher expression and had an increased melting temperature (Tm50) of 5.5 °C compared with the short variant (Fig. 4g). Moreover, extension of HR2 improved the trimer-to-monomer ratio in a semi-stabilized RSV-B F Δfoldon variant (Supplementary Fig. 4d). Finally, both Δfoldon trimer variants were in the prefusion conformation based on their antigenicity profiles (Fig. 4h).
In conclusion, the introduction of stabilizing head domain substitutions in combination with HR2 stem elongation and interface optimization resulted in stable, trimeric preF RSV-A and RSV-B proteins that do not require a foldon.
Structural characterization of RSV preF-Δfoldon by cryo-EM
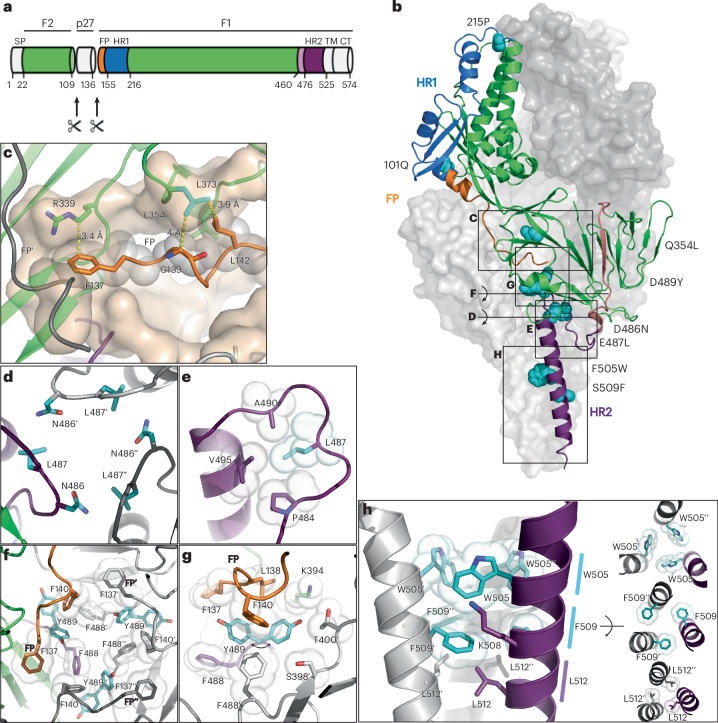
To confirm the native prefusion conformation and better understand the mechanism underlying the applied stabilizing substitutions, we determined the structure of RSV-A preF-Δfoldon containing P101Q, S215P, Q354L, D486N, E487L, D489Y, F505W and S509F using single-particle cryo-electron microscopy (cryo-EM) (Fig. 5a,b, Supplementary Fig. 5 and Supplementary Table 1). A 3.28 Å resolution three-dimensional (3D) reconstruction was obtained, which showed the side chains of all stabilizing substitutions. Notably, the extended HR2 stem up to residue S521 was resolved, thereby making it the most complete RSV structure currently available. Stabilization of the fusion peptide was provided by the Q354L substitution, which stabilizes the region of positive charge repulsion (Fig. 5c and Supplementary Fig. 6). The 354–357 loop, including the side chain of wild-type Q354, has a high B-factor in all RSV preF structures solved to date14,15,29. In our structure, L354 actually engages in hydrophobic interactions with L142 and G139, thereby stabilizing a loop in the fusion peptide. Another interaction that stabilizes the fusion peptide was observed for R339. Although in the wild-type preF R339 forms the positive repulsive cluster together with R106 and the N-terminus of F137 (Fig. 3d), in the stabilized structure, R106 is not visible and the aliphatic side chain of R339 interacts with the phenyl ring of F137 (Fig. 5c). The substitutions of all six negative charges in the ring of negative repulsion (D486N and E487L) resulted in stable interprotomeric interactions of N486 to Q494 and K498 and hydrophobic contacts of L487 with HR2 residues P484, A490 and V495 (Fig. 5d,e). The D489Y substitution in the same region of instability engages in an interaction with the fusion peptide (Fig. 5f,g). The stem region is stabilized by intraprotomeric interactions (F509–L513) and layers of interprotomeric interactions (Fig. 5h).
Fig. 5. Cryo-EM analysis of stabilized RSV-A preF-Δfoldon.
a, Schematic overview of RSV F protein in green, fusion peptide (FP) in orange, HR1 in blue, loop 460–476 in salmon and HR2 in purple. The soluble F ectodomain used in experiments consists of residues 1–524 and lacks the transmembrane (TM) and cytoplasmic (CT) regions. b, Structure of the stabilized trimer, with two monomers shown as surface representation (grey) and one monomer as ribbons with colours according to a, and side chains of stabilizing substitutions shown as cyan spheres. Letters C–H refer to the view of panels c–h relative to b. c, Docking site shown in transparent surface of fusion peptide (orange) with yellow dotted lines showing the interactions of R339 with F137 and substitution 354L (cyan) stabilizing the turn in the fusion peptide. Grey spheres are shown for interacting residues F137, G139 and L142. d, Top view of the neutralized charged ring around the trimeric symmetry axis. e, Side view of the top HR2 with intraprotomeric hydrophobic interactions of L487 (cyan). f,g, Top view around the trimeric symmetry axis (f) and side view (g) of the aromatic cluster showing interactions of both observed 489Y rotamers (cyan). Two different densities were observed for Y489, interacting either with F140 or with both F140 and F137, the N-terminal residue of the fusion peptide. Van der Waals surfaces are shown for the other aromatics. h, Side view (left) of the stabilized HR2 stem and top views (right) with the top layer showing the stabilizing W505 residues (cyan), the middle layer showing the stabilizing F509 residues (cyan) and the bottom layer with L512.
