Abstract
Background
Pediatric non-alcoholic fatty liver disease (NAFLD) is a chronic steatosis of the liver associated with energy metabolism in children and adolescents, failure to intervene promptly can elevate the risk of developing hepatocellular carcinoma. Therefore, this study aimed to understand the underlying mechanism of pediatric NAFLD and investigate potential biomarkers and therapeutic targets.
Methods
We investigated genes using the GSE185051 data set related to energy metabolism from the GeneCards database, constructed protein–protein interaction network, identified hub genes and established networks representing interactions between these hub genes and miRNA, RNA-binding proteins, transcription factors, and drugs. Subsequently, we performed Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, Gene Set Enrichment Analysis (GSEA), and immune infiltration analysis.
Results
Our analysis identified 9 hub genes through the PPI network. The target molecules were identified through the interaction network between hub genes and miRNAs, RNA-binding proteins, transcription factors, and drugs. GO analysis revealed that hub genes were associated with oxidative stress responses and other pathways. KEGG analysis highlighted their involvement in pathways such as insulin resistance, among others. GSEA revealed that hub genes were highly enriched in pathways related to Omega-9 fatty acid synthesis, among others. Immune infiltration analysis suggested that mast cells and T follicular helper cells play significant roles in the pathogenesis of NAFLD.
Conclusion
We identified the hub genes in pediatric NAFLD closely related to energy metabolism. These findings offer the potential for identifying potential novel diagnostic biomarkers, and establishing therapeutic targets for pediatric NAFLD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-024-08702-4.
Keywords: Pediatric non-alcoholic fatty liver disease, Chronic steatosis, Integrative analyses, Genes, Energy metabolism, Hepatocellular carcinoma
Introduction
Pediatric non-alcoholic fatty liver disease (NAFLD) is a clinicopathologic syndrome characterized by chronic steatosis of the liver in children and adolescents under the age of 18 years. It involves the accumulation of fat in the liver, affecting more than 5% of the liver cells, except in cases where chronic fat deposition in the liver is attributed to alcohol consumption or other definitive pathogenic factors. Over the past two decades, the prevalence of NAFLD among American children and adolescents has increased by over 100 percent, making it the predominant cause of chronic liver disease in many nations [1]. In European children (aged 3–18 years), prevalence varies from 1.3 to 22.5%, with an average of 11% in children (average age of 12.4 ± 2.6 years) [2]. In China, an estimated 45% of obese adolescents are affected by NAFLD [3]. Without timely intervention, NAFLD poses a risk of hepatocellular carcinoma in the future. Presently, research on NAFLD primarily focuses on adults, despite the unique characteristics of pediatric NAFLD. The incidence of NAFLD in pediatrics is alarming, and there are presently no biological indicators or measures that can be used to properly diagnose and track the course of the condition [4], emphasizing the urgent need to determine whether uniform and distinct pathogenic mechanisms exist in pediatrics. Furthermore, there are currently no authorized therapies for NAFLD in adults or children and no medications in phase 3 trials for pediatric patients. Therefore, elucidating the mechanisms of pediatric NAFLD and the investigation of new biomarkers and therapeutic targets is crucial for the precise diagnosis and management of NAFLD [5].
There exists substantial evidence supporting the notion that metabolic dysfunction plays a pivotal role in the development and progression of NAFLD [6]. In 2021, an international panel of experts renamed NAFLD as “metabolism-associated fatty liver disease” [4]. Research has shown that the only effective therapies for mitigating hepatic steatosis and improving the metabolic phenotype in adults include weight loss achieved through dietary modifications or bariatric surgery, increased physical activity, and reduced fructose consumption [7, 8]. Although NAFLD is closely associated with metabolic dysfunction, the causal relationships and underlying pathogenesis have yet to be fully elucidated. Therefore, further research into the relationship between energy metabolism and NAFLD is warranted.
Effective bioinformatics analysis can provide valuable insights into the molecular-level origins and progression of diseases. Therefore, utilizing differentially expressed genes (DEGs) associated with NAFLD and energy metabolism in pediatrics, we created a Protein–Protein Interaction (PPI) network, identified key genes (referred to as “hub genes”), and established mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-drugs interactions networks involving these hub genes. We also conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses, Gene Set Enrichment Analysis (GSEA), and immune infiltration analysis to study the pathogenesis of pediatric NAFLD from the perspective of energy metabolism, as well as to identify potential novel diagnostic biomarkers and therapeutic targets.
Methods
Data Download
The expression profile data set GSE185051 [9] of pediatric NAFLD was downloaded from the GEO database [10]using the R package GEOquery [11]. The data set GSE185051 pertains to Homo sapiens and comprises expression profiles of samples of the GSE185051 obtained from high-throughput sequencing of liver biopsies from 52 pediatric NAFLD cases and 5 corresponding healthy liver control samples. The data platform utilized was GPL24676 Illumina NovaSeq 6000.
The expression profile data of 52 pediatric NAFLD samples (NAFLD group) and 5 healthy liver control samples (Control group) from the data set GSE185051, measured by RNA-seq, were included in subsequent analyses. Any references to probe annotations were related to earlier steps or additional comparative analyses, not the RNA-seq data itself. Energy metabolism is an important factor affecting biological metabolic activities. Energy Metabolism Related Genes (EMRGs) were identified through the GeneCards database [12] (https://www.GeneCards.org/). GeneCards is a comprehensive database containing extensive information on human genomes. We used the term “Energy metabolism” as a search keyword and only retained 69 EMRGs with “Protein Coding” and “Relevance score > 6.000.” In addition, we also used “Energy metabolism” as the search keyword in Molecular Signatures Database (MSigDB) [13] (https://www.gsea-msigdb.org/), resulting in two reference gene sets REACTOME_INTEGRATION_OF_ENERGY_METABOLISM and WP_ENERGY_METABOLISM totaling 153 EMRGs. Overall, 137 EMRGs were obtained after merging and deduplication in the GSE185051 data set. Specific gene names were provided in Table S1.
DEGs Related to Pediatric NAFLD
To elucidate the potential mechanisms, biological characteristics, and pathways related to differential gene expression in pediatric NAFLD, we first standardized the data set GSE185051 using the limma package [14] and performed differential analysis to obtain DEGs between the NAFLD group and the Control group. DEGs were selected based on the criteria of |logFC|> 1.5 and P. adj. < 0.05 as the difference for our further research. Among them, genes with logFC > 1.5 and P. adj. < 0.05 were considered up-regulated DEGs, and genes with logFC < 1.5 and P. adj. < 0.05 were considered down-regulated DEGs.
To identify Energy Metabolism Related Differentially Expressed Genes (EMRDEGs) in pediatric NAFLD, we determined the intersection of DEGs and EMRGs using a Venn diagram. The outcomes of the differential analysis were further visualized through heatmaps generated with the R package heatmap and volcanic plots created using the R program ggplot2.
ROC Curve
The Receiver Operating Characteristic (ROC) [15] curve is a graphical tool used to assess model performance, select optimal models, or establish ideal thresholds. It reflects the connection between sensitivity and specificity. The area under the curve (AUC) of ROC typically ranges from 0.5 to 1. The diagnostic impact increases as the AUC gets closer to 1. AUC values between 0.5 and 0.7 suggest low accuracy, while those between 0.7 and 0.9 indicate moderate accuracy, and AUC values above 0.9 represent high accuracy. To assess the diagnostic potential of EMRDEGs in pediatric NAFLD, we computed the AUC using the pROC package of R to create the ROC curve of EMRDEGs.
Differential Gene Function Enrichment GO Analysis and Pathway Enrichment KEGG Analysis
GO [16] analysis is a widely adopted approach for examining functional enrichment on a large scale, including biological processes (BP), molecular functions (MF), and cellular components (CC). KEGG [17] is an extensively used repository that contains comprehensive data related to genomes, BP, and medications, among others. The R package clusterProfiler [18] was utilized to perform GO and KEGG analysis on EMRDEGs. The Benjamini–Hochberg approach was used for P-value correction. P.adj. is directly utilized for filtering significant enriched terms, while FDR measures the rate of false positives introduced by multiple testing. We first screened potential enriched terms using p.adj. and then validated their significance using FDR. This approach provides a double check that ensures both statistical significance and reproducibility of the enriched pathways we have identified. The statistical significance of the entry screening criterion was determined using P. adj. < 0.1 and an FDR < 0.05.
GSEA
GSEA [19] is a computational method used to assess the distribution pattern of genes within a predetermined gene set in a sorted gene table based on their association with a specific phenotype. This analysis enables the determination of the potential contribution of the gene set to the observed phenotype. In this study, genes from the GSE185051 data set were categorized into two distinct groups based on positive and negative logFC values. Subsequently, the clusterProfiler tool was used to conduct enrichment analysis on all DEGs within the positive and negative logFC value cohorts. The GSEA was conducted with the following parameters: seed value set to 2023, 1000 calculations, a minimum of 10 genes per gene set, and a maximum of 500 genes per gene set. The P value correction method employed was Benjamini–Hochberg method. The c2.cp.all.v2022.1.Hs.symbols.gmt gene set was acquired from the MSigDB. The criteria used to determine substantial enrichment was a P.adj. value < 0.05 and an FDR value (q.value) < 0.25.
