Abstract
Certain types of primary tumor, particularly triple primary tumors with germline mutations, are rare. The present study reports a novel case of the metachronous occurrence of three pathological conditions, namely, non-small-cell lung cancer (NSCLC), early T cell precursor acute lymphoblastic leukemia (ETP-ALL) and SCLC. The present study used next-generation sequencing to aid diagnosis. A 44-year-old male patient presented to The First Affiliated Hospital Zhejiang University School of Medicine (Hangzhou, China) in September 2016.) with a nodule in the right lower lung during an annual checkup. Then, the patient was diagnosed with poorly differentiated NSCLC (T1N2M0; stage IIIA) and underwent surgical resection and biopsy. In September 2018, the patient was diagnosed with ETP-ALL with superficial lymphadenopathy. Germline testing demonstrated germ cell variants of ERCC excision repair 6, chromatin remodeling factor (ERCC6; c.1322A>G) and LYL1 basic helix-loop-helix family member (LYL1; c.587T>A). In November 2020, the patient was diagnosed with SCLC by bronchoscopic biopsy following allogeneic hematopoietic stem cell transplantation. The patient was diagnosed with lung cancer in October 2016 and the treatment were: surgery, chemotherapy, radiotherapy, and targeted therapy. In October 2018, the patient was diagnosed with ETP-ALL and the treatment were: chemotherapy and allogeneic hematopoietic stem cell transplantation. In November 2020, the patient was diagnosed with small cell lung cancer and received chemotherapy and radiotherapy. The patient died at September 2022. The present case highlighted the importance of monitoring germline mutations in patients and their families to facilitate early diagnosis, appropriate treatment and prognostic evolution in the face of rapid recurrent cancer.
Keywords: triple primary tumor, gene mutation, multiple primary tumors, high-throughput sequencing, non-small-cell lung cancer
Introduction
Tumors that occur at different sites and/or belong to different histological or morphological groups are considered to be a multi-primary cancer (1). A study in 1921 reported that 4.7% of 3,000 patients with malignant tumors had multiple tumors (2). Epidemiological studies have reported that the frequency of multiple primary cancer was 2–17% in 2014 (3–7). Weir et al (5) reported that following Surveillance, Epidemiology and End Results guidelines (8), the incidence of multiple primary cancer was 19.7% in patients with colon cancer [16.9% as per International Association of Cancer Registries (IACR) guidelines (9)] and 21% in those with lung cancer (19.9% according to the IACR guidelines). The epidemiological factors contributing to occurrence of multiple primary cancer include host factors, such as genetic factors, hormones and tumor history, lifestyle factors, such as smoking and alcohol consumption, and environmental factors, such as occupation, pathogen exposure and geographical location (10). Among these, genetic factors have attracted increasing attention from researchers (3,11–16). It is estimated that between 5 and 10% of all breast cancer cases and ~20% of ovarian cancer cases are caused by an inherited pathogenic variant associated with hereditary breast and ovarian cancer syndrome (17–21). First- and second-degree relatives and first cousins have a 12.5–50.0% probability of inheriting the respective cancer predisposition variants (22,23). Therefore, timely identification of genetic variants decreases morbidity and mortality in individuals with inherited cancer risk and facilitates targeted therapy for patients with cancer (24). Table I provides a brief overview of germline mutations in patients with cancer from larger sequencing studies (25–32). Genetic testing serves as a robust and efficient auxiliary examination tool that provides information on molecular subtypes and therapeutic targets and aids in the development of potential treatment strategies and selection of appropriate drugs (33).
Table I.
Common genes with germline mutations in patients with cancer.
| Gene | Prevalence of mutation (globally) | Disease | Clinical characteristics | (Refs.) |
|---|---|---|---|---|
| BRCA1 | 1/500-1/300 | Breast, ovary, prostate and pancreas cancer | Sensitivity to platinum-containing therapy or PARP inhibitors | (25) |
| p53 | 1/5,000-1/500 | Li-Fraumeni syndrome | Breast cancer, soft tissue sarcoma and osteosarcoma | (26) |
| Adenomatous polyposis coli | 1/10,000-1/8,000 | Colorectal cancer | Familial occurrence, a younger age of onset, and is commonly seen in children. | (27–29) |
| MutL homolog 1, MSH2, MSH6 and PMS2 | 1/3,000 | Lynch syndrome | Colorectal, endometrial and ovarian cancer and gastric and ureteral carcinoma | (30) |
| Von Hippel-Lindau tumor suppressor | 1/36,000 | Von Hippel-Lindau syndrome | Retinal angioma, hemangioblastoma, pancreatic cyst and renal carcinoma | (31) |
| SDHB | 6/100-8/100 | SDHB-deficient paraganglioma/pheochromocytoma syndrome | Hypertension, headache, palpitations, hyperhidrosis and catecholamines in blood or urine | (32) |
SDBH, succinate dehydrogenase complex iron sulfur subunit B; MSH, mutS homolog; PMS2, PMS1 homolog 2, mismatch repair system component.
The 5-year survival rate of patients with 27 common types(including pancreas to testis) of cancer in the UK ranges from 7 to 88% (34). Depending on the type of combined tumor, survival times vary among patients with different recurring types of cancer, especially those with hematological disease (8), who exhibit rapid progression, high degree of malignancy, difficulty in treatment and a low survival rate. A number of patients with multiple primary tumors of hematological disease) carry germline driver gene mutations (CEBPA OR TP53 and so on) associated with a poor prognosis (35). Previous studies (36,37) have shown that a small number of patients carrying two germline mutations, ERCC excision repair 6, chromatin remodeling factor (ERCC6) and LYL1 basic helix-loop-helix family member (LYL1), develop non-small-cell lung cancer (NSCLC), early T cell precursor acute lymphoblastic leukemia (ETP-ALL) and SCLC. The present study describes a patient with ERCC6(+) and LYL1(+) mutations with triple primary tumors.