PreF-Δfoldon protects mice against RSV challenge
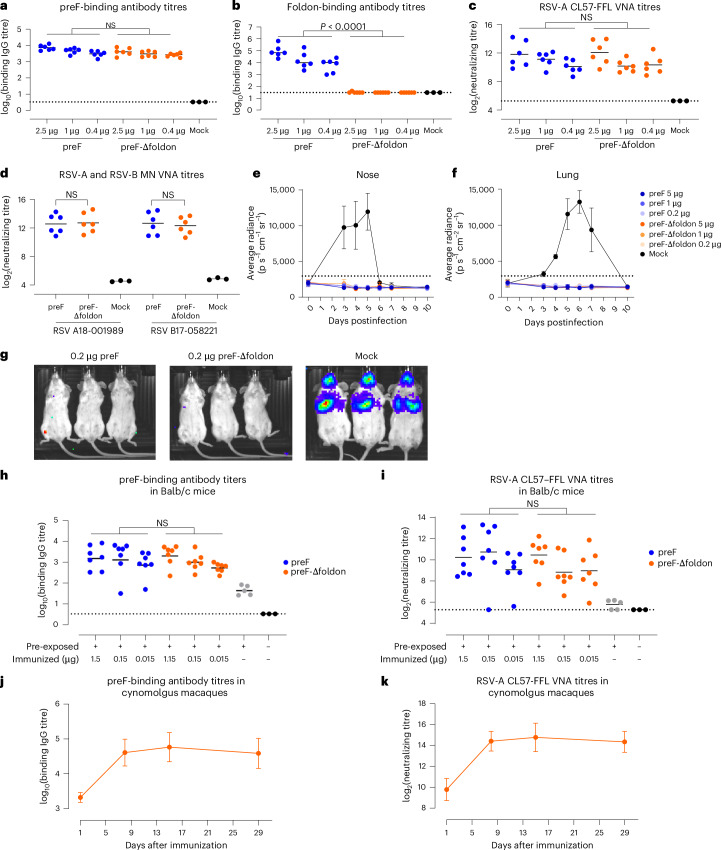
For preclinical evaluation, we generated stabilized RSV-A and RSV-B preF-Δfoldon proteins based on the sequences of circulating strains of 2023 (Supplementary Fig. 7). To investigate whether the RSV-A preF-Δfoldon protein would be an attractive alternative for the clinically evaluated foldon-containing preF protein14,27,36, but without inducing off-target foldon responses, the level and quality of the humoral immunogenicity were assessed in mice (Fig. 6a–g). Balb/c mice were immunized twice with different doses of RSV-A preF protein or RSV-A preF-Δfoldon protein adjuvanted with AS01E. Both proteins induced preF-binding antibodies, but foldon-binding antibodies were present only in mice immunized with foldon-containing preF (Fig. 6a,b). Dose-dependent induction of virus-neutralizing antibody titres (VNTs) against RSV-A CL57 was observed for both preF proteins without a statistically significant difference (Fig. 6c). To examine whether the preF and preF-Δfoldon proteins were capable of inducing neutralizing antibodies against clinical isolates, microneutralization (MN) assays were performed on serum samples of the groups that received the highest protein dose. VNTs against both RSV-A 18-001989 and RSV-B 17-058221 were induced by both proteins without statistically significant differences (Fig. 6d). Both preF and preF-Δfoldon induced similar preF/postF binding antibody ratios (Supplementary Fig. 8a,b), indicating that both proteins induce quantitatively and qualitatively similar levels of binding and neutralizing antibodies in naive mice. Next, the protective efficacy of RSV-A preF + foldon and preF-Δfoldon protein was assessed in mice that were immunized twice with AS01B-adjuvanted protein. Three weeks after the last immunization, mice were intranasally challenged with RSV-A strain CL57 expressing firefly luciferase (RSV-CL57–FFL). FFL signals in the nose and lung areas were monitored before and after the challenge. Viral replication in mock animals peaked between day 3 and 5 in the upper respiratory tract and between day 5 and 7 in the lower respiratory tract. Mice immunized with preF or preF-Δfoldon were completely protected against infection in the upper and lower tracts as evidenced by the absence of FFL signal (Fig. 6e–g).
Fig. 6. Immunogenicity and protective efficacy of RSV preF-Δfoldon.
a–d, Balb/c mice were immunized at week 0 and week 4 with formulation buffer (Mock, n = 3) or with AS01E-adjuvanted preF or preF-Δfoldon protein (see Supplementary Fig. 7 for preF designs) at indicated doses (n = 6 per group). Two weeks after the second immunization, preF-binding antibody titres (a), foldon-binding antibody titres (b) and RSV-A CL57–FFL VNT (c) were measured in all mice; VNTs against RSV-A 18-001989 and RSV-B 17-058221 were measured in the highest dose groups and mock animals (d). Black bars represent the mean response of each group, and dotted lines refer to the LLOD or lower limit of qualification (LLOQ). All measurements under the LLOD and LLOQ were set at the LLOD and LLOQ, respectively. e,f, To assess protective efficacy, Balb/c mice (n = 3 per group) were immunized with the indicated doses of AS01B-adjuvanted preF or preF-Δfoldon protein or mock control at week 0 and week 4 and intranasally challenged at day 49 with RSV CL57–FFL. Luciferase expression was measured in the nose (e) and lungs (f) before the challenge (day 0) and on days 3, 4, 5, 6, 7 and 10 after the challenge. Means and standard deviations are indicated. The background is indicated with a dotted line. g, Luciferase expression within the animals on day 5. h,i, Immunogenicity of RSV preF-Δfoldon in pre-exposed Balb/c mice. Mice were immunized with unadjuvanted preF or preF-Δfoldon 20 weeks after pre-exposure (n = 7 per group) or were mock immunized (n = 5). Naive mice (n = 3) were added as controls. Serum samples were isolated 6 weeks after immunization. PreF-binding antibody titres (h) and RSV-A CL57–FFL VNT (i) were determined. j,k, Cynomolgus macaques (n = 4) with pre-existing immunity were immunized with 50 µg RSV-A preF-Δfoldon. PreF-binding antibody titres (j) and RSV-A CL57–FFL VNT (k) were determined in serum isolated at the indicated time points. Mean titres (log2 for VNT and log10 binding IgG titres) with standard deviation are plotted. No LOD and LLOQ are defined. Statistical testing (two sided) was performed across doses using a Tobit model (a–d) or a Tobit model with Dunnett correction for multiple comparisons (h,i).