PPI Network
The PPI network comprises interactions between various proteins, playing pivotal roles in numerous BP, including the transmission of biological signals, regulation of gene expression, production of energy and materials, and cell cycle control. Understanding the operating principles of proteins within biological systems, the response mechanism of biological signals and the mechanisms governing biological signal responses, and the role of energy-related substances in specific physiological states like diseases all depend on a systematic analysis of the interactions among a large number of proteins within these biological systems. The STRING database [20] is a comprehensive and searchable database that contains information on both confirmed and hypothesized PPI. In this study, we established a PPI network from the screened EMRDEGs using the STRING database (minimum required interaction score: medium confidence (0.400)). To visualize the PPI network model, we utilized Cytoscape (version 3.9.1) [21] and designated these EMRDEGs as the central hub genes associated with pediatric NAFLD.
Construction of mRNA-miRNA, mRNA-RBP, mRNA-TF, mRNA-Drugs Interaction Network
The MiRDB database [22] serves as a platform for the prediction of target genes and functional annotation of miRNAs. We predicted miRNAs that interacted with hub genes and created a network of mRNA-miRNA interactions using summarized data from MiRDB database with a Target Score > 80.
The ENCORI database (version 3.0) [23], accessible at https://starbase.sysu.edu.cn/, is an extension of the starBase database. It encompasses interactions involving miRNA-ncRNA, miRNA-mRNA, ncRNA-RNA, RNA-RNA, RBP-ncRNA, and RBP-mRNA all of which are derived from CLIP-seq and degradome sequencing (for plants) data mining. ENCORI offers multiple visualization interfaces for exploring miRNA targets. In addition, we utilized the ENCORI database to predict interactions between RNA-binding proteins (RBPs) and hub genes. We screened mRNA-RBP interaction pairs using the criteria of clusterNum > 4 and clipExpNum > 4 and subsequently constructed mRNA-RBP interaction network.
CHIPBase database (version 3.0) [24], accessible at https://rna.sysu.edu.cn/chipbase/, is capable of identifying binding motif matrices and their corresponding binding sites by analyzing ChIP-seq data obtained from DNA-binding proteins. Additionally, it can predict the transcriptional regulatory connections between numerous transcription factors (TFs) and genes. The hTFtarget database [25], accessible at http://bioinfo.life.hust.edu.cn/hTFtarget, is a comprehensive repository containing information on human TFs and their corresponding regulatory targets. Utilizing the CHIPBase and hTFtarget databases, we conducted a search for TFs that bind to hub genes and subsequently presented our findings using Cytoscape.
Additionally, using the comparative toxicogenomics database CTD [26], available at http://ctdbase.org/, we further predicted potential medications or small molecule compounds interacting with hub genes. Subsequently, we visualized mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-drugs interaction networks using the Cytoscape software.
Immune Infiltration Assay (CIBERSORT)
To determine the composition and number of immune cells within mixed cell populations, we used the immune infiltration analysis algorithm CIBERSORT [27]. This algorithm deconvolutes the transcriptome expression matrix based on the principles of linear support vector regression. In order to filter out data with an enrichment score of immune cells greater than zero, we integrated the pediatric NAFLD data set matrix data using the CIBERSORT package [28] in conjunction with the LM22 characteristic gene matrix. The integration yielded precise results regarding the abundance matrix of infiltrating immune cells. The Spearman correlation method was used to determine the associations among various immune cell types within samples from the pediatric NAFLD data set, and the ggplot2 R package was used to display the results. Furthermore, using the R package ggplot2, we merged the gene expression matrix from the pediatric NAFLD data set to determine the associations between immune cells and hub genes in various groups, ultimately creating a correlation dot map.
Statistical Analysis
The R program (Version 4.1.2) was used for all data processing and analysis in this study. Continuous variables were presented as mean ± standard deviation. The Wilcoxon rank sum test was used to compare the two groups. Unless stated otherwise, the results represened the Spearman correlation analysis' correlation coefficients between various molecules, with a significance threshold set at a P value of less than 0.05 to identify significant differences.
Results
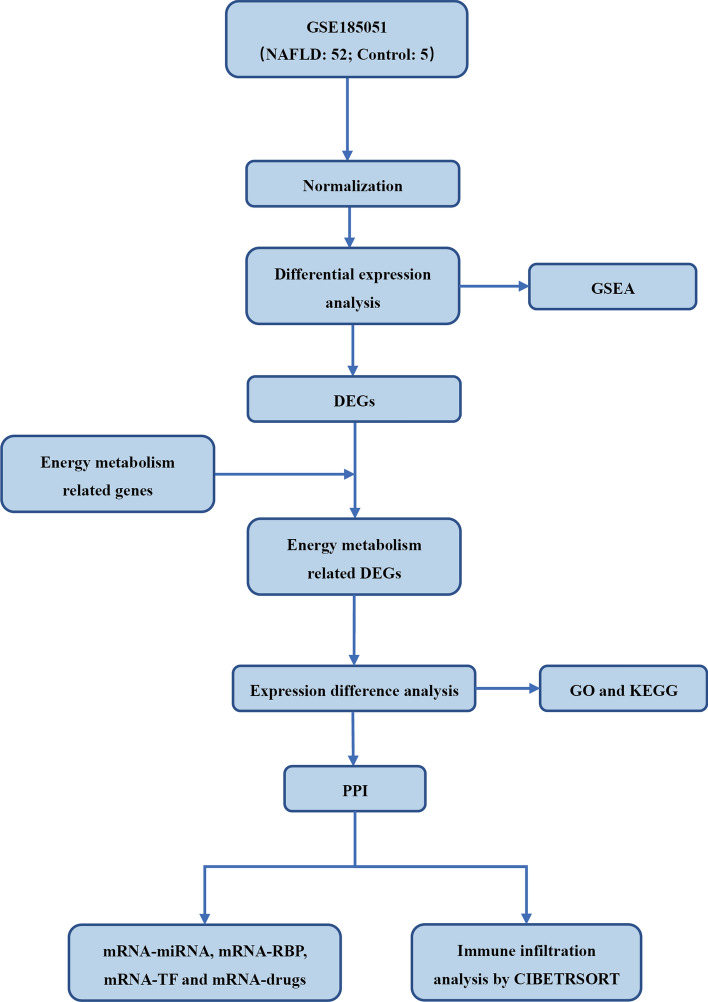
This study utilized the GSE185051 data set, which contains 52 NAFLD samples and 5 control samples. The data were first standardized to ensure consistency among samples. Through differential expression analysis, significantly DEGs between NAFLD and control samples were identified, and GSEA was performed to identify gene sets and pathways associated with NAFLD. After identifying the DEGs, the focus shifted to genes related to energy metabolism, screening these genes and conducting detailed expression difference analysis. Subsequently, GO analysis and KEGG pathway enrichment analysis were performed on the EMRDEGs to reveal their roles in biological processes and signaling pathways. PPI analysis was conducted to identify the interaction relationships between these genes, to further understand their role in NAFLD. To gain deeper insights into the regulatory network of genes and potential drug targets, correlation analyses were also performed between mRNA and miRNA, mRNA and RBP, mRNA and TF, as well as mRNA and drugs. Finally, CIBERSORT was used to analyze immune cell infiltration, providing insights into the changes in the immune microenvironment in NAFLD. The workflow of bioinformatics analysis of the present study was shown in Fig. 1.
Fig. 1.
The workflow of bioinformatics analysis
Standardization and Differential Analysis of Pediatric NAFLD
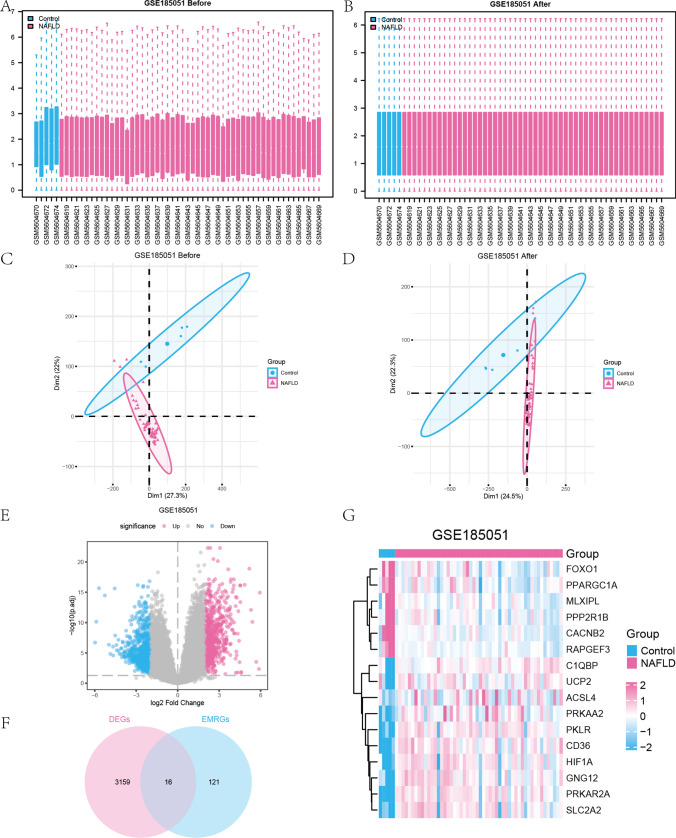
While all the samples in the GSE185051 dataset originate from the same cohort (pediatric NAFLD and healthy controls), technical variations during high-throughput sequencing data processing can introduce batch effects, we standardized the pediatric NAFLD data set GSE185051 using the R package “limma” (Fig. 2A–D). The dataset consisted of 57 samples in the GSE185051 data set: including 5 healthy liver samples in the Control group and 52 samples from pediatric NAFLD patients in the NAFLD group (Fig. 2A–B). As illustrated in Fig. 2A–B, the expression profile data of the pediatric NAFLD data set GSE185051 were effectively normalized, resulting in consistent expression patterns of the sample data. Next, we assessed the effectiveness of batch effect removal using principal component analysis (PCA). As evident from comparing the two figures (Fig. 2C–D), the corrected plot showed a tighter clustering of samples across different experimental groups, indicating that batch effects have been mitigated to a certain extent. This ensured the accuracy of subsequent differential expression analyses.