Case report
A 40-year-old male patient (healthy and non-smoker) was found to have a right lower lung space during an annual routine checkup in September 2016 (Fig. 1A) at The First Affiliated Hospital Zhejiang University School of Medicine (Hangzhou, China). The patient's father, who smoked for 40 years, had also been diagnosed with lung cancer but refused genetic testing. Radical resection of the lower right lung cancer was performed in October 2016. The pathological diagnosis was adenocarcinoma of the lower right lung (T1N2M0, stage IIIA) (38). Postoperative concurrent chemoradiotherapy included four cycles of pemetrexed + platinum, with a total radiotherapy dose of 50 Gy in 25 fractions (50 Gy/25 f). In March 2018, routine chest computed tomography (CT) scan demonstrated a new nodule near the pleura in the middle lobe of the right lung (Fig. 1B), which indicated local recurrence of adenocarcinoma. Gefitinib (250 mg, once daily) was administered orally and the nodules subsequently disappeared by July 2018, as confirmed by chest CT scan (data not shown).
Figure 1.
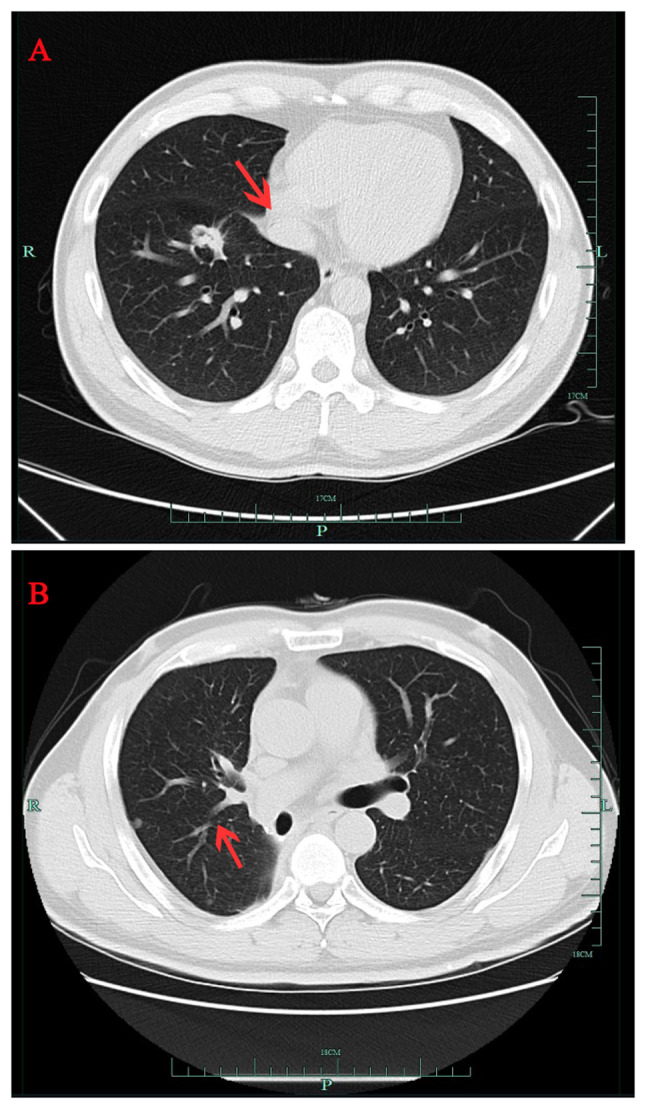
Serial chest CT scans of patient with non-small cell lung cancer. (A) CT scan from September 2016 showing a right lung mass (red arrow). (B) CT scan from March 2018 showing a small nodule (red arrow) near the pleura in the middle lobe of the right lung. CT, computed tomography.
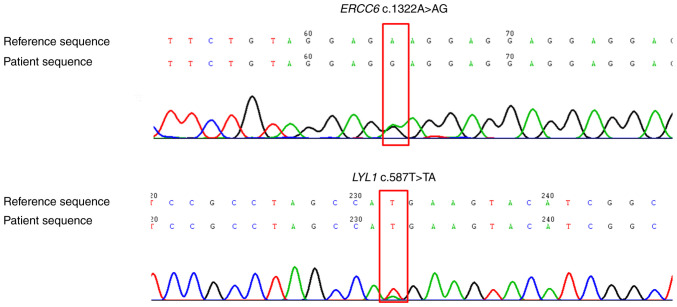
In September 2018, at the Tongde Hospital of Zhejiang Province (Hangzhou, China) for the first presentation, the patient developed submental lymph node enlargement, which was diagnosed as ETP-ALL based on lymph node biopsy and bone marrow tests [CD7(++), CD1α(−), CD8(−), CD5(dim), CD34(+), CD2(+), cyCD3 (weakly positive) and CD4(+). Gefitinib was discontinued and hyper-cyclophosphamide, vindesine, liposomal doxorubicin and dexamethasone/methotrexate and cytarabine was initiated (Table II); however, this proved ineffective. Germline gene sequencing of skin tissue demonstrated ERCC6 variant c.1322A>G and LYL1 variant c.587T>A (Fig. 2).
Table II.
Timeline of early T cell precursor acute lymphoblastic leukemia treatment.
| Date | Chemotherapy regimen | Drug |
|---|---|---|
| October 2018 | Hyper CVAD-part A | Cyclophosphamide, 0.55 g q12h d-3; dexamethasone, 40 mg qd d1-4; liposomal doxorubicin, 60 mg qd d4; vindesine, 4 mg qd d4 |
| November 2018 | Hyper CVAD-part B | Methotrexate, 2 g qd d1; cytarabine, 1g q12h d2-3 |
| November 2018 | V + CAG | Venetoclax, 400 mg qd d1-28; cytarabine, 25 mg q12h d1-14; aclarubicin, 20 mg qd d1-4; G-CSF, 300 µg d1-14 |
| January 2019 | V + CAG | Venetoclax, 400 mg qd d1-28; cytarabine, 100 mg q12h d1-7; aclarubicin, 20 mg qd d1-4; G-CSF, 300 µg d1-14 |
G-CSF, granulocyte colony-stimulating factor; CVAD, cyclophosphamide, dexamethasone, liposomal doxorubicin and vindesine; V + CAG, venetoclax, cytarabine, aclarubicin and G-CSF; qd, once daily; q12h, once every 12 h; d, day.