Immunogenicity was additionally assessed in mice and cynomolgus macaques with RSV pre-existing immunity to better reflect the clinical situation in human adults. Mice were intranasally inoculated with RSV-A before immunization. RSV-A preF-binding antibodies in serum isolated 8 weeks later were measured and confirmed successful RSV inoculation (Supplementary Fig. 8c). Twenty weeks after pre-exposure, animals were immunized with different doses of RSV-A preF or RSV-A preF-Δfoldon. Antibody responses and VNTs were measured 6 weeks after immunization and showed that both proteins induced similar levels of preF- and postF-binding and RSV-neutralizing antibodies (Fig. 6h,i and Supplementary Fig. 8d), resulting in a similar preF-to-postF binding antibody ratio (Supplementary Fig. 8e). We also confirmed the immunogenicity of the RSV-B-based preF-Δfoldon protein in this mouse pre-exposure model (Supplementary Fig. 8f).
Finally, RSV-A preF-Δfoldon potency was evaluated in cynomolgus macaques with pre-existing RSV immunity. Antibody titres sharply increased after immunization by 20- to 25-fold for preF-binding antibody titres and 24- to 32-fold for neutralizing antibodies at 8 days after vaccination and remained at this level for 4 weeks (Fig. 6j,k).
Altogether, the humoral immune response and protective efficacy induced by the RSV-A preF-Δfoldon protein was comparable to that of clinically tested foldon-containing preF protein in mice, and was shown to be immunogenic in RSV-experienced cynomolgus macaques without inducing non-desirable off-target anti-foldon responses.
Discussion
RSV causes severe respiratory disease in infants and elderly adults, and only recently, efficacious subunit vaccines have been approved. These are based on prefusion-stabilized F proteins fused to foldon14,15,24,37. An undesired side effect of the foldon is the induction of off-target antibodies, which could negatively impact vaccine potency after repeated administrations, and potentially the potency of future foldon-containing vaccines or therapeutics. We showed that such antibodies are indeed induced upon administration of foldon-containing vaccines in mice, non-human primates (NHPs) and humans upon repeated immunizations. It will be of considerable interest to study whether the limited VNT boosting in humans upon the second vaccination observed for both AREXVY and ABRYSVO, with geometric mean fold rises ranging from only 1 to 2.3 compared with pre-revaccination levels38, is the result of immune skewing towards the immunogenic foldon domain.
The activation of RSV F occurs at a late stage and involves double cleavage by furin-like proteases, resulting in the release of the bulky p27 domain that impedes trimerization. Here we showed that a wild-type ectodomain expressed as a monomer remained stable in the prefusion conformation39. However, upon forced trimerization by the addition of a foldon, the wild-type F protein destabilized and quickly transformed to the postfusion conformation.
The unwanted immune response to foldon and the realization that trimerization of RSV preF triggers destabilization prompted us to improve RSV preF stability and develop a foldon-free preF trimer. Two regions of instability that form upon trimerization were identified using MD: (1) a ring of negative charges between the top of the HR2 stem and the fusion peptide and (2) a repulsive cluster in the plane of the fusion peptide that involves the positively charged N-terminus of the fusion peptide. MD showed how the negatively charged cluster is unstable and highly mobile. The tip of the fusion peptide also shows increased mobility upon trimerization, and within the central cavity, the 354–357 loop shows mobility; as a result, Q354 is not able to form a stable interaction with the fusion peptide. Optimizing these two regions, as well as the interface of the HR2 stem, resulted in a preF trimer that was no longer dependent on foldon.
In mice, the preF-Δfoldon protein generated comparable levels of preF and postF binding antibodies, and neutralizing antibody responses against clinical isolates of RSV-A and RSV-B when compared with a clinically tested foldon-containing preF14,36. Furthermore, the preF-Δfoldon protein showed protection against both upper and lower respiratory tract infections. To closely mimic the clinical situation in adults, immunogenicity was also assessed in mice and cynomolgus macaques with RSV pre-existing immunity. In pre-exposed mice, the magnitude and quality of the antibody responses were comparable between the preF and preF-Δfoldon proteins. PreF-Δfoldon was also able to boost pre-existing antibody responses in cynomolgus macaques with VNTs of 24- to 32-fold above the pre-immunization levels, which is in line with the human data regarding protein-based RSV vaccines that showed approximately 5–20-fold increase after a single immunization38,40.
In conclusion, RSV preF trimerization results in several charged regions in the lower portion of the trimer that destabilize the prefusion conformation. These insights resulted in a next-generation preF design that encompasses the complete ectodomain, is independent of a heterologous trimerization domain, and has high stability, good manufacturability characteristics and excellent immunogenic properties, offering protection against RSV infection without inducing off-target anti-foldon responses.
Methods
Ethical statement
The clinical study was approved by the ethics committee or institutional review board at each participating centre as described in protocol VAC18193RSV2001. This study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. All participants provided written informed consent that allows use of the samples in this study. An independent data and safety monitoring committee monitored safety outcomes throughout the trials. The African green monkey study was conducted at the Wake Forest School of Medicine test facility and approved by the IACUC of Wake Forest University. The cynomolgus macaque study was performed by Alpha Genesis and approved by the Institutional Animal Care and Use Committee of Alpha Genesis. Mouse studies were conducted at Janssen Vaccines and Prevention B.V. according to the Dutch Animal Experimentation Act and the Guidelines on the Protection of Animals for scientific purposes by the Council of the European Committee after approval by the Centrale Commissie Dierproeven and the Dier Experimenten Commissie of Janssen Vaccines and Prevention B.V. Project license: AVD213002020-10024. Approval codes: RSV23MM01-10024, RSV23MM09-10024 and RSV23MM12-10024.