Fig. 2.
Standardization processing of pediatric NAFLD dataset. A–B Boxplot diagrams of GSE185051 data set before (A) and after (B) normalization. The Y-axis represented the gene expression values (after log2 transformation). C–D PCA plots of GSE185051 before (C) and after (D) batch effect removal. E Volcano plot of DEGS between the NAFLD group and the Control group of the GSE185051 data set. F Venn diagram of DEGs and EMRGs in the dataset. G Complex numerical heatmap of EMRDEGs in the GSE185051 data set
To examine the disparities in gene expression between the NAFLD group and the Control group, we used the “limma” software to conduct differential analysis on the GSE185051 data set, resulting in the identification of DEGs.
Overall, 22,808 genes were obtained from the data set GSE185051, among which 3175 genes matched the threshold of |logFC|> 1.5 and P. adj. < 0.05. Under this threshold, 1439 genes exhibited higher expression in the NAFLD group (with positive logFC indicating lower expression in the Control group), while 1736 genes showed lower expression in the NAFLD group (with negative logFC, indicating higher expression in the Control group). A volcano plot was displayed to visualize the outcomes of the differential analysis of the data sets. (Fig. 2E).
We identified 16 EMRDEGs in pediatric NAFLD, and a Venn diagram was constructed to illustrate their overlap (Fig. 2F). These 16 EMRDEGs included ACSL4, C1QBP, CACNB2, CD36, FOXO1, GNG12, HIF1A, MLXIPL, PKLR, PPARGC1A, PPP2R1B, PRKAA2, PRKAR2A, RAPGEF3, SLC2A2, and UCP2.
According to the findings from the Venn diagram, we examined the differential expression of these 16 EMRDEGs in the NAFLD group compared to the Control group within the GSE185051 data set (Fig. 2G). Subsequently, we created a heatmap using the R package “pheatmap” to visualize the expression patterns of these 16 EMRDEGs.
Expression Difference Analysis of EMRDEGs
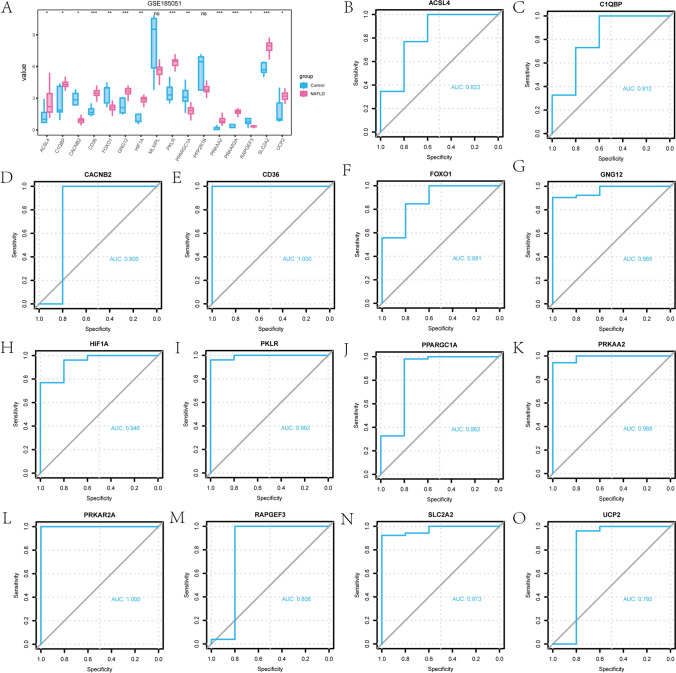
To further investigate the expression differences of the 16 EMRDEGs, we conducted an analysis of their expression levels in the GSE185051 dataset and their association with the relationship between the concentrated expression level of EMRDEGs in the GSE185051 data set and the NAFLD group and Control group (Fig. 3A).
Fig. 3.
Expression difference analysis of EMRDEGs. A Group comparison chart of the expression difference analysis of EMRDEGs in the GSE185051 data set. The “value” on the Y-axis represented the normalized expression value of the gene. B–O ROC curve analysis results for ACSL4 (B), C1QBP (C), CACNB2 (D), CD36 (E), FOXO1 (F), GNG12 (G), HIF1A (H), PKLR (I), PPARGC1A (J), PRKAA2 (K), PRKAR2A (L), RAPGEF3 (M), SLC2A2 (N), UCP2 (O) in the GSE185051 data set. The symbol ns was equal to P ≥ 0.05, not statistically significant; the symbol * was equal to P < 0.05, statistically significant; the symbol ** was equal to P < 0.01, highly statistically significant; the symbol *** was equal to P < 0.001, very statistically significant
Firstly, we assessed the expression differences of the 16 EMRDEGs in the GSE185051 data set between the NAFLD group and the Control group using the Wilcoxon signed rank test. The results showed that 14 EMRDEGs (ACSL4, C1QBP, CACNB2, CD36, FOXO1, GNG12, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, RAPGEF3, SLC2A2, and UCP2) in the GSE185051 data set exhibited statistically significant differences in expression between the NAFLD group and the Control group (Fig. 3A) (Symbol * was equivalent to P < 0.05, symbol ** was equivalent to P < 0.01, symbol *** was equivalent to P < 0.001. The significant differential expression of these genes in NAFLD may be associated with the occurrence and progression of the disease, deserving further investigation and research).
Overall, 14 EMRDEGs exhibited statistically significant expression differences in the GSE185051 data set and the results were displayed (Fig. 3B–O). The ROC curve analysis revealed the diagnostic accuracy of these EMRDEGs for pediatric NAFLD. Notably, the expression levels of CACNB2 (AUC = 0.965, Fig. 3D), CD36 (AUC = 1.000, Fig. 3E), GNG12 (AUC = 0.965, Fig. 3G), HIF1A (AUC = 0.946, Fig. 3H), PKLR (AUC = 0.992, Fig. 3I), PRKAA2 (AUC = 0.988, Fig. 3K), PRKAR2A (AUC = 1.000, Fig. 3L), and SLC2A2 (AUC = 0.973, Fig. 3N) demonstrated high diagnostic accuracy for pediatric NAFLD. Additionally, ACSL4 (AUC = 0.823, Fig. 3B), C1QBP (AUC = 0.881, Fig. 3C), FOXO1 (AUC = 0.881, Fig. 3F), PPARGC1A (AUC = 0.862, Fig. 3J), RAPGEF3 (AUC = 0.808, Fig. 3M), UCP2 (AUC = 0.792, Fig. 3O) demonstrated a reasonable level of diagnostic accuracy for pediatric NAFLD.
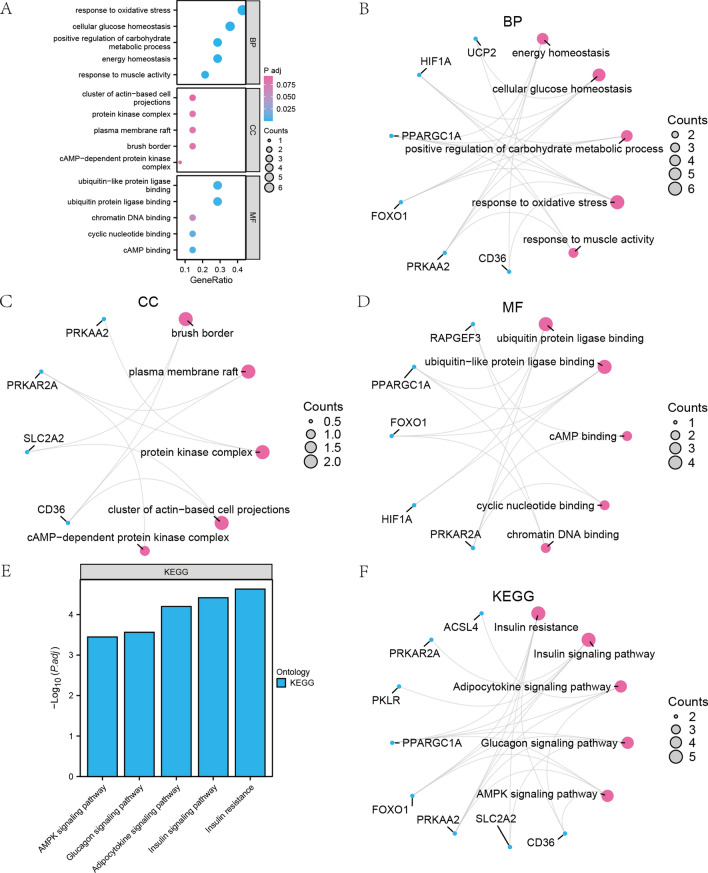
EMRDEGs GO and KEGG Analysis
To elucidate the relationship between BP, MF, CC, and other biological pathways associated with 14 EMRDEGs (ACSL4, C1QBP, CACNB2, CD36, FOXO1, GNG12, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, RAPGEF3, SLC2A2, and UCP2) and pediatric NAFLD, we conducted a GO analysis on the 14 EMRDEGs (Table 1). Enrichment items with a P. adj. < 0.1 and an FDR value (q.value) < 0.05 were considered statistically significant. The results indicated that these 14 EMRDEGs were primarily enriched in BP related to energy homeostasis, cellular glucose homeostasis, positive regulation of carbohydrate metabolic process, responses to oxidative stress, responses to muscle activity, and other BP in pediatric NAFLD. Furthermore, they were associated with CC, such as the brush border, plasma membrane raft, protein kinase complex, cluster of actin-based cell projections, and cAMP-dependent protein kinase complex, among others. Additionally, these genes were found to be involved in MF, including enrichment in ubiquitin protein ligase binding, ubiquitin-like protein ligase binding, cAMP binding, cyclic nucleotide binding, and chromatin DNA binding, among others. The results of the GO analysis were visually presented in a bubble plot (Fig. 4A) and network maps (Fig. 4B–D), illustrating the results of the BP, CC, and MF pathways.