Figure 2.
Next-generation sequencing demonstrated ERCC6 and LYL1 mutations (red box). ERCC6, ERCC6 excision repair 6, chromatin remodeling factor; LYL1, LYL1 basic helix-loop-helix family member.
The primers were as follows: ERCC6-E5 forward (F), 5′-GAGGAAGATGACGAGGTGGA-3′ and reverse (R), 5′-GGCTGCAGAAATCCAACCTC-3′ and LYL1-E4 F, 5′-CAGACCCATGAGTACACCCA-3′ and R, 5′-CTGACGTCTTCACTGGTCCT-3′. The high-throughput sequencing was Aligent SureSelect. The method used to verify the quality/integrity of the processed samples was A 2200 bioanalyzer for genomic DNA or RNA. The type of sequencing was 300 bp for length and paired end for direction of sequencing. The loading concentration of the final library, including how concentrations were measured:5 pM for DNA sequencing). i) The patient received two courses of venetoclax combined with granulocyte colony-stimulating factor, cytarabine and aclacinomycin chemotherapy (Table II). February 2019 bone marrow reexamination indicated complete remission (data not shown).
Modified busulfan-cyclophosphamide pretreatment (Cytarabine 7.3 g-10d,-9d; busulfan 54 mg q6h8d,-7d,-6d; cyclophosphamide 3.3 g-5d,-4d; oral semustine 450 mg-3d. Cytarabine-busulfan and cyclophosphamide intravenous infusion. Semustine was Oral administration, the purpose of this were to fully eliminate or suppress the patient's immune system to prevent graft rejection; reduce the number of tumor cells to a minimum; remove the patient's hematopoietic stem cells from the bone marrow niche to provide sufficient space for the engrafted donor hematopoietic stem cells to support proliferation and differentiation. ii) was performed in March 2019. Haploidentical hematopoietic stem cell transplantation was performed following stem cell donations from the patient's son (March 2019). During transplantation, anti-thymocyte globulin (ATG; total dose 700 mg), cyclosporin A and short-course methotrexate(cyclosporin A 75 mg q12h qd Intravenous infusion; methotrexate 10mg +1d,+3d,+6,+11d, were Intravenous infusion) were administered on days 1, 3, 5 and 11 following transplantation, to prevent acute graft vs. host disease. After ATG was administered, leukocyte count (Table III) was monitored. In April 2019, bone marrow examination showed that the ETP-ALL was in remission, the short tandem repeat was of the complete donor type and the right lung lesion was smaller, as observed through CT scan (data not shown). During the cyclosporin anti-rejection treatment, routine Positron Emission Tomography-CT examination indicated multiple small nodules in both lungs, which prompted clinical consideration of lung adenocarcinoma recurrence. Gefitinib (250 mg, once daily) targeted therapy was re-administered for 10 months to alleviate the increase in the number of lung nodules.-we decided to switch from gefitinib to osimertinib (80 mg/d). The number of nodules in both lungs decreased following this treatment.
Table III.
Leukocyte levels following anti-thymocyte globulin treatment.
| Day | 1 | 4 | 7 | 9 | 10 | 11 | 12 | 14 | 15 | 17 | 22 | 24 | 26 | 28 | 30 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| White blood cell count, ×109/l | 3.4 | 0.1 | 0.2 | 0.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 2.0 | 2.8 | 2.4 | 2.8 | 3.4 |
| Absolute neutrophil count 109/l | 3.3 | 0.1 | 0.2 | 0.4 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 1.5 | 1.8 | 2.0 | 2.6 |
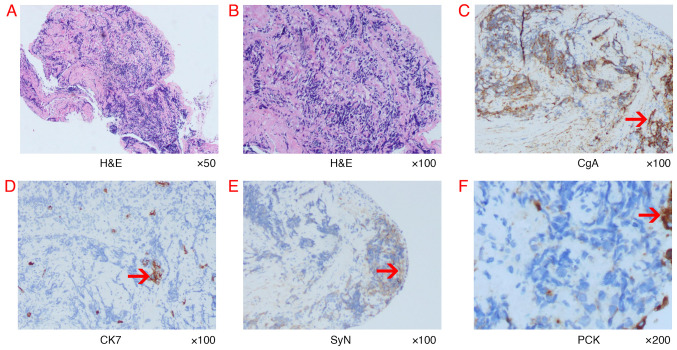
Sudden hemoptysis occurred in November 2020 and a new obstruction in the lumen was noted following bronchoscopy (Fig. 3). Pathological examination of brush cytology smear under a microscope demonstrated SCLC (Fig. 4). Immunohistochemistry was negative for CK7 and positive for pancytokeratin (PCK), chromogranin A) and SyN(Synaptophysin). A marked decrease in foreign body count was observed after combining chemoradiotherapy with osimertinib treatment through chest CT (data not shown). A timeline of the three types of cancer diagnosed is presented in Table IV. No metastases in the skull, bones or gastrointestinal area were observed during treatment. In August 2022, the patient succumbed to progressive lung cancer and no autopsy was performed.
Figure 3.