Foldon responses in humans
To investigate the induction of foldon responses in humans, a subset of clinical trial samples of a randomized, double-blind, placebo-controlled phase 2b study to assess the efficacy, immunogenicity and safety of an Ad26.RSV.preF-based regimen in the prevention of reverse transcriptase-PCR-confirmed RSV-mediated lower respiratory tract disease in adults aged 65 years and older (NCT03982199) were used. The subset contained serum samples of participants that first received placebo, followed by an immunization with 5 × 1011 vector particles (vp) Ad26.RSV.preF + 150 µg RSV-A preF protein 1 year later (week 0 of the data shown) and a second immunization 1 year later (week 52) with 5 × 1011 Ad26.RSV.preF or 150 µg RSV-A preF protein or 5 × 1011 Ad26.RSV.preF + 150 µg RSV-A preF protein. Foldon-binding antibody titres were measured in serum samples isolated before immunization (week 0 and week 52) and 2 weeks after immunization (week 2 and week 54).
Foldon responses in African green monkeys
A detailed description of the study in African green monkeys (Chlorocebus aethiops sabaeus) used to measure foldon responses is given in ref. 27. In short, adult and elderly female animals (9–26 years of age at the start of the study) were pre-exposed to 7.5 × 105 plaque-forming units (pfu) of RSV-A Memphis 37, 19 weeks before intramuscular immunization at week 0 and week 60 with 5 × 1010 vp of Ad26.RSV.preF + 150 µg RSV-A preF protein. At week 189, the animals were boosted with 5 × 1010 vp of Ad26.RSV.preF + 150 µg RSV-A or RSV-B preF protein (both proteins containing a foldon domain).
Serum isolated from 10 animals at weeks −5, 4, 60, 64, 189 and 193 were analysed for foldon-binding antibody responses. The aim of this study was initially as described in ref. 27, and therefore, statistical power analysis to detect differences in foldon-binding antibody titres was not performed upfront.
Immunogenicity assessment in cynomolgus macaques
Four cynomolgus macaques (two males and two females, approximately 7 years of age at the start of the study) were rolled over from a vaccination study in which the animals were intranasally pre-exposed to RSV-A and subsequently immunized with RSV-A-based vaccine candidates. The animals were all confirmed to be RSV serum converted at the start of the study to assess the immunogenicity of preF-Δfoldon. The animals received a single immunization with 50 µg preF-Δfoldon protein via the intramuscular route. Blood was collected on the day of immunization and 8, 15 and 29 days after immunization, from which serum was isolated. The serum samples were analysed for preF-binding and CL57-neutralizing antibody titres. As the results were purely descriptive and explorative, statistical power analysis was not performed upfront.
Immunogenicity assessment in naive and RSV-pre-exposed mice
RSV-naive mice, female Balb/c mice (Charles River Laboratories; 6–8 weeks old) were randomly assigned to the different groups and immunized intramuscularly twice with a 4 week interval with 0.4, 1 or 2.5 µg preF, preF-Δfoldon or AREXVY (GSK) adjuvanted with AS01E (10.9 µl per animal). Two weeks after the second immunization, serum was isolated and tested for foldon-binding antibody titres, preF- and postF-binding antibody titres and VNTs against RSV A strain CL57, RSV A 18-001989 and RSV B 17-058221.
In the pre-exposure model, 20 weeks before immunization, female Balb/c mice (Charles River Laboratories; 6–8 weeks old) were randomly assigned to the different groups and intranasally pre-exposed to 5 × 105 pfu of RSV A 18-001989. At week 20, the mice were immunized intramuscularly with different dose levels of RSV-A preF or preF-Δfoldon or RSV-B preF-Δfoldon protein without adjuvant. Serum samples were collected for assessments of preF- and postF-binding antibody titres and VNT against strains RSV A CL57.
Group sizes were chosen based on power calculations that used historical internal results of animal studies in naive and pre-exposed mice, and aimed to show a difference in VNT between the groups immunized with preF and preF-Δfoldon using an across-dose statistical test.
RSV mouse challenge model
Mice (female Balb/c mice; Charles River Laboratories; 6–8 weeks old) were randomly assigned to the different groups and immunized intramuscularly with different doses of AS01B-adjuvanted preF or preF-Δfoldon or formulation buffer. Forty-nine days later, mice were intranasally inoculated with 1 × 106 pfu of RSV-A strain CL57 containing a firefly luciferase gene (RSV CL57–FFL). Mice were imaged before the challenge and at days 3–7 and at day 10 after the challenge for FFL expression. Therefore, mice received 200 µl of d-luciferin potassium salt in PBS (15 mg ml−1) through subcutaneous administration in the scruff of the neck. After administration of luciferin, mice were kept awake for 5 min to allow distribution of the substrate before being imaged under anaesthesia (isoflurane) using the IVIS Lumina II (Perkin Elmer). Regions of interest (nose and lung) were selected for calculation of signal intensity. Light emission was measured in photons (p) s−1 cm−2 sr−1 (photon flux). Acquisition and analysis were performed with Living Image Software, version 4.5 (Caliper Life Sciences). The background, defined as average + 3 × standard deviation of all mice imaged before the challenge, is indicated with a dotted line in Fig. 6e,f.
No statistical power calculation was performed to determine group sizes as our study represents an exploratory study. Group sizes were chosen based on operational feasibility, and results were not used for statistical analysis.