Table 1.
GO enrichment analysis results of hub genes
| Ontology | ID | Description | Gene ratio | Bg ratio | p value | p.adjust |
|---|---|---|---|---|---|---|
| BP | GO:0097009 | Energy homeostasis | 4/14 | 40/18800 | 1.73e-08 | 1.93e-05 |
| BP | GO:0001678 | Cellular glucose homeostasis | 5/14 | 154/18800 | 6.52e-08 | 3.62e-05 |
| BP | GO:0045913 | Positive regulation of carbohydrate metabolic process | 4/14 | 77/18800 | 2.52e-07 | 9.35e-05 |
| BP | GO:0006979 | Response to oxidative stress | 6/14 | 433/18800 | 3.7e-07 | 9.65e-05 |
| BP | GO:0014850 | Response to muscle activity | 3/14 | 21/18800 | 4.34e-07 | 9.65e-05 |
| CC | GO:0005903 | Brush border | 2/14 | 102/19594 | 0.0023 | 0.0821 |
| CC | GO:0044853 | Plasma membrane raft | 2/14 | 113/19594 | 0.0029 | 0.0821 |
| CC | GO:1902911 | Protein kinase complex | 2/14 | 115/19594 | 0.0030 | 0.0821 |
| CC | GO:0098862 | Cluster of actin-based cell projections | 2/14 | 158/19594 | 0.0055 | 0.0929 |
| CC | GO:0005952 | cAMP-dependent protein kinase complex | 1/14 | 10/19594 | 0.0071 | 0.0929 |
| MF | GO:0031625 | Ubiquitin protein ligase binding | 4/14 | 298/18410 | 5.93e-05 | 0.0043 |
| MF | GO:0044389 | Ubiquitin-like protein ligase binding | 4/14 | 317/18410 | 7.53e-05 | 0.0043 |
| MF | GO:0030552 | cAMP binding | 2/14 | 23/18410 | 0.0001 | 0.0051 |
| MF | GO:0030551 | Cyclic nucleotide binding | 2/14 | 37/18410 | 0.0004 | 0.0100 |
| MF | GO:0031490 | Chromatin DNA binding | 2/14 | 105/18410 | 0.0028 | 0.0591 |
Fig. 4.
GO analysis and KEGG analysis of EMRDEGs. A Bubble diagram display of GO functional enrichment analysis results of EMRDEGs. B–D Circular network diagrams of BP pathway (B), CC pathway (C), and MF pathway (D) in the GO functional enrichment analysis results of EMRDEGs. E–F The results of KEGG pathway enrichment analysis of EMRDEGs were shown in bar graphs (E) and network graphs (F). The ordinate in the bubble diagram (A) was GO terms, and the bubble color indicated the P. adj. value of the pathway. Blue dots in network diagrams (B, C, D, F) represented specific genes, and red circles represented specific pathways. The screening criteria for GO and KEGG enrichment items were P.adj. < 0.1 and FDR value (q.value) < 0.05
Next, we conducted a KEGG analysis (Table 2) on these 14 EMRDEGs. The analysis revealed significant enrichment of these genes in pathways such as Insulin resistance, Insulin signaling, Adipocytokine signaling, Glucagon signaling, and AMPK signaling pathways. We used histograms (Fig. 4E) and network diagrams (Fig. 4F) to display the gene expression in these five enriched KEGG pathways.
Table 2.
KEGG enrichment analysis results of EMRDEGs
| Ontology | ID | Description | Gene ratio | Bg Ratio | p value | p. adjust |
|---|---|---|---|---|---|---|
| KEGG | hsa04931 | Insulin resistance | 5/12 | 108/8164 | 2.72e-07 | 2.34e-05 |
| KEGG | hsa04910 | Insulin signaling pathway | 5/12 | 137/8164 | 8.91e-07 | 3.83e-05 |
| KEGG | hsa04920 | Adipocytokine signaling pathway | 4/12 | 69/8164 | 2.2e-06 | 6.3e-05 |
| KEGG | hsa04922 | Glucagon signaling pathway | 4/12 | 107/8164 | 1.27e-05 | 0.0003 |
| KEGG | hsa04152 | AMPK signaling pathway | 4/12 | 121/8164 | 2.07e-05 | 0.0004 |
GSEA of Pediatric NAFLD Data Set
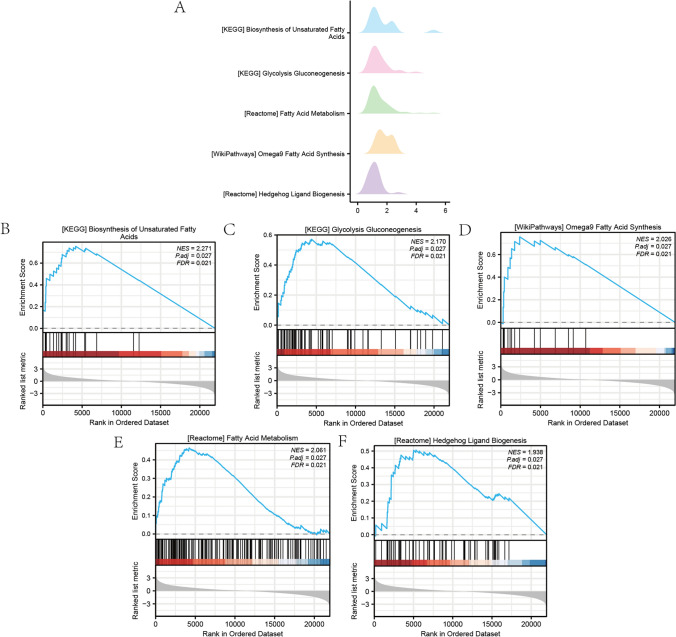
To investigate the influence of gene expression levels on the risk of pediatric NAFLD, we used GSEA to assess the BP, affected cell types, and the expression of individual genes within the GSE185051 data set. We employed stringent screening criteria, requiring a P. adj. < 0.05 and an FDR value (q. value) < 0.25. According to the findings, the GSEA of the GSE185051 data set had five main biological characteristics (Fig. 5A), the DEGs were considerably enriched in Biosynthesis of Unsaturated Fatty Acids (Fig. 5B), Glycolysis Gluconeogenesis (Fig. 5C), Omega-9 Fatty Acid Synthesis (Fig. 5D), Fatty Acid Metabolism (Fig. 5E), Hedgehog Ligand Biogenesis (Fig. 5F) and various other pathways (Fig. 5B–F, Table 3). These findings offer insights into how specific biological processes and pathways may contribute to the risk of pediatric NAFLD based on gene expression patterns.
Fig. 5.
GSEA of pediatric NAFLD data set GSE185051. A The GSEA of the GSE185051 dataset revealed five main biological characteristics. The X-axis represented the LogFC (Log2 Fold Change) value of gene expression changes. B–E The DEGs in the GSE185051 data set were significantly enriched in Biosynthesis of Unsaturated Fatty Acids (B), Glycolysis Gluconeogenesis (C), Omega-9 Fatty Acid Synthesis (D), Fatty Acid Metabolism (E), Hedgehog Ligand Biogenesis (F) pathway. The significant enrichment screening criteria for GSEA enrichment analysis were P. adj. < 0.05 and FDR value (q.value) < 0.25
Table 3.