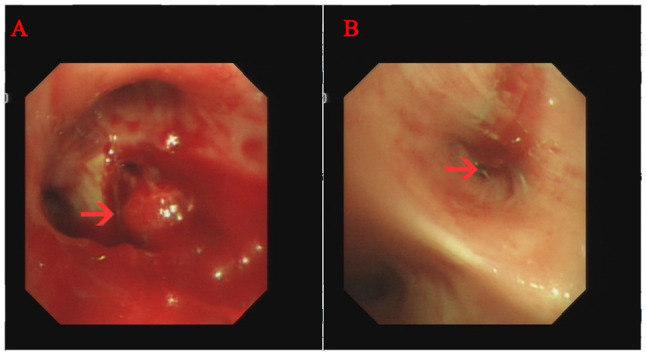
Bronchoscopic findings at time of diagnosis of small cell lung cancer. (A) Polypoid endobronchial mass (red arrow) blocked the right upper lobe bronchus (red arrow). (B) Tracheal occlusion after resection of the right lower lobe bronchus.
Figure 4.
Histopathological findings of bronchoscopic biopsy and representative IHC-labelled tumor tissue. (A and B) Representative images of tumor morphology. Magnification, ×50 and ×100, respectively. The histological results of (C) CgA, (D) CK7, (E) SyN and (F) PCK. The red arrows indicate positive IHC labelling. IHC, immunohistochemistry; PCK, Pancytokeratin; H&E, hematoxylin and eosin; CgA, Chromogranin A; SyN, Synaptophysin; CK7, Cytokeratin 7.
Table IV.
Timeline of triple cancer progression.
| Cancer | Date of diagnosis | Key pathological features | Molecular findings | Treatment |
|---|---|---|---|---|
| Non-small cell lung carcinoma | October 2016 | Surgery, radiotherapy, chemotherapya and targeted therapyb | ||
| Early T cell precursor acute lymphoblastic leukemia | October 2018 | CD7(+), CD1α(−), CD8(−)c, CD5<75%, CD34(+), CD2(+), CD3(+) and CD4(+) | LYL1 and ERCC6d | Chemotherapye and haplo-hematopoietic stem cell transplantation |
| Small cell lung carcinoma | November 2020 | PCK(+), CgA(+), SyN(+), CK7(−) | Not applicable | Radiotherapy and chemotherapy |
Pemetrexed and platinum.
Gefitinib.
Method of detection was flow cytometry.
Tissue sample taken from skin of the leg.
Hyper cyclophosphamide, dexamethasone, liposomal doxorubicin and vindesine, methotrexate, cytarabine;V+CAG, venetoclax, cytarabine, aclarubicin and G-CSF. ERCC6, ERCC6 excision repair 6, chromatin remodeling factor; LYL1, LYL1 basic helix-loop-helix family member; PCK, pancytokeratin; CgA, Chromogranin A; SyN, Synaptophysin; CK7, cytokeratin-7.
Discussion
Germline gene mutations implicated in development of multiple primary tumors warrant investigation. Paired tumor germline genomic analyses have identified at least one pathogenic or potentially pathogenic germline variant of cancer susceptibility genes in 8–18% of patients with cancer (39–41). Oncologists increasingly recommend genetic testing for pathogenic germline variants in patients with cancer to advise on individual risk of developing cancer and the likelihood of family members carrying the same genetic predisposition to cancer (42,43). For example, inactivating mutations in the cancer suppressor genes BRCA1 and BRCA2 predict response to drugs, such as PARP inhibitors docetaxel and cisplatin, and are associated with increased genetic susceptibility (44).
ERCC6 encodes a DNA-binding protein key for excision and repair during transcriptional coupling. The protein exhibits ATP-stimulated ATPase activity, interacts with numerous transcription and excision repair proteins and may promote complex formation at DNA repair sites. ERCC excision repair 6, chromatin remodeling factor (alternatively named Cockayne syndrome complementation group B), encoded by ERCC6, recruits nucleotide excision repair factors to the DNA damage site and serves a key role in the repair process. Mutations in ERCC6 are commonly observed in patients with Cockayne syndrome type B and cerebro-oculofacial bone syndrome type I (45,46). Several population-based studies have shown that ERCC6 polymorphisms markedly affect risk of certain types of cancers, including lung cancer (47–49), and the recurrence of superficial bladder cancer (50). In a case-control study of lung cancer in a Chinese population, Ma et al (51) reported that each single nucleotide polymorphism may serve only a small role individually, and multiple loci in ERCC6 may collectively contribute to lung cancer susceptibility.
LYL1 encodes a basic helix-loop-helix (bHLH) transcription factor that is hypothesized to serve roles in vascular maturation and hematopoietic processes (52,53). LYL1 was first reported to be associated with the translocation t(7;19)(q35;p13) in patients with T cell acute lymphoblastic leukemia (T-ALL) (54). As a super enhancer (55,56), LYL1 induces transcription of target genes and shows closer association with oncogenes compared with typical enhancers (such as PARP inhibitors for breast cancer (BRCA1 mutation), oncogenes LYL1 is associated with DNMT3A/IDH2) (57). It has been hypothesized that the antitumor activity of GNE-987 drug that is still under development and has not yet been released to the market.) targeting bromodomain-containing protein 4 in acute myeloid leukemia (AML) involves downregulation of various super enhancers and associated oncogenes, including LYL1 (58). Furthermore, LYL1 expression levels in bone marrow of patients with AML is reported to be higher compared with that in normal bone marrow (44). Structurally, both LYL1 and its homolog T cell acute lymphocytic leukemia protein 1 (TAL1) that form DNA-binding heterodimers with E proteins (such as E2A:transcription factor 3 and HEB: transcription factor 12), are bHLH factors (59). For example, TAL1 forms a complex with E2A, LDB1 (LIM domain-binding protein 1), LMO2 (LIM-domain-only protein 2), GATA3 (endothelial transcription factor 3) and RUNX1 to mediate a core transcriptional regulatory circuit in T-ALL (60). LYL1 expression levels are associated with poor prognosis of AML (61). In the present case of a patient harboring mutations in ERCC6 and LYL1, three tumors developed within a short period (1.5–2 years). These two germline gene mutations could potentially influence cancer recurrence.