Detecting foldon-binding antibody responses by ELISA
IgG antibodies binding to the foldon domain were measured in duplicate by ELISA. White 96-well half-area plates were coated with streptavidin (0.66 µg ml−1) and incubated overnight at 4 °C. After the wells were washed, they were incubated with biotinylated-foldon peptide (Biotin-Ttds-GYIPEAPRDGQAYVRKDGEWVLLSTFL-OH; JPT Peptide Technologies) for 1 h at room temperature (RT). After the wells were washed, serially diluted serum was added and incubated for 1 h at RT. After the wells were washed, foldon-binding IgG antibodies were detected by HRP-labelled anti-mouse IgG or anti-human (also for NHP) (1 h at RT), and after they were washed, the wells were developed with Lumiglo. The luminescence signal was measured, and end-point titres were calculated.
Detecting preF and postF binding antibody responses by ELISA
IgG antibodies binding to RSV preF and RSV postF were measured in duplicate by ELISA. White 96-well plates were coated with streptavidin (5 µg ml−1) and incubated for 2 h at 37 °C. After the wells were washed, they were blocked with casein in PBS for 1 h at RT and washed again, followed by the addition of biotinylated preF or postF protein and incubation for 1 h at RT. After the wells were washed, serially diluted serum was added and incubated for 1 h at RT. After the wells were washed, RSV F-specific antibodies were detected using HRP-labelled anti-mouse IgG or HRP-labelled anti-human IgG (1 h at RT), and after the wells were washed, they were developed with electrochemiluminescence. The luminescence signal was measured. Titres were calculated and expressed as relative potency to a standard serum sample taken along on each plate.
Detecting RSV-A strain CL57–FFL-neutralizing antibody titres
Neutralizing antibody titres in mouse and cynomolgus macaque serum were determined in duplicate by an automated microneutralization assay using FFL-labelled RSV-A CL57 propagated on A549 cells. Serially diluted heat-inactivated serum samples were mixed with 25 × 103 pfu RSV CL57 in half area white tissue culture plates and incubated for 1 h at RT. Subsequently, 5 × 103 A549 cells per well were added, and plates were incubated for 20 h at 37 °C and 10% CO2. After 20 h, Neolite substrate was added. The luminescence signal was determined with the EnVision plate reader. VNA titres were calculated as the antibody concentration that caused a 90% reduction in luminescence, expressed as IC90 titres.
Detecting RSV-A 18-001989-and RSV-B 17-058221-neutralizing antibody titres by MN VNA
RSV-neutralizing antibody titres were measured in duplicate by a microneutralization assay using RSV-susceptible Vero cells. Mouse serum samples, which had been heat inactivated, were serially diluted and then mixed with 1,000 pfu of clinical isolates of RSV-A 18-001989 or RSV-B 17-058221 strains. Following 1 h of incubation at room temperature, Vero cells were introduced, and the plates were allowed to incubate for 4 days at 37 °C. Subsequently, the cell monolayers were washed and fixed using 80% cold acetone. The extent of RSV replication was determined by measuring F protein expression, using a biotin-conjugated anti-F monoclonal antibody known as MAb8262 (clone 133-1H, Merck), which has equal affinity for both RSV-A and RSV-B F proteins. The plates were then subjected to incubation with streptavidin–horseradish peroxidase, and after the plates were washed, a chemiluminescent substrate (LumiGLO, SeraCare) was applied. The luminescent signal was quantified using the Biotek Synergy Neo plate reader, and inhibitory concentration 50% (IC50) titres were subsequently calculated.
Statistical analysis
Data collection and analysis were not performed blinded to the conditions of the experiments. No data points or animals were excluded from the analysis. Binding antibody titres were log10 transferred, and VNTs were log2 transformed and compared using a two-sided paired t-test, ANOVA or ANOVA for potentially censored values (Tobit model). This model contained group as explanatory factor. Bonferroni or Dunnett adjustment was applied to correct for multiple testing. P values (corrected P values in case multiple testing correction was applied) ≤ 0.05 were considered statistically significant.
Protein expression and purification
Plasmids encoding RSV-A and RSV-B F proteins were codon optimized, synthesized and cloned into a pCDNA2004 vector at Genscript. For expression screening experiments, Expi293F cells were transfected at a 200 μl scale using ExpiFectamine (Gibco, Thermo Fisher Scientific) according to the manufacturer’s instructions in Expi293F Expression medium [+] GlutaMAX (Gibco, Thermo Fisher Scientific) with 100 U ml−1 Pen-Strep (Gibco) in 96-half deep-well plates. Transfected cells were cultured for 3 days at 37 °C, 75% humidity, 250 rpm and 8.0% CO2, after which the supernatants were collected by centrifugation and subsequent filtration using a 0.2 µm filter (Pall Life Sciences). For protein purification experiments, Expi293F cells were transfected at 300 ml scale using ExpiFectamine according to the manufacturer’s instructions in Expi293F Expression medium [+] GlutaMAX and cultured for 5 days at 37 °C, 75% humidity, 125 rpm and 8.0% CO2. Supernatants were collected by centrifugation and clarified by sterile filtration using a 0.22 µm PES vacuum filter (Nalgene). RSV F trimers were purified using a two-step protocol on an ÄKTA Avant 25 system (Cytiva). Clarified supernatant was diluted with 1.33 volume of Milli-Q water and conditioned to pH 5 by adding 200 mM NaOAc, pH 5.0, to a final concentration of 40 mM NaOAc. The conditioned supernatant was applied to a 5 ml Capto SP Impress column (Cytiva) equilibrated in 20 mM NaOAc, pH 5.0. A mixture of 5% buffer B (20 mM NaOAc, 1 M NaCl, pH 5.0) in buffer A (20 mM NaOAc, pH 5.0) was used to wash the column, after which the protein was eluted stepwise with 15%, 30%, 50% and 100% buffer B with a length of 4 column volumes per step. RSV F trimer that eluted in the 15% buffer B step was pooled and concentrated using an Amicon Ultra 15 30 kDa cutoff filter (Millipore) before further purification via size exclusion. The protein was applied on a HiLoad Superdex 200 16/600 column (Cytiva) equilibrated in 20 mM Tris and 75 mM NaCl, pH 7.5, and monodisperse fractions were pooled and filtered 0.22 µm to form the final product.