GSEA analysis results of dataset GSE185051
| Description | Set size | Enrichment score | NES | p.adjust | q value |
|---|---|---|---|---|---|
| KEGG_BIOSYNTHESIS_OF_UNSATURATED_FATTY_ACIDS | twenty two | 0.754905 | 2.271131 | 0.027132 | 0.020635 |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | 61 | 0.571754 | 2.170466 | 0.027132 | 0.020635 |
| REACTOME_FATTY_ACID_METABOLISM | 174 | 0.465028 | 2.060954 | 0.027132 | 0.020635 |
| WP_OMEGA9_FATTY_ACID_SYNTHESIS | 14 | 0.756823 | 2.026126 | 0.027132 | 0.020635 |
| REACTOME_HEDGEHOG_LIGAND_BIOGENESIS | 64 | 0.50621 | 1.938256 | 0.027132 | 0.020635 |
| REACTOME_TNFR2_NON_CANONICAL_NF_KB_PATHWAY | 100 | 0.455465 | 1.884371 | 0.027132 | 0.020635 |
| WP_GLYCOLYSIS_AND_GLUCONEOGENESIS | 44 | 0.524923 | 1.860042 | 0.027132 | 0.020635 |
| WP_FATTY_ACID_OMEGAOXIDATION | 14 | 0.689731 | 1.84651 | 0.027132 | 0.020635 |
| WP_OMEGA3_OMEGA6_FATTY_ACID_SYNTHESIS | 15 | 0.667767 | 1.82177 | 0.027132 | 0.020635 |
| REACTOME_SIGNALING_BY_TGFB_FAMILY_MEMBERS | 122 | 0.34902 | 1.486172 | 0.038322 | 0.029146 |
EMRDEGs PPI Network, mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-Drugs Interaction Network
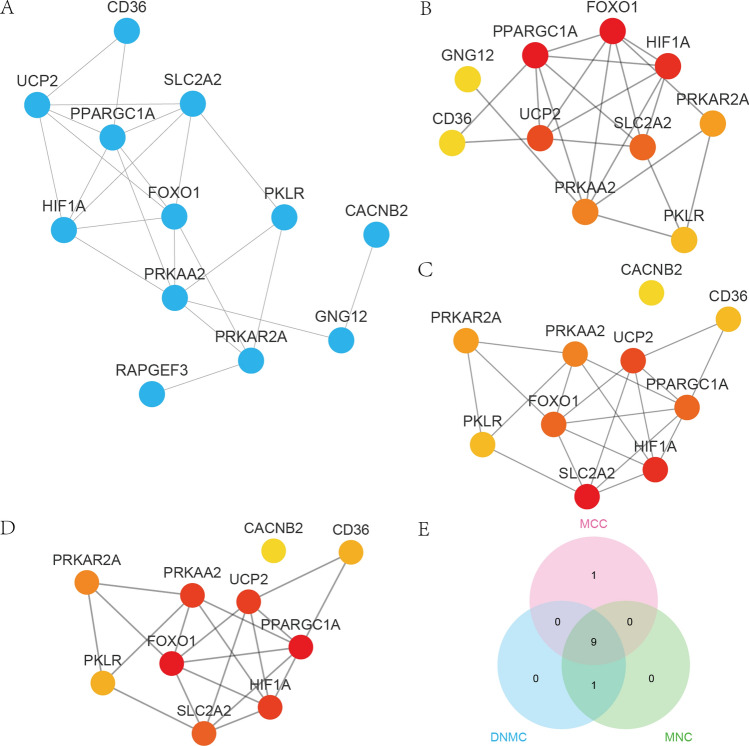
We used the STRING database to analyze 14 EMRDEGs (ACSL4, C1QBP, CACNB2, CD36, FOXO1, GNG12, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, RAPGEF3, SLC2A2, and UCP2) for PPI network (minimum required interaction score: medium confidence (0.400)). We retained only EMRDEGs connected to other nodes, resulting in a PPI network comprising 12 EMRDEGs (CACNB2, CD36, FOXO1, GNG12, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, RAPGEF3, SLC2A2, and UCP2) all considered as hub genes. We visualized this PPI network using Cytoscape software (Fig. 6A).
Fig. 6.
EMRDEGs PPI network. A PPI network of EMRDEGs. B–E MCC (B), DMNC (C), and MNC (D) algorithm top 10 node networks of the PPI network of EMRDEGs. E The top 10 node Venn diagram results of the four algorithms MCC, DMNC, MNC, and Degree in the EMRDEGs PPI network were displayed
To identify the hub genes within the PPI network, we analyzed the nodes connected to other nodes through the cytoHubba plug-in in the Cytoscape software. The cytoHubba [29] plug-in is used to identify hub genes in PPI networks. This plug-in implements various algorithms to evaluate the importance of genes in PPI networks. Through the cytoHubba plug-in, we used three algorithms: Matthews correlation coefficient metric (Fig. 6B), differential metabolic network construction (Fig. 6C), maximal neighborhood coefficient (Fig. 6D). Matthews Correlation Coefficient is a statistical metric that takes into account true positives, true negatives, false positives, and false negatives to measure the performance of binary classification models in classification tasks. It is also a measurement method to evaluate the importance of nodes in a network. Its value ranges from -1 to 1, where 1 represents complete correlation, 0 indicates no correlation, and -1 represents negative correlation. Through ranking by scores, genes with high centrality and importance in PPI networks can be identified. Differential metabolic network reconstruction is a method that identifies significantly changed network nodes and edges in a metabolic network by comparing different conditions (such as healthy and pathological groups) to determine the changes that occur under specific diseases or physiological states. In this study, it was utilized to analyze and identify hub genes involved in the pathological process of NAFLD. The maximal neighborhood coefficient is an indicator used to evaluate the importance of network nodes. It is calculated based on the neighborhood density surrounding a node. Maximal neighborhood coefficient measures the degree of interconnection between a node and its neighboring nodes. A high maximal neighborhood coefficient indicates that the node has higher connectivity in the network, potentially playing a more important biological function. By combining these three methods, we were able to identify hub genes with biological importance under specific conditions in the PPI network. The top 10 EMRDEGs with the highest scores were selected for each algorithm (Fig. 6B–D). These scores gradually increased from yellow to red in the figures. We then identified the intersection of the top 10 EMRDEGs obtained by each algorithm (Matthews correlation coefficient, differential metabolic network construction, maximal neighborhood coefficient), respectively, to acquire the hub genes of the EMRDEGs PPI network and displayed the results in a Venn diagram (Fig. 6E). Overall, we identified 9 hub genes: CD36, FOXO1, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, and UCP2.
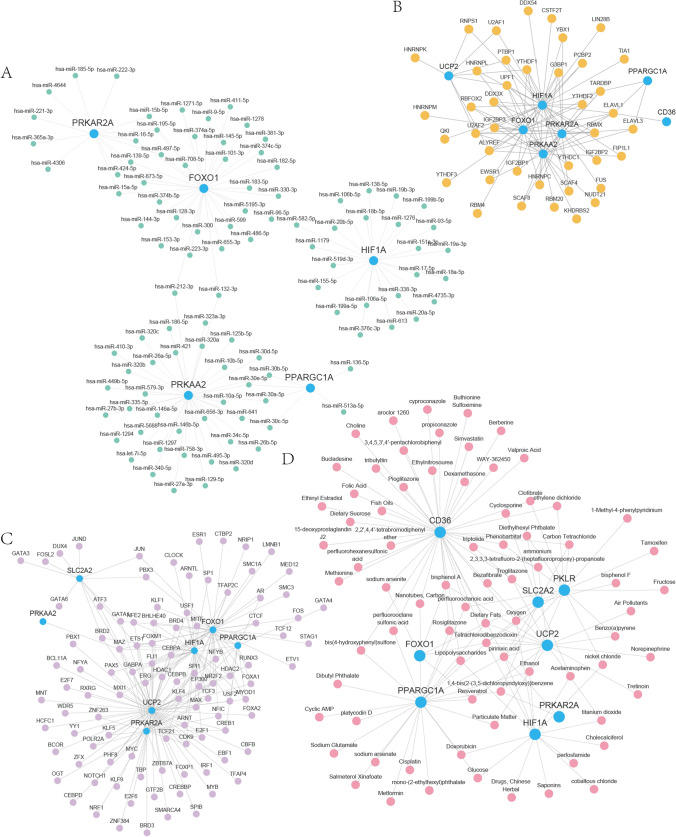
For miRNA interactions with these 9 hub genes (CD36, FOXO1, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, UCP2), we utilized the mRNA-miRNA data from the miRDB database. The mRNA-miRNA interaction network was then displayed using Cytoscape software (Fig. 7A). The sky blue circles in the mRNA-miRNA interaction network represented mRNAs, and the green circles represented miRNAs. The mRNA-miRNA interaction network revealed that our network included 5 hub genes (FOXO1, HIF1A, PPARGC1A, PRKAA2, and PRKAR2A), 117 pairs of mRNA-miRNA interactions, and 101 miRNA molecules. The results were shown in Table S2.
Fig. 7.
Hub genes mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-drugs interaction network. A–D mRNA-miRNA (A), mRNA-RBP (B), mRNA-TF (C), mRNA-drugs (D) interaction network of hub genes. The sky blue circles in the mRNA-miRNA (A) interaction network were mRNAs; the green circles were miRNAs. In the mRNA-RBP (B) interaction network, the sky blue circles were mRNAs; the orange circles were RBPs. The sky blue circles in the mRNA-TF (C) interaction network were mRNAs; the purple circles were TFs. In the mRNA-drugs (D) interaction network, the sky blue circles were mRNAs; the pink circles were drugs. RBP RNA-Binding Protein, TF Transcription Factors
Additionally, we predicted RBPs interacting with 9 hub genes (CD36, FOXO1, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, UCP2) via the ENCORI database, retaining mRNA-RBP interaction pairs with clusterNum > 4 and clipExpNum > 4, then visualized the mRNA-RBP interaction network by Cytoscape software (Fig. 7B). This network included 7 hub genes (CD36, FOXO1, HIF1A, PPARGC1A, PRKAA2, PRKAR2A, and UCP2), 40 RBP molecules and 102 pairs of mRNA-RBP interaction relationships. Notably, hub gene HIF1A had interactions with 31 RBP molecules. The specific mRNA-RBP interaction relationships were shown in Table S3.
We identified TFs that bind to hub genes through the CHIPBase database (version 2.0) and the hTFtarget database. These interactions overlapped with 9 hub genes, resulting in 7 hub genes (FOXO1, HIF1A, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, and UCP2) and 100 TFs, which we mapped out using Cytoscape (Fig. 7C). The sky blue circles were mRNAs and the purple circles were TFs. In the mRNA-TF interaction network, we found 65 pairs of significant mRNA-TF interactions particularly involving the hub gene UCP2. Table S4 displayed the precise interactions between mRNAs and TFs.
To find potential medications or chemical compounds targeting 9 hub genes, as shown in the mRNA-drugs interaction network (Fig. 7D), we searched 76 potential drugs or molecular compounds corresponding to 8 hub genes (CD36, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, and UCP2) through the CTD database. The sky blue circles represented mRNAs, and the pink circles represented drugs. We identified 43 medications or chemical substances that target the CD36 gene among them. Various mRNA-drug interaction correlations were shown in Table S5.