In addition to driver genes, radiotherapy and chemotherapy are considered risk factors for recurring types of cancer (62). In a previous study, four patients with heterochronous manifestations received chemotherapy and/or radiation therapy for lung cancer before developing AML (63). In all four patients, lung cancer preceded AML by 5–10 years and the patients died within 2 months of being diagnosed with AML. In the present case, the patient initially developed NSCLC and ETP-ALL occurred within 2 years of treatment with pemetrexed combined with cisplatin chemotherapy and radiotherapy, which progressed rapidly. The patient harbored the driver genes ERCC6(+) and LYL1(+), which promoted the occurrence of acute leukemia following radiotherapy and chemotherapy. Later, owing to the high degree of malignancy of ETP-ALL, the patient received allogeneic hematopoietic stem cell transplantation, requiring strong immunosuppression to prevent rejection. These anti-rejection drugs act by removing T cells, resulting in T cell exhaustion (42). According to Chan et al (64), a recurrent SCLC subpopulation may exist in an immunosuppressed tumor microenvironment characterized by exhausted CD8+ T cells, as described by Guo et al (65). Research has shown that the genetic profile of activated tumor regulatory T cells is associated with a poor prognosis in lung adenocarcinoma: Chan et al (64) has shown that SCLC exhibits increased immune isolation and decreased immune infiltration compared with lung adenocarcinoma. Here, 1.5 years after transplantation, the patient developed SCLC, which was possibly linked to use of immunosuppressants such as ATG.
In the present study, the patients father's long-term smoking may have exposed the patient to tobacco smoke for numerous years and may represent a carcinogenic exposure factor. Epidemiological studies of exposure to environmental tobacco smoke, along with the detection of tobacco-specific carcinogens in blood and urine of non-smokers in such environments, have indicated that long-term inhalation of tobacco smoke is a cause of lung cancer (66,67) Subsequent studies (68–70) have similarly confirmed that exposure to environmental tobacco smoke significantly increases risk of lung cancer for non-smokers.
The National Comprehensive Cancer Network guidelines recommend genetic screening for breast, ovarian, pancreatic, lung, colorectal and prostate cancer based on previous studies (71). Genetic screening is performed according to American College of Medical Genetics and Genomics, which offers the advantage of early identification of tumors and effective treatment (72,73). For example (74,75), positive screening of commonly inherited breast cancer gene BRCA1/2 mutations can prompt appropriate treatment measures such as surgery or enhanced monitoring of patients who refuse surgery. However, screening faces a number of challenges, such as cost, invasiveness of the procedure. Therefore, in clinical decision-making, patients should receive information on the advantages and disadvantages, with their preferences respected.
Management of multiple primary tumors poses challenges, with implications for overall survival and quality of life, such as decreasing infections, avoiding transfusions and shorter hospital stays. Therefore, multidisciplinary collaboration to develop a personalized treatment plan is essential. In the present case, when the ETP-ALL diagnosis for the second tumor was made, multiple multidisciplinary discussions with oncology, respiratory and radiotherapy departments were conducted regarding the choice of chemotherapy regimen for ETP-ALL and the decision on whether to proceed with a transplant. The patient and their family were also consulted. Finally, a chemotherapy regimen for AML that combined venetoclax with cytarabine, aclarubicin and G-CSF was chosen, which achieved complete remission. Subsequently, one consolidation cycle was administered, followed by a related donor allogeneic hematopoietic stem cell transplant. Treatment plans for lung cancer post-transplant due to emergence of the third SCLC tumor were similarly coordinated with oncology, respiratory and radiotherapy specialists.
The present study had several strengths, such as genetic testing of germline genes upon development of a second tumor, successful management of ETP-ALL with hematopoietic stem cell transplantation and sustained remission. However, there were also limitations, which included the absence of sample testing from the patient's father, inability to verify the genetic pattern and the need for case reports.
In summary, the present study described a patient with NSCLC harboring mutations in the germline genes ERCC6 and LYL1 who developed ETP-ALL and SCLC shortly after remission. Considering the rapid progression of recurring types of cancer, clinicians should prioritize screening for germline mutations in patients and their family members to facilitate early diagnosis, treatment and prognosis assessment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CT
computed tomography
- ETP-ALL
early T cell precursor acute lymphoblastic leukemia
- NSCLC
non-small cell lung cancer
- ATG
anti-thymocyte globulin
- IACR
International Association of Cancer Registries
Funding Statement
The present study was supported by Zhejiang Traditional Chinese Medicine Administration (grant no. 2021ZQ020) and Key Specialties of Zhejiang Administration of Traditional Chinese Medicine in the 13th Five-year Plan.
Availability of data and materials
The sequencing data generated in the present study may be found in the National Centre of Biotechnology database under accession number PRJNA1146555 or at the following URL: https://dataview.ncbi.nlm.nih.gov/?archive=bioproject.
Authors' contributions
XFX conceived the study and revised the manuscript. HFJ interpreted the radiological findings. YXJ and MXH interpreted the pathological findings. HYW interpreted the genetic findings and wrote the manuscript. XFX and HYW confirm the authenticity of all the raw data All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Review Committee of Tongde Hospital of Zhejiang Province (approval no. 106-JY.2022; Hangzhou, China).