Analytical size-exclusion chromatography
An ultra-high-performance liquid chromatography system (Vanquish, Thermo Scientific) and µDAWN TREOS instrument (Wyatt) coupled to an Optilab µT-rEX Refractive Index Detector (Wyatt), in combination with an in-line Nanostar DLS reader (Wyatt), was used for performing analytical SEC experiments. The cleared crude cell culture supernatants were applied to a Unix-C SEC-300 15 cm column (Sepax Technologies) with the corresponding guard column (Sepax Technologies) equilibrated in running buffer (150 mM sodium phosphate, 50 mM NaCl, pH 7.0) at 0.35 ml min−1. When analysing supernatant samples, µMALS detectors were offline and analytical SEC data were analysed using Chromeleon 7.2.8.0 software package (Thermo Fisher Scientific). The signal of supernatants of non-transfected cells was subtracted from the signal of supernatants of RSV F-transfected cells. When purified proteins were analysed using size exclusion chromatography with multi-angle static light scattering (SEC-MALS), µMALS detectors were inline and data were analysed using Astra 8.0.0.19 software package (Wyatt). For the protein component, a differential change in refractive index with respect to concentration (dn/dc in ml g−1) value of 0.1850 was used, and for the glycan component, a value of 0.1410 was used. Molecular weights were calculated using the RI detector as source for concentration and mass recoveries using UV as source for concentration.
Heat stress analytical SEC
Cleared crude cell culture supernatant was heated at 60 °C, 65 °C or 70 °C for 15 min, whereas the control was kept at 4 °C. Subsequently, the samples were centrifuged at 18,000 × g for 10 min. Analytical SEC was performed and analysed as described above.
Biolayer interferometry
An Octet RED384 system (FortéBio) was used to perform BLI experiments at a shaking speed of 1,000 rpm at 30 °C in 96-well black flat-bottom polypropylene microplates (FortéBio). Anti-hIgG (AHC) sensors (FortéBio) were activated for 600 s in 1× kinetics buffer (Sartorius). Next, anti-RSV F antibodies CR9501 (ref. 28), RSV90, RSD5-GL, ADI-15644 and CR9506 were immobilized for 1,800 s on the AHC sensors at a concentration of 5 μg ml−1 in 1× kinetics buffer. For RSV F protein analysis in clarified supernatant, a blocking step with the supernatant of non-transfected cells was applied for 600 s. For purified RSV F protein, a 300 s baseline step in 1× kinetics buffer was used. Following this step, binding of RSV F protein either in undiluted supernatant or as purified protein at a concentration of 20 μg ml−1 in 1× kinetics buffer was measured during a 900 s association step. The initial binding rate and R-Equilibrium was calculated using FortéBio Data Analysis 12.0 software (FortéBio).
Differential scanning fluorimetry
In a 96-well optical qPCR plate, 20 μg purified protein in 90 μl PBS pH 7.4 (Gibco) was mixed with 10 μl of 50-times-diluted SYPRO orange fluorescent dye (5,000 × stock, Invitrogen). Triplicates of 30 μl of the mixture were dispensed in a MicroAmp Fast Optical 96-well plate (Thermo Fisher Scientific) and sealed with MicroAmp Optical Adhesive Film (Thermo Fisher Scientific). A negative control sample containing only the dye was used for reference subtraction. The measurement was performed in a qPCR instrument (Applied Biosystems ViiA 7) using a temperature ramp from 25 °C to 95 °C with a rate of 0.015 °C s−1. Data were collected continuously, measuring reporter ROX. The melting temperatures were derived from the negative first derivative that was plotted as a function of temperature. The lowest point in the curve indicates the melting temperature.
Native PAGE western blot
The oligomeric state of RSV F protein in clarified supernatant was assessed by blue native PAGE according to the manufacturer’s instructions (Thermo Fisher Scientific). Briefly, the inner chamber of a Novex XCell SureLock gel running system (Thermo Fisher Scientific) was filled with 1× native page running buffer supplemented with 0.1× cathode additive. The outer chamber was filled with 1× native page running buffer. Samples were supplied with sample buffer and loaded, and the gel was run for 120 min at 150 V. After running, the gel was incubated in 1× transfer buffer for 10 min. Semi-dry blotting was performed according to manufacturers’ recommendations (Thermo Fisher Scientific). Briefly, the samples were transferred onto a PVDF membrane using the iBlot system (Thermo Fisher Scientific). The membrane was blocked for 1 h in Odyssey Blocking Buffer (Li-Cor), followed by overnight incubation with anti-RSV F CR9506 antibody at 1:10,000 dilution. After the blot was washed three times with TBS–0.05% Tween-20, it was incubated with secondary anti-human CW800 antibody for 1 h at 1:10,000, followed by washing three times with TBS–0.05% Tween-20 and a final wash with PBS. The blot was visualized by scanning on an Odyssey scanner (Li-Cor), using both the 700CW and 800CW channel.