Immune Infiltration Analysis of Pediatric NAFLD Data Set (CIBERSORT)
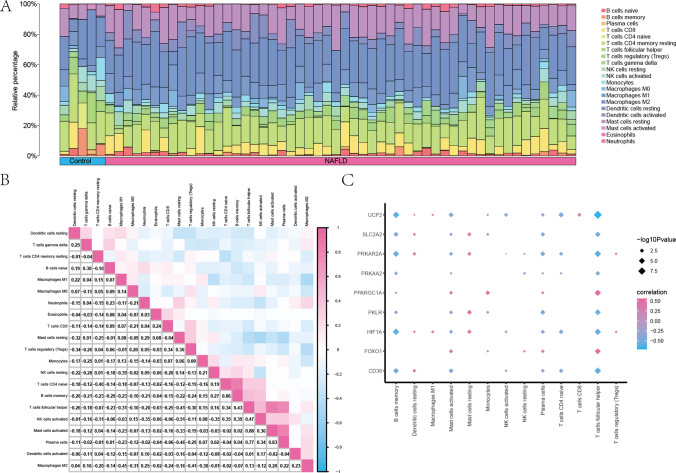
We used the CIBERSORT package and the Pearson algorithm [30] to evaluate the correlation between 22 distinct immune cell types and the expression profile data of pediatric NAFLD data set GSE185051. A graphical representation of the immune cell infiltration in each sample of the GSE185051 data set was created (Fig. 8A), focusing on 21 immune cell types where the cumulative infiltration abundance was greater than 0.
Fig. 8.
Immune infiltration analysis of pediatric NAFLD (CIBERSORT). A Histogram display of immune infiltration results of immune cells in the pediatric NAFLD data set GSE185051. B Correlation heatmap analysis results of immune cell infiltration abundance in the GSE185051 data set. C Correlation dot plots of immune cells and hub genes in the GSE185051 data set. The absolute value of the correlation coefficient (r) in the correlation heat map was strong if the absolute value was above 0.8; if the absolute value was 0.5–0.8, it was moderately correlated; if the absolute value was 0.3–0.5, it was weak; if the absolute value was below 0.3, it was weak or not relevant
The association between the infiltration abundance of 21 immune cell types and the expression profiles within the pediatric NAFLD data set GSE185051 was computed and presented (Fig. 8B). The findings revealed that the majority of the infiltration abundances among these 21 immune cell categories exhibited a negative correlation.
Simultaneously, we investigated the relationships between the expression levels of 9 hub genes (CD36, FOXO1, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, and UCP2) and the infiltration abundance of 21 immune cell types (Fig. 8C). Notably, mast cells had a significantly positive correlation with CD36 and PRKAR2A, while T follicular helper cells exhibited positive correlation with FOXO1 and PPARGC1A. Conversely, B cells memory had a significant negative correlation with CD36, HIF1A, PRKAR2A, and UCP2, and T follicular helper cells showed negative correlation with CD36, HIF1A, PKLR, PRKAR2A, and UCP2.
Discussion
Pediatric NAFLD is closely linked to disruptions in energy metabolism. Numerous studies [31, 32] have shown that abnormalities in energy metabolic processes, particularly mitochondrial dysfunction and oxidative stress, are crucial factors in the pathogenesis of NAFLD. Therefore, investigating EMRDEGs can help us gain a deeper understanding of the underlying pathological mechanisms of NAFLD. Furthermore, energy metabolism plays a pivotal role in the interplay of multiple systems and organs. In the liver, its dysregulation not only affects hepatic metabolic functions but can also trigger other metabolic syndromes, including obesity and insulin resistance. Therefore, a focused study of these genes provides a systematic and biologically plausible integration perspective, revealing the complex pathophysiological mechanisms of NAFLD. In this study, we comprehensively analyzed the data set GSE185051, collected EMRGs through the GeneCards database, identified EMRDEGs, and extracted 9 hub genes through the PPI network. Furthermore, we determined target molecules through mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-drugs interaction networks. GO analysis revealed associations with functions such as response to oxidative stress, and KEGG analysis involved pathways related to insulin resistance. GSEA showed gene sets enriched in pathways such as omega-9 fatty acid synthesis. Immune infiltration analysis suggested that mast cells and immune cells such as T follicular helper cells played an important part in pediatric NAFLD. Our research provided new insights into the pathogenesis of pediatric NAFLD, offering potential novel diagnostic biomarkers and therapeutic targets from an energy metabolic perspective.
We identified 14 EMRDEGs from the GSE185051 data set and GeneCards database and plotted the ROC curves to demonstrate that the AUCs of CACNB2, HIF1A, PKLR, PRKAA2, PRKAR2A, and SLC2A2 were all greater than 0.9. The results not only showcase the differential expression of EMRDEGs between NAFLD group and the control group, but also provide the possibility of using these genes as potential biomarkers for NAFLD diagnosis. These findings offer new perspectives and promising directions for further clinical research and the identification of therapeutic targets. We compared the results with adult NAFLD studies, and a bioinformatics analysis proposed that NDUFA4, TFAM, and CDKN1B have significant diagnostic value for NAFLD [33]. In another study, the author proposed that the SIRT family plays a crucial role in regulating mitochondrial function during the development of NAFLD [34]. Govaere [35] established a model for identifying risky steatohepatitis that includes four proteins: ADAMTSL2, AKR1B10, CFHR4, and TREM2. From the above studies, it is apparent that the genes related to NAFLD in adults are significantly different from those in children. In our study, we did not find that these genes played a role in pediatric NAFLD, which further highlights the necessity of conducting research on pediatric NAFLD. Another adult study [36] found that hepatic ACSL4 levels were elevated in patients with NAFLD. Suppressing ACSL4 expression promoted mitochondrial respiration, thereby enhancing the capacity of hepatocytes to mediate β-oxidation of fatty acids and minimize lipid accumulation by up-regulating peroxisome proliferator-activated receptor coactivator-1 alpha. Our study also found that ACSL4 is closely related to pediatric NAFLD, but it may play different biological roles in different age groups. The significant expression of ACSL4 in pediatric NAFLD may be related to the special metabolic needs during the growth and development stage, while in adults, these genes may be more related to lifestyle. We need to conduct further experiments to validate this viewpoint. In a study of genes related to pediatric NAFLD, PNPLA3 I148M variant has been shown to increase susceptibility to the whole spectrum of liver damage related to NAFLD and to be a general modifier of liver disease progression [37]. Carriage of the I148M variant increases the risk of liver disease, particularly in children (< 18 years), and interacts with dietary factors such as intake of fructose-enriched drinks. Another study showed variants in the genes encoding glucokinase regulator (GCKR) and membrane bound O-acyl transferase 7 (MBOAT7) also contribute to the risk of pediatric NAFLD [37]. The EMRDEGs in this study are all distinct from those reported previously, the novel genes we have identified will provide new insights into the pathogenesis of NAFLD in children.
In our network analysis, we utilized the STRING database and Cytoscape software for PPI network analysis, identifying 12 EMRDEGs with close interactions. Through the MCC, DMNC, and MNC algorithms, we further identified 9 hub genes (CD36, FOXO1, HIF1A, PKLR, PPARGC1A, PRKAA2, PRKAR2A, SLC2A2, UCP2), which are likely to play significant roles in the crucial regulation of NAFLD. Additionally, our mRNA-miRNA interaction network analysis revealed 117 interactions between five of these hub genes and 101 miRNA molecules, indicating that these miRNAs may represent novel gene regulatory layers in the pathological process of NAFLD. Among them, the increased activity of hub gene FOXO1, on the one hand, promotes the activity of genes involved in gluconeogenesis, on the other hand, leads to lipid metabolism disorder, which is characterized by hyperglycemia and hypertriglyceremia. The upregulation of FOXO1 expression by long non-coding Gomafu has been shown to contribute to the promotion of hepatic insulin resistance via the mechanism of miR-139-5p sponging, suggesting that FOXO1 and miRNA interactions could promote insulin resistance [38]. This is consistent with our findings and could be a potential target for the treatment of insulin resistance in pediatric NAFLD.
Our mRNA-RBP interaction network analysis identified 102 interactions between 7 hub genes and 40 RBP molecules. Notably, the close interaction between the HIF1A gene and 31 RBP molecules suggests its central regulatory role in NAFLD, among which HIF1A was most closely associated with RBPs. HIF1A is an important transcription factor for cellular response to hypoxia and regulates angiogenesis, cell adhesion, energy metabolism, apoptosis, and other important physiological processes. Increased expression of HIF1A could lead to liver injury, whereas its inhibitor, genistein, could protect against liver failure by inhibiting cellular reactive oxygen species production, reducing necrosis, and decreasing permeability transition pores in mitochondria [39]. It has been shown that insulin-like growth factor binding protein-3 (IGF2BP3), acting as a m6A reader, recognizes m6A modification in HIF1A mRNA, enhancing HIF1A production by improving RNA stability [40]; this is consistent with our results. Additionally, a study in adolescents also demonstrated that IGF2BP3 was elevated in pediatric NAFLD, unchanged in adult NAFLD, and could serve as a unique biomarker for pediatric NAFLD [9].