Patient consent for publication
Written informed consent was obtained from the patient for the publication of this report and any accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shah SA, Riaz U, Zahoor I, Jalil A, Zubair M. Carcinoma multiplex. J Coll Physicians Surg Pak. 2013;23:290–292. [PubMed] [Google Scholar]
- 2.Owen LJ. Multiple malignant neoplasms. JAMA. 1921;76:1329–1333. doi: 10.1001/jama.1921.02630200001001. [DOI] [Google Scholar]
- 3.Coyte A, Morrison DS, McLoone P. Second primary cancer risk-The impact of applying different definitions of multiple primaries: Results from a retrospective population-based cancer registry study. BMC Cancer. 2014;14:1–11. doi: 10.1186/1471-2407-14-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buiatti E, Crocetti E, Acciai S, Gafà L, Falcini F, Milandri C, La Rosa M. Incidence of second primary cancers in three Italian population-based cancer registries. Eur J Cancer. 1997;33:1829–1834. doi: 10.1016/S0959-8049(97)00173-1. [DOI] [PubMed] [Google Scholar]
- 5.Weir HK, Johnson CJ, Thompson TD. The effect of multiple primary rules on population-based cancer survival. Cancer Causes Control. 2013;24:1231–1242. doi: 10.1007/s10552-013-0203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosso S, De Angelis R, Ciccolallo L, Carrani E, Soerjomataram I, Grande E, Zigon G, Brenner H, Eurocare Working Group Multiple tumours in survival estimates. Eur J Cancer. 2009;45:1080–1094. doi: 10.1016/j.ejca.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 7.Second primary cancers, corp-author. Victoria Karaholios E, English D, Thursfield V, Simpson J. http://www.cancervic.org.au/downloads/cec/Second-Primary-Cancers.pdf. [ August; 2009 ];2009 [Google Scholar]
- 8.Tsikitis VL, Wertheim BC, Guerrero MA. Trends of incidence and survival of gastrointestinal neuroendocrine tumors in the United States: A seer analysis. J Cancer. 2012;3:292. doi: 10.7150/jca.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch SM, Heeran AB, Burke C, Lynam-Lennon N, Eustace AJ, Dean K, Robson T, Rahman A, Marcone S. Precision oncology, artificial intelligence, and novel therapeutic advancements in the diagnosis, prevention, and treatment of cancer: Highlights from the 59th Irish Association for Cancer Research (IACR) Annual conference. Cancers (Basel) 2024;16:1989. doi: 10.3390/cancers16111989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogt A, Schmid S, Heinimann K, Frick H, Herrmann C, Cerny T, Omlin A. Multiple primary tumours: Challenges and approaches, a review. ESMO Open. 2017;2:e000172. doi: 10.1136/esmoopen-2017-000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood ME, Vogel V, Ng A, Foxhall L, Goodwin P, Travis LB. Second malignant neoplasms: Assessment and strategies for risk reduction. J Clin Oncol. 2012;30:3734–3745. doi: 10.1200/JCO.2012.41.8681. [DOI] [PubMed] [Google Scholar]
- 12.Bajdik CD, Abanto ZU, Spinelli JJ, Brooks-Wilson A, Gallagher RP. Identifying related cancer types based on their incidence among people with multiple cancers. Emerg Themes Epidemiol. 2006;3:17. doi: 10.1186/1742-7622-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaskin HS, Hardy RE, Fletcher RL. Multiple primary malignancies in black patients. J Natl Med Assoc. 1981;73:1065–1068. [PMC free article] [PubMed] [Google Scholar]
- 14.Donin N, Filson C, Drakaki A, Tan HJ, Castillo A, Kwan L, Litwin M, Chamie K. Risk of second primary malignancies among cancer survivors in the United States, 1992 through 2008. Cancer. 2016;122:3075–3086. doi: 10.1002/cncr.30164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.AIRTUM Working Group, corp-author. Italian cancer figures, report 2010: Cancer prevalence in Italy. Patients living with cancer, long-term survivors and cured patients. Epidemiol Prev. 2010;34((5–6 Suppl 2)):S1–S188. (In English, Italian) [PubMed] [Google Scholar]
- 16.Hauben EI, Arends J, Vandenbroucke JP, van Asperen CJ, Van Marck E, Hogendoorn PC. Multiple primary malignancies in osteosarcoma patients. Incidence and predictive value of osteosarcoma subtype for cancer syndromes related with osteosarcoma. Eur J Hum Genet. 2003;11:611–618. doi: 10.1038/sj.ejhg.5201012. [DOI] [PubMed] [Google Scholar]
- 17.Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, Gao C, Lilyquist J, Yadav S, Boddicker NJ, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chubb D, Broderick P, Frampton M, Kinnersley B, Sherborne A, Penegar S, Lloyd A, Ma YP, Dobbins SE, Houlston RS. Genetic diagnosis of high-penetrance susceptibility for colorectal cancer (CRC) is achievable for a high proportion of familial CRC by exome sequencing. J Clin Oncol. 2015;33:426–432. doi: 10.1200/JCO.2014.56.5689. [DOI] [PubMed] [Google Scholar]
- 19.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: Setting paradigms in personalized cancer care and prevention. Science. 2014;343:1466–1470. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of embrace. J Natl Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg S, Buys SS, Edwards SL, Espinel W, Fraser A, Gammon A, Hafen B, Herget KA, Kohlmann W, Roundy C, et al. Population prevalence of individuals meeting criteria for hereditary breast and ovarian cancer testing. Cancer Med. 2019;8:6789–6798. doi: 10.1002/cam4.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grosse SD, Rogowski WH, Ross LF, Cornel MC, Dondorp WJ, Khoury MJ. Population screening for genetic disorders in the 21st century: Evidence, economics, and ethics. Public Health Genom. 2010;13:106–115. doi: 10.1159/000226594. [DOI] [PubMed] [Google Scholar]