MD of the wild-type RSV F
The input structure for the MD simulations was based on a stabilized RSVF trimer structure, closely resembling the structure of RSV F Variant Cav1 (PDB: 4MMS) with a root mean square deviation (RSMD) of 0.4 Å (ref. 15). The structure contains a complete F2 C-terminus extending to residue 506. For generating the wild-type F trimer configuration, all stabilizing mutations were reverted to their original wild-type residues using Rosetta’s fixed backbone methodology41. In addition, the F1 C-terminus was elongated to residue L513, using the trimeric structure from PDB entry 5TPN for the missing residues29. The structure of the D486N mutant was prepared from the wild-type monomer and trimer by mutating the D486 residue to asparagine using Rosetta’s fixed backbone method. MD simulations were conducted with ACEMD42, using the AMBER14SB force-field, TIP3P water model and Langevin thermostat to ensure accurate representation of the physical interactions42. The system was surrounded by a solvation layer of water molecules extending 10 Å from the protein structure’s surface, and periodic boundary conditions were used. Simulations were carried out at a temperature of 37 °C to reflect the relevant biological or experimental context. Before the dynamics simulations, the structures underwent energy minimization to remove potential steric clashes and relax the system. Constraints on heavy atoms were gradually lifted over the initial 60 ns of simulation to allow natural system equilibration. A total of one 500 ns simulation for the monomer and five separate 500 ns simulations for the trimer were completed, providing a comprehensive dataset for analysis. Subsequent analysis focused on the RMSFs of the atomic positions, calculated using the Gromacs suite to quantify the dynamic behaviour and stability of the protein structures across the simulation trajectories.
Cryo-EM
Grid preparation and data acquisition
Purified RSV-A preF-Δfoldon, 3.5 µl at 1.6 mg ml−1, was applied to the plasma-cleaned (Gatan Solarus) Quantifoil 1.2/1.3 UltraAuFoil holey gold grid, and subsequently vitrified using a Vitrobot Mark IV (FEI Company). To overcome an orientation bias, n-octyl-β-d-glucopyranoside (BOG, Anatrace) was added to the sample before freezing. Cryo grids were loaded into a Glacios transmission electron microscope (Thermo Fisher Scientific) operating in nanoprobe at 200 keV with a Falcon direct electron detector. Images were recorded with EPU (Thermo Fisher Scientific) in counting mode. Images were recorded in an electron-event representation (EER) format corresponding to a total dose of 40.0 electrons per Å2. All details corresponding to individual datasets are summarized in Supplementary Table 1.
Electron microscopy data processing
The videos were subjected to beam-induced motion correction, contrast transfer function parameter estimation, automated reference particle picking, particle extraction with a box size of 320 pixels and two-dimensional classification in cryoSPARC 3.2.0 (ref. 43) live during the data acquisition. Particle images with clear prefusion features were merged and subjected to ab initio 3D reconstruction with C3 symmetry in cryoSPARC. Multiple rounds of optimized 3D heterogeneous refinement yielded two classes with a clear prefusion density. The particles were refined using non-uniform and local refinements within cryoSPARC with C3 symmetry. Local resolution was calculated using ResMap44. All resolutions were estimated by applying a soft mask around the protein complex density and based on the gold-standard (two halves of data refined independently) Fourier shell correlation (FSC) = 0.143 criterion. Before visualization, all density maps were sharpened by applying different negative temperature factors using automated procedures and, along with the half maps, were used for model building. The number of particles in each dataset and other details related to data processing are summarized in Supplementary Fig. 2 and Supplementary Table 1.
Model building and refinement
The initial template of the RSV prefusion complex was derived from a homology-based model calculated by SWISS-MODEL45. The model was docked into the EM density map using ChimeraX version 1.6.1 (ref. 46) and followed by manual adjustment using COOT version 1.2 (ref. 47). Each model was independently subjected to global refinement and minimization in real space using the module phenix.real_space_refine in PHENIX version 1.20 (ref. 48) against separate EM half maps with default parameters. The model was refined into a working half map, and improvement of the model was monitored using the free half map. The geometry parameters of the final models were validated in COOT version 1.2 and using MolProbity version 4.5 (ref. 49) and EMRinger (ref. 50). These refinements were performed iteratively until no further improvements were observed. The final refinement statistics were provided in Supplementary Table 1. Model overfitting was evaluated through its refinement against one cryo-EM half map. FSC curves were calculated between the resulting model and the working half map as well as between the resulting model and the free half and full maps for cross-validation. Figures were produced using PyMOL51 and ChimeraX version 1.6.1 (ref. 46).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1–8 and Table 1.
Source data for supplementary figures.
Source data
Statistical source data.
Acknowledgements
We thank H. Kuipers, H. van Diepen and J. Serroyen of Janssen Vaccines & Prevention BV, Leiden, The Netherlands, for assistance. We received no specific funding for this work.
Author contributions
M.J.G.B. and J.P.M.L. conceptualized the study. M.J.G.B., A.K., D.v.O., L.L. and R.V. planned and/or performed biochemical assays and purifications. M.J.G.B., F.C., W.v.d.H., J.V., A.T., L.T., L.v.d.F., R.Z. and J.P.M.L. planned and/or performed preclinical experiments. J.J. performed MD simulations. X.Y. performed the cryo-EM characterization. M.J.G.B., F.C., A.K., X.Y., J.J. and J.P.M.L. wrote the paper with input from all other authors.
Peer review
Peer review information
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The coordinates and EM maps generated in this study have been deposited in the Protein Data Bank and the Electron Microscopy Data Bank under accession codes PDB 9B2X and EMD-44117. Source data are provided with this paper.
Competing interests
The authors declare no competing financial interests. M.J.G.B., A.K., J.J. and J.P.M.L. are co-inventors on related vaccine patents. M.J.G.B., F.C., X.Y., D.v.O., L.L., W.v.d.H., J. V., A.T., R.V., J.J., L.v.d.F., R.Z. and J.P.M.L. are current or former employees of Johnson and Johnson.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mark J. G. Bakkers, Freek Cox.
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-024-01860-1.