The mRNA-TF interaction network analysis identified interactions between 7 hub genes and 100 transcription factors. The UCP2 gene exhibited the most interactions with transcription factors, indicating that it may be an important transcriptional regulatory node. UCP2 is an inner mitochondrial membrane ion carrier that controls insulin secretion, free fatty acid concentration, and lipid metabolism, all of which are crucial to maintaining stable energy levels. It has been reported that HNRNPK is linked to UCP2 mRNA via locations in the transcript's 3'-untranslated region, promoting the production of mitochondrial UCP2 in response to insulin [41]. It is hypothesized that UCP2 and HNRNPK interaction contributes to the progression of NAFLD through an insulin resistance mechanism. Furthermore, we also identified a total of 65 pairs of interaction relationships between UCP2 and TFs. Among these, ATF3, E2F1, FOXA1, FOXA2, HDAC1, MYC, NOTCH1, USF1, and YY1 could be involved in lipid metabolism in NAFLD; however, no interaction with UCP2 has been reported in the previous studies. Our study suggested that the expression of TFs regulatory UCP2 might be involved in the pathogenesis of pediatric NAFLD; however, the specific regulatory mechanism requires further exploration.
Currently, there are no medications with definitive efficacy against pediatric NAFLD; therefore, we searched for 76 possible medications or chemical compounds that matched to the 8 hub genes through the CTD database and found 43 drugs or molecular compounds targeting CD36, including thiazolidinediones such as rosiglitazone, pioglitazone, and troglitazone. CD36 is a fatty acid transporter protein that is involved in transport and/or acts as a regulator of fatty acid transport. Studies have found that rosiglitazone, pioglitazone, and troglitazone could be used to improve insulin resistance and lipid metabolic abnormalities in NAFLD [42–44], offering new options for the treatment of pediatric NAFLD. Another drug, Resveratrol, regulates SIRT1-FoxO1 pathway-mediated cholesterol metabolism [45] and also inhibits ethanol-induced insulin production in INS-1 cells through the SIRT1-UCP2 axis [46]. Several studies [47, 48] have demonstrated the therapeutic potential of Resveratrol in NAFLD, and further research is needed to explore its use in children.
The GO analysis showed that the key biological processes related to NAFLD include energy homeostasis, cellular glucose homeostasis, positive regulation of carbohydrate metabolism, response to oxidative stress, and response to muscle activity. Among them, Oxidative stress may play a crucial role in the occurrence and development of NAFLD. Oxidative stress-induced cellular senescence may affect hepatocyte function and metabolism, contributing to the progression of NAFLD from mild steatosis to inflammation, fibrosis, or hepatocellular cancer [49]; thus, various drugs that inhibit oxidative stress could be potential therapeutic targets for NAFLD. Regarding the identification of “response to muscle activity” and “brush border” pathways in the GO analysis of EMRDEGs from liver biopsy samples, these findings likely reflect the multifaceted functions of these genes. Although these samples originate from liver biopsies, it is worth noting that the genes identified can potentially play a role in other tissues or physiological processes. For instance, the expression and function of certain genes in muscle activity or cellular membrane structures may also influence their detection in liver samples [50, 51]. KEGG analysis showed that the majority of EMRDEGs were linked to insulin resistance pathways. Prior research has suggested that activating the PI3K/AKT pathway might reduce inflammation and modulate insulin sensitivity in NAFLD patients [52], which was consistent with our result. Many traditional Chinese medicines have also been verified to improve NAFLD through PI3K/AKT pathways [52, 53]. Many biomarkers in mRNA-miRNA, mRNA-RBP, mRNA-TF, and mRNA-drugs interaction networks in this study were also associated with insulin resistance. Thus, insulin resistance is an important mechanism of pediatric NAFLD and should be a key target for NAFLD treatment. GSEA of EMRDEGs was significantly enriched in the biosynthesis of unsaturated fatty acids, glycolysis gluconeogenesis, fatty acid metabolism, omega9 fatty acid synthesis, and hedgehog ligand biogenesis. Lipid metabolism plays a central role in the pathogenesis of NAFLD. A study in adults found that NAFLD severity was associated with reduced liver Omega-3 polyunsaturated fatty acids [54], Omega-3 dietary supplements showed the potential to be employed as a treatment for NAFLD [55], but there were no studies about Omega-9. Omega-9 are monounsaturated and non-essential fatty acids that can be synthesized in vivo, and whether NAFLD has the same potential to treat NAFLD as Omega-3 in children requires further investigation.
Several studies have shown that inflammation exacerbates metabolic disturbances in NAFLD and that immune cell infiltration is critical in the establishment and progression of adipose tissue inflammation in NAFLD [56, 57]. In this study, it was found that mast cells were strongly linked with CD36 and PRKAR2A. In a mouse study, mast cells promoted hepatobiliary injury and might cause microvesicular steatosis development during the transition from NAFLD to NASH via miR-144-3p/ALDH1A3 signaling [58]. Additionally, mast cells from the portal vein region and fiber spacing were implicated in the pathophysiology of NAFLD-related liver fibrosis [59]. Thus, inhibition of mast cell activation might be a therapeutic approach for NAFLD treatment. T follicular helper cells express the chemokine receptor CXCR5, which controls the differentiation and clonal selection of memory and antibody-secreting B cells, hence regulating the development of antibody memory and affinity. In this study, T follicular helper cells were positively correlated with FOXO1 and PPARGC1A and negatively correlated with CD36, HIF1A, PKLR, and PRKAR2A. T follicular helper cells played a significant role in the process of liver damage. In acetaminophen-induced acute liver failure, it was found that the number of CD4 naive T cells, CD8 T cells, and T follicular helper cells increased [60]. T follicular helper cells infiltration and enhanced IL-21 production may be linked to the development of AIH in patients with high blood IgG4 [61]. The absence of previous studies on T follicular helper cells in NAFLD suggests a new direction for future research in NAFLD.
Although our study initially identified EMRDEGs in pediatric NAFLD, further research is needed to comprehensively understand the pathological mechanisms of pediatric NAFLD and confirm the roles of these genes. Future studies can be conducted in several directions. Firstly, the same analytical methods should be applied to RNA-seq datasets from adult NAFLD to conduct systematic comparative studies, aiming to clarify the differences and similarities in gene expression and pathological mechanisms between pediatric and adult NAFLD. Secondly, to improve the reliability and generalizability of the research results, future studies should expand the sample size and conduct large-scale multi-center studies by combining RNA-seq data from multiple centers for pediatric NAFLD. Additionally, in vitro and in vivo experiments are necessary to validate the specific roles and mechanisms of these key energy metabolism genes in NAFLD. These experiments can include gene knockout, overexpression, and pharmacological interventions. Finally, future research should focus on the potential clinical applications of the biomarkers we have discovered in the diagnosis and treatment of NAFLD. Exploring their feasibility and effectiveness as diagnostic tools or therapeutic targets will facilitate the translation of research findings into practical clinical applications. Through these research directions, we will gain a deeper and more comprehensive understanding of the pathological mechanisms of pediatric NAFLD and provide new scientific evidence and strategies for its diagnosis and treatment.
However, there are some limitations in this study. Firstly, obtaining pathologic specimens from young children for invasive liver puncture was clinically challenging, and performing liver puncture on healthy children would be ethically questionable, making experimental validation difficult. Secondly, our current study completed the later stage of critical data analysis and validation, and we have not included new pediatric NAFLD studies as a validation cohort for the time being. This may impact the external validation of our results. However, we recognize the importance of introducing more similar pediatric studies for validation and plan to expand our research scope in future studies, continuing to search for and integrate similar pediatric datasets to further validate and consolidate our findings.
Conclusion
Our study combined the GEO data set and GeneCards database of EMRGs to identify hub genes and interacting miRNAs, RBPs, TFs, and drugs. Through GO, KEGG analyses, and GSEA and immune infiltration analyses, we have provided new insights, biomarkers, and therapeutic targets for common molecular mechanisms in pediatric NAFLD, focusing on the energy metabolism perspective.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- ROC
Receiver operating characteristic
- AUC
Area under curve
- NAFLD
Pediatric non-alcoholic fatty liver disease
- PPI
Protein–protein interaction
- RBPs
RNA-binding proteins
- TFs
Transcription factors
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- GSEA
Gene set enrichment analysis
- DEGs
Differentially expressed genes
- EMRGs
Energy metabolism related genes
- MSigDB
Molecular signatures database
- EMRDEGs
Energy metabolism related differentially expressed genes
- BP
Biological processes
- MF
Molecular functions
- CC
Cellular components
- TFs
Transcription factors
- CTD
Comparative toxicogenomics database
- PCA
Principal component analysis
Author’s contribution
YL wrote the draft; RZ conceptualized the study, reviewed the draft and collected data; YC, YC and WO helped in data analysis; HY supervised the study. All the authors have read and approved the final manuscript.
Funding
This study was funded by Joint Funds for the innovation of science and Technology, Fujian province (Grant number: 2023Y9386), Provincial-level special subsidy funds for health in Fujian Province of China (Grant number: Fujian Finance Index (2020) 467).