- 24.Schienda J, Stopfer J. Cancer genetic counseling-current practice and future challenges. Cold Spring Harb Perspect Med. 2020;10:a036541. doi: 10.1101/cshperspect.a036541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng M, Chen HH, Zhu X, Luo M, Zhang K, Xu CJ, Hu KM, Cheng P, Zhou JJ, Zheng S, Chen YD. Prevalence and clinical outcomes of germline mutations in BRCA1/2 and PALB2 genes in 2769 unselected breast cancer patients in China. Int J Cancer. 2019;145:1517–1528. doi: 10.1002/ijc.32184. [DOI] [PubMed] [Google Scholar]
- 26.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 27.Clark SK. Management of genetically determined colorectal cancer. Surgeon. 2019;17:165–171. doi: 10.1016/j.surge.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Aghabozorgi AS, Ebrahimi R, Bahiraee A, Tehrani SS, Nabizadeh F, Setayesh L, Jafarzadeh-Esfehani R, Ferns GA, Avan A, Rashidi Z. The genetic factors associated with Wnt signaling pathway in colorectal cancer. Life Sci. 2020;256:118006. doi: 10.1016/j.lfs.2020.118006. [DOI] [PubMed] [Google Scholar]
- 29.Short E, Sampson J. The role of inherited genetic variants in colorectal polyposis syndromes. Adv Genet. 2019;103:183–217. doi: 10.1016/bs.adgen.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Sekine Y, Iwasaki Y, Hakozaki N, Endo M, Kamatani Y, Matsuda K, Murakami Y, Sano T, Akamatsu S, Kobayashi T, et al. Prevalence and risk estimation of cancer-predisposing genes for upper urinary tract urothelial carcinoma in Japanese. Jpn J Clin Oncol. 2022;52:1441–1445. doi: 10.1093/jjco/hyac141. [DOI] [PubMed] [Google Scholar]
- 31.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 32.Baysal BE, Willett-Brozick JE, Lawrence EC, Drovdlic CM, Savul SA, McLeod DR, Yee HA, Brackmann DE, Slattery WH, III, Myers EN, et al. Prevalence of SDHB, SDHC, and SDHD germline mutations in clinic patients with head and neck paragangliomas. J Med Genet. 2002;39:178–183. doi: 10.1136/jmg.39.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbina-Jara LK, Rojas-Martinez A, Martinez-Ledesma E, Aguilar D, Villarreal-Garza C, Ortiz-Lopez R. Landscape of germline mutations in DNA repair genes for breast cancer in Latin America: Opportunities for PARP-like inhibitors and immunotherapy. Genes. 2019;10:786. doi: 10.3390/genes10100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer survival rates, 2023, corp-author. https://www.nuffieldtrust.org.uk/resource/cancer-survival-rates. [ June 27; 2023 ];Nuffieldtrust. [Google Scholar]
- 35.Zhao Z, Sun K, Yan T, Wei R, Guo W. Multiple primary tumors: A case report and review of the literature. BMC Musculoskelet Disord. 2020;21:394. doi: 10.1186/s12891-020-03426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Pooter RF, Dias S, Chowdhury M, Bartom ET, Okoreeh MK, Sigvardsson M, Kee BL. Cutting Edge: Lymphomyeloid-primed progenitor cell fates are controlled by the transcription factor tal1. J Immunol. 2019;202:2837–2842. doi: 10.4049/jimmunol.1801220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H, Hu Z, Wang H, Jin G, Wang Y, Sun W, Chen D, Tian T, Jin L, Wei Q, et al. ERCC6/CSB gene polymorphisms and lung cancer risk. Cancer Lett. 2009;273:172–176. doi: 10.1016/j.canlet.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Rami-Porta R, Bolejack V, Crowley J, Ball D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF, Jr, Travis WD, et al. The lASLC lung cancer staging project: Proposals for the Revisions of the T Descriptors in the Forthcoming Eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10:990–1003. doi: 10.1097/JTO.0000000000000559. [DOI] [PubMed] [Google Scholar]
- 39.Mandelker D, Zhang L, Kemel Y, Stadler ZK, Joseph V, Zehir A, Pradhan N, Arnold A, Walsh MF, Li Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318:825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, Hedges D, Ma X, Zhou X, Yergeau DA, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, Ni A, Thomas T, Benayed R, Ashraf A, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2:104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domchek SM. Germline genetic testing for breast cancer: Which patients? What genes? Genet Med. 2020;22:698–700. doi: 10.1038/s41436-019-0721-9. [DOI] [PubMed] [Google Scholar]
- 43.Konstantinopoulos PA, Norquist B, Lacchetti C, Armstrong D, Grisham RN, Goodfellow PJ, Kohn EC, Levine DA, Liu JF, Lu KH, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. 2020;38:1222–1245. doi: 10.1200/JCO.19.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarty D, Solit DB. Clinical cancer genomic profiling. Nat Rev Genet. 2021;22:483–501. doi: 10.1038/s41576-021-00338-8. [DOI] [PubMed] [Google Scholar]
- 45.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-X. [DOI] [PubMed] [Google Scholar]
- 46.Meira LB, Graham JM, Jr, Greenberg CR, Busch DB, Doughty AT, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC. Manitoba aboriginal kindred with original Cerebro-Oculo-Facio-Skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am J Hum Genet. 2000;66:1221–1228. doi: 10.1086/302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ. Genetic variation in the nucleotide excision repair pathway and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:2263–2269. doi: 10.1158/1055-9965.EPI-06-0449. [DOI] [PubMed] [Google Scholar]