References
- 1.Rha, B. et al. Respiratory syncytial virus-associated hospitalizations among young children: 2015–2016. Pediatrics146, e20193611 (2020). [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin, J. M. et al. Respiratory syncytial virus-associated hospitalization rates among US infants: a systematic review and meta-analysis. J. Infect. Dis.225, 1100–1111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenmoe, S. & Nair, H. The disease burden of respiratory syncytial virus in older adults. Curr. Opin. Infect. Dis.37, 129–136 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandya, M. C., Callahan, S. M., Savchenko, K. G. & Stobart, C. C. A contemporary view of respiratory syncytial virus (RSV) biology and strain-specific differences. Pathogens8, 67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouzas, M. L. et al. Respiratory syncytial virus a and b display different temporal patterns in a 4-year prospective cross-sectional study among children with acute respiratory infection in a tropical city. Medicine95, e5142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLellan, J. S., Ray, W. C. & Peeples, M. E. Structure and function of RSV surface glycoproteins. Curr. Top. Microbiol. Immunol.372, 83–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison, S. C. Viral membrane fusion. Virology479–480, 498–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLellan, J. S. et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science340, 1113–1117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, P. L. & Mottet, G. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol.72, 3095–3101 (1991). [DOI] [PubMed] [Google Scholar]
- 10.González-Reyes, L. et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl Acad. Sci. USA98, 9859–9864 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmer, G., Conzelmann, K.-K. & Herrler, G. Cleavage at the furin consensus sequence RAR/KR(109) and presence of the intervening peptide of the respiratory syncytial virus fusion protein are dispensable for virus replication in cell culture. J. Virol.76, 9218–9224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crank, M. C. et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science365, 505–509 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Ngwuta, J. O. et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci. Transl. Med.7, 309ra162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krarup, A. et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun.6, 8143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLellan, J. S. et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science342, 592–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papi, A. et al. Respiratory syncytial virus prefusion F protein vaccine in older adults. N. Engl. J. Med.388, 595–608 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Walsh, E. E. et al. Efficacy and safety of a bivalent RSV prefusion F vaccine in older adults. N. Engl. J. Med.388, 1465–1477 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Sacconnay, L. et al. The RSVPreF3-AS01 vaccine elicits broad neutralization of contemporary and antigenically distant respiratory syncytial virus strains. Sci. Transl. Med.15, eadg6050 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Verwey, C., Dangor, Z. & Madhi, S. A. Approaches to the prevention and treatment of respiratory syncytial virus infection in children: rationale and progress to date. Paediatr. Drugs26, 101–112 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipp, M. A. & Empey, K. M. Recent advances in the prevention of respiratory syncytial virus in pediatrics. Curr. Opin. Pediatr.36, 182–189 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letarov, A. V., Londer, Y. Y., Boudko, S. P. & Mesyanzhinov, V. V. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry64, 817–823 (1999). [PubMed] [Google Scholar]
- 22.Górski, A. et al. Bacteriophages targeting intestinal epithelial cells: a potential novel form of immunotherapy. Cell. Mol. Life Sci.75, 589–595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne, P. O. & McLellan, J. S. Principles and practical applications of structure-based vaccine design. Curr. Opin. Immunol.77, 102209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Che, Y. et al. Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine. Sci. Transl. Med.15, eade6422 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Güthe, S. et al. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol.337, 905–915 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, K. J. et al. A general computational design strategy for stabilizing viral class I fusion proteins. Nat. Commun.15, 1335 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeland, E. et al. Combination Ad26.RSV.preF/preF protein vaccine induces superior protective immunity compared with individual vaccine components in preclinical models. NPJ Vaccines8, 45 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilman, M. S. A. et al. Transient opening of trimeric prefusion RSV F proteins. Nat. Commun.10, 2105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mousa, J. J., Kose, N., Matta, P., Gilchuk, P. & Crowe, J. E. A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat. Microbiol.2, 16271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, H. G. et al. Alternative conformations of a major antigenic site on RSV F. PLoS Pathog.15, e1007944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilman, M. S. A. et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci. Immunol.1, eaaj1879 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casalino, L. et al. Breathing and tilting: mesoscale simulations illuminate influenza glycoprotein vulnerabilities. ACS Cent. Sci.8, 1646–1663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langedijk, J. P. M. et. al. Stabilized pre-fusion RSV FB antigens. WO2022175477 (2022).
- 34.Langedijk, J. P. M. et al. Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein. Nat. Commun.15, 4629 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakkers, M. J. G. et al. Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer. Nat. Commun.15, 6270 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falsey, A. R. et al. Efficacy and safety of an Ad26.RSV.preF-RSV preF protein vaccine in older adults. N. Engl. J. Med.388, 609–620 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Graham, B. S. Vaccine development for respiratory syncytial virus. Curr. Opin. Virol.23, 107–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leroux-Roels, I. et al. Safety and immunogenicity of a revaccination with a respiratory syncytial virus prefusion F vaccine in older adults: a phase 2b study. J. Infect. Dis.229, 355–366 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson, K. A. et al. A monomeric uncleaved respiratory syncytial virus F antigen retains prefusion-specific neutralizing epitopes. J. Virol.88, 11802–11810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leroux-Roels, I. et al. Safety and immunogenicity of a respiratory syncytial virus prefusion F (RSVPreF3) candidate vaccine in older adults: phase 1/2 randomized clinical trial. J. Infect. Dis.227, 761–772 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science302, 1364–1368 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Harvey, M. J., Giupponi, G. & Fabritiis, G. D. ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput.5, 1632–1639 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res.46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.PyMOL by Schrödinger (Schrodinger, 2021).
- 52.McLellan, J. S., Yang, Y., Graham, B. S. & Kwong, P. D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol.85, 7788–7796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–8 and Table 1.
Source data for supplementary figures.
Statistical source data.
Data Availability Statement
The coordinates and EM maps generated in this study have been deposited in the Protein Data Bank and the Electron Microscopy Data Bank under accession codes PDB 9B2X and EMD-44117. Source data are provided with this paper.