Data availability
No datasets were generated or analyzed during the current study.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sarin SK, Kumar M, Eslam M et al. Liver diseases in the Asia-Pacific region: a lancet gastroenterology & hepatology commission. Lancet Gastroenterol Hepatol. 2020;5:167–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiegand S, Keller KM, Röbl M et al. Obese boys at increased risk for nonalcoholic liver disease: evaluation of 16,390 overweight or obese children and adolescents. Int J Obes (Lond). 2010;34:1468–1474. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Hou DQ, Duan JL et al. Prevalence of nonalcoholic fatty liver disease and metabolic abnormalities in 387 obese children and adolescents in Beijing, China. Zhonghua Liu Xing Bing Xue Za Zhi. 2013;34:446–450. [PubMed] [Google Scholar]
- 4.Eslam M, Alkhouri N, Vajro P et al. Defining paediatric metabolic (dysfunction)-associated fatty liver disease: an international expert consensus statement. Lancet Gastroenterol Hepatol. 2021;6:864–873. [DOI] [PubMed] [Google Scholar]
- 5.Mann JP, Tang GY, Nobili V, Armstrong MJ. Evaluations of lifestyle, dietary, and pharmacologic treatments for pediatric nonalcoholic fatty liver disease: a systematic review. Clin Gastroenterol Hepatol. 2019;17:1457-1476.e7. [DOI] [PubMed] [Google Scholar]
- 6.Cariou B, Byrne CD, Loomba R, Sanyal AJ. Nonalcoholic fatty liver disease as a metabolic disease in humans: a literature review. Diabetes Obes Metab. 2021;23:1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basaranoglu M, Basaranoglu G, Sabuncu T, Sentürk H. Fructose as a key player in the development of fatty liver disease. World J Gastroenterol. 2013;19:1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor R, Al-Mrabeh A, Sattar N. Understanding the mechanisms of reversal of type 2 diabetes. Lancet Diabetes Endocrinol. 2019;7:726–736. [DOI] [PubMed] [Google Scholar]
- 9.Yao K, Tarabra E, Sia D et al. Transcriptomic profiling of a multiethnic pediatric NAFLD cohort reveals genes and pathways associated with disease. Hepatol Commun. 2022;6:1598–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett T, Troup DB, Wilhite SE et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics. 2007;23:1846–1847. [DOI] [PubMed] [Google Scholar]
- 12.Stelzer G, Rosen N, Plaschkes I et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform. 2016;54:1–30. [DOI] [PubMed] [Google Scholar]
- 13.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315–1316. [DOI] [PubMed] [Google Scholar]
- 16.Yu G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol Biol. 2020;2117:207–215. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v20: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou KR, Liu S, Sun WJ et al. ChIPBase v20: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Liu W, Zhang HM et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genom Proteom Bioinform. 2020;18:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis AP, Grondin CJ, Johnson RJ et al. Comparative toxicogenomics database (CTD): update 2021. Nucleic Acids Res. 2021;49:D1138–D1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Liu CL, Green MR et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8:S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson K. VII note on regression and inheritance in the case of two parents. Proc Royal Soc Lond. 1985. 10.1098/rspl.1895.0041. [Google Scholar]
- 31.Mansouri A, Gattolliat CH, Asselah T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology. 2018;155:629–647. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Tian R, She Z, Cai J, Li H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic Biol Med. 2020;152:116–141. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Cheng W, Hu P et al. Integrative analysis identifies oxidative stress biomarkers in non-alcoholic fatty liver disease via machine learning and weighted gene co-expression network analysis. Front Immunol. 2024;15:1335112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng C, Chen M. Progress in nonalcoholic fatty liver disease: SIRT family regulates mitochondrial biogenesis. Biomolecules. 2022;12:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Govaere O, Hasoon M, Alexander L et al. A proteo-transcriptomic map of non-alcoholic fatty liver disease signatures. Nat Metab. 2023;5:572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan J, Wang Z, Duan R et al. Therapeutic targeting of hepatic ACSL4 ameliorates NASH in mice. Hepatology. 2022;75:140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobili V, Alisi A, Valenti L, Miele L, Feldstein AE, Alkhouri N. NAFLD in children: new genes, new diagnostic modalities and new drugs. Nat Rev Gastroenterol Hepatol. 2019;16:517–530. [DOI] [PubMed] [Google Scholar]
- 38.Yan C, Li J, Feng S, Li Y, Tan L. Long noncoding RNA gomafu upregulates Foxo1 expression to promote hepatic insulin resistance by sponging miR-139-5p. Cell Death Dis. 2018;9:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie F, Dong J, Zhu Y et al. HIF1a inhibitor rescues acute-on-chronic liver failure. Ann Hepatol. 2019;18:757–764. [DOI] [PubMed] [Google Scholar]
- 40.Cheng K, Liu S, Li C, Zhao Y, Wang Q. IGF2BP3/HIF1A/YAP signaling plays a role in driving acute-on-chronic liver failure through activating hepatocyte reprogramming. Cell Signal. 2023;108:110727. [DOI] [PubMed] [Google Scholar]
- 41.Ostrowski J, Klimek-Tomczak K, Wyrwicz LS, Mikula M, Schullery DS, Bomsztyk K. Heterogeneous nuclear ribonucleoprotein K enhances insulin-induced expression of mitochondrial UCP2 protein. J Biol Chem. 2004;279:54599–54609. [DOI] [PubMed] [Google Scholar]
- 42.Katoh S, Hata S, Matsushima M et al. Troglitazone prevents the rise in visceral adiposity and improves fatty liver associated with sulfonylurea therapy–a randomized controlled trial. Metabolism. 2001;50:414–417. [DOI] [PubMed] [Google Scholar]
- 43.Mulder P, Morrison MC, Verschuren L et al. Reduction of obesity-associated white adipose tissue inflammation by rosiglitazone is associated with reduced non-alcoholic fatty liver disease in LDLr-deficient mice. Sci Rep. 2016;6:31542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Du H, Zhao Y et al. Response to pioglitazone in non-alcoholic fatty liver disease patients with vs. without type 2 diabetes: a meta-analysis of randomized controlled trials. Front Endocrinol (Lausanne). 2023;14:1111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang C, Xing H, Wang C, Xu X, Hao Y, Qiu B. Resveratrol improves the progression of osteoarthritis by regulating the SIRT1-FoxO1 pathway-mediated cholesterol metabolism. Mediators Inflamm. 2023;2023:2936236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo G, Xiao L, Wang D et al. Resveratrol protects against ethanol-induced impairment of insulin secretion in INS-1 cells through SIRT1-UCP2 axis. Toxicol Vitro. 2020;65:104808. [DOI] [PubMed] [Google Scholar]
- 47.Guo J, Wang P, Cui Y, Hu X, Chen F, Ma C. Alleviation effects of microbial metabolites from resveratrol on non-alcoholic fatty liver disease. Foods. 2022;12:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarahmadi S, Farahmandian N, Fadaei R et al. therapeutic potential of resveratrol and atorvastatin following high-fat diet uptake-induced nonalcoholic fatty liver disease by targeting genes involved in cholesterol metabolism and miR33. DNA Cell Biol. 2023;42:82–90. [DOI] [PubMed] [Google Scholar]
- 49.Anastasopoulos NA, Charchanti AV, Barbouti A et al. The role of oxidative stress and cellular senescence in the pathogenesis of metabolic associated fatty liver disease and related hepatocellular carcinoma. Antioxidants (Basel). 2023;12:1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Zhu S, Jia Y et al. Positive selection of somatically mutated clones identifies adaptive pathways in metabolic liver disease. bioRxiv. 2023. 10.1016/j.cell.2023.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savant JD, Betoko A, Meyers KE et al. Vascular stiffness in children with chronic kidney disease. Hypertension. 2017;69:863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao L, Hao P, Jiang M, Chu W. Liquiritigenin regulates insulin sensitivity and ameliorates inflammatory responses in the nonalcoholic fatty liver by activation PI3K/AKT pathway. Chem Biol Drug Des. 2023. 10.1111/cbdd.14292. [DOI] [PubMed] [Google Scholar]
- 53.Wang S, Yang FJ, Shang LC, Zhang YH, Zhou Y, Shi XL. Puerarin protects against high-fat high-sucrose diet-induced non-alcoholic fatty liver disease by modulating PARP-1/PI3K/AKT signaling pathway and facilitating mitochondrial homeostasis. Phytother Res. 2019;33:2347–2359. [DOI] [PubMed] [Google Scholar]
- 54.Spooner MH, Jump DB. Nonalcoholic fatty liver disease and omega-3 fatty acids: mechanisms and clinical use. Annu Rev Nutr. 2023. 10.1146/annurev-nutr-061021-030223. [DOI] [PubMed] [Google Scholar]
- 55.Musazadeh V, Karimi A, Malekahmadi M, Ahrabi SS, Dehghan P. Omega-3 polyunsaturated fatty acids in the treatment of non-alcoholic fatty liver disease: an umbrella systematic review and meta-analysis. Clin Exp Pharmacol Physiol. 2023;50:327–334. [DOI] [PubMed] [Google Scholar]
- 56.Song Y, Zhang J, Wang H et al. A novel immune-related genes signature after bariatric surgery is histologically associated with non-alcoholic fatty liver disease. Adipocyte. 2021;10:424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo S, Wang Y, Yuan X et al. Identification of common signature genes and pathways underlying the pathogenesis association between nonalcoholic fatty liver disease and atherosclerosis. Front Cardiovasc Med. 2023;10:1142296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennedy L, Meadows V, Sybenga A et al. Mast cells promote nonalcoholic fatty liver disease phenotypes and microvesicular steatosis in mice fed a western diet. Hepatology. 2021;74:164–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewandowska E, Wosiak A, Zieliński A et al. Role of mast cells in the pathogenesis of liver fibrosis in nonalcoholic fatty liver disease. Pol J Pathol. 2020;71:38–45. [DOI] [PubMed] [Google Scholar]
- 60.Shi M, Zhou Z, Zhou Z et al. Identification of key genes and infiltrating immune cells among acetaminophen-induced acute liver failure and HBV-associated acute liver failure. Ann Transl Med. 2022;10:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Zhang Y, Jiang D, Zhou L, Wang B. Clinical characteristics and potential mechanisms in patients with abnormal liver function indices and elevated serum IgG4. Can J Gastroenterol Hepatol. 2022;2022:7194826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.