- 48.Chiu CF, Tsai MH, Tseng HC, Wang CL, Tsai FJ, Lin CC, Bau DT. A novel single nucleotide polymorphism in ERCC6 gene is associated with oral cancer susceptibility in Taiwanese patients. Oral Oncol. 2008;44:582–586. doi: 10.1016/j.oraloncology.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Lin Z, Zhang X, Tuo J, Guo Y, Green B, Chan CC, Tan W, Huang Y, Ling W, Kadlubar FF, et al. A variant of the Cockayne syndrome B gene ERCC6 confers risk of lung cancer. Hum Mutat. 2008;29:113–122. doi: 10.1002/humu.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu J, Zhao H, Dinney CP, Zhu Y, Leibovici D, Bermejo CE, Grossman HB, Wu X. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408–1415. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 51.Ma H, Hu Z, Wang H, Jin G, Wang Y, Sun W, Chen D, Tian T, Jin L, Wei Q, et al. ERCC6/CSB gene polymorphisms and lung cancer risk. Cancer Lett. 2009;273:172–176. doi: 10.1016/j.canlet.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mellentin JD, Smith SD, Cleary ML. Lyl-1, a novel gene altered by chromosomal translocation in T cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989;58:77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- 55.Meng YS, Khoury H, Dick JE, Minden MD. Oncogenic potential of the transcription factor LYL1 in acute myeloblastic leukemia. Leukemia. 2005;19:1941–1947. doi: 10.1038/sj.leu.2403836. [DOI] [PubMed] [Google Scholar]
- 56.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, Hammer AS, Hamilton Dutoit S, Lossos IS, Levy R. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sang X, Zhang Y, Fang F, Gao L, Tao Y, Li X, Zhang Z, Wang J, Tian Y, Li Z, et al. BRD4 Inhibitor GNE-987 exerts anticancer effects by targeting super-enhancer-related gene LYL1 in acute myeloid leukemia. J Immunol Res. 2022;2022:7912484. doi: 10.1155/2022/7912484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyamoto A, Cui X, Naumovski L, Cleary ML. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol Cell Biol. 1996;16:2394–2401. doi: 10.1128/MCB.16.5.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thoms JAI, Truong P, Subramanian S, Knezevic K, Harvey G, Huang Y, Seneviratne JA, Carter DR, Joshi S, Skhinas J, et al. Disruption of a GATA2-TAL1-ERG regulatory circuit promotes erythroid transition in healthy and leukemic stem cells. Blood. 2021;138:1441–1455. doi: 10.1182/blood.2020009707. [DOI] [PubMed] [Google Scholar]
- 62.Griesinger F, Metz M, Trümper L, Schulz T, Haase D. Secondary leukaemia after cure for locally advanced NSCLC: Alkylating type secondary leukaemia after induction therapy with docetaxel and carboplatin for NSCLC IIIB. Lung Cancer. 2004;44:261–265. doi: 10.1016/j.lungcan.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 63.Varadarajan R, Ford L, Sait SNJ, Block AW, Barcos M, Wallace PK, Ramnath N, Wang ES, Wetzler M. Metachronous and synchronous presentation of acute myeloid leukemia and lung cancer. Leuk Res. 2009;33:1208–1211. doi: 10.1016/j.leukres.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O, Masilionis I, Egger J, Chow A, Walle T, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39:1479–1496. doi: 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 66.Hackshaw AK, Law MR, Wald NJ. The accumulated evidence on lung cancer and environmental tobacco smoke. BMJ. 1997;315:980–988. doi: 10.1136/bmj.315.7114.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang HJ, Min HY, Kang YP, Boo HJ, Kim J, Ahn JH, Oh SH, Jung JH, Park CS, Park JS, et al. Tobacco-induced hyperglycemia promotes lung cancer progression via cancer cell-macrophage interaction through paracrine IGF2/IR/NPM1-driven PD-L1 expression. Nat Commun. 2024;15:4909. doi: 10.1038/s41467-024-49199-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong L, Goldberg MS, Parent ME, Hanley JA. Exposure to environmental tobacco smoke and the risk of lung cancer: A meta-analysis. Lung Cancer. 2000;27:3–18. doi: 10.1016/S0169-5002(99)00093-8. [DOI] [PubMed] [Google Scholar]
- 69.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 70.Adler S, Yip R, Chan H, Cai Q, Zhu Y, Triphuridet N, Kaufman A, Taioli E, Flores R, Henschke CI, et al. Comparison of lung cancer aggressiveness in patients who never smoked compared to those who smoked. Lung Cancer. 2022;171:90–96. doi: 10.1016/j.lungcan.2022.07.002. [DOI] [PubMed] [Google Scholar]
- 71.Gradishar WJ, Moran MS, Abraham J, Abramson V, Aft R, Agnese D, Allison KH, Anderson B, Burstein HJ, Chew H, et al. NCCN Guidelines® insights: Breast cancer, version 4. J Natl Compr Canc Netw. 2023;21:594–608. doi: 10.6004/jnccn.2023.0031. [DOI] [PubMed] [Google Scholar]
- 72.Parsons MT, de la Hoya M, Richardson ME, Tudini E, Anderson M, Berkofsky-Fessler W, Caputo SM, Chan RC, Cline MS, Feng BJ, et al. Evidence-based recommendations for gene-specific ACMG/AMP variant classification from the ClinGen ENIGMA BRCA1 and BRCA2 Variant Curation expert panel. Am J Hum Genet. 2024;111:2044–2058. doi: 10.1016/j.ajhg.2024.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pal T, Agnese D, Daly M, La Spada A, Litton J, Wick M, Klugman S, Esplin ED, Jarvik GP, Professional Practice Guidelines Committee Points to consider: Is there evidence to support BRCA1/2 and other inherited breast cancer genetic testing for all breast cancer patients? A statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2020;22:681–685. doi: 10.1038/s41436-019-0712-x. [DOI] [PubMed] [Google Scholar]
- 74.Lucas AL, Fu Y, Labiner AJ, Dimaio CJ, Sethi A, Kastrinos F. Frequent Abnormal pancreas imaging in patients with pathogenic ATM, BRCA1, BRCA2, and PALB2 breast cancer susceptibility variants. Clin Gastroenterol Hepatol. 2023;21:2686–2688. doi: 10.1016/j.cgh.2022.08.040. [DOI] [PubMed] [Google Scholar]
- 75.Narod SA, Giannakeas V. In response to ‘Pregnancy after breast cancer in patients with germline BRCA mutations’. J Clin Oncol. 2020;38:4352. doi: 10.1200/JCO.20.02253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data generated in the present study may be found in the National Centre of Biotechnology database under accession number PRJNA1146555 or at the following URL: https://dataview.ncbi.nlm.nih.gov/?archive=bioproject.