Abstract
The SSY1 gene of Saccharomyces cerevisiae encodes a member of a large family of amino acid permeases. Compared to the 17 other proteins of this family, however, Ssy1p displays unusual structural features reminiscent of those distinguishing the Snf3p and Rgt2p glucose sensors from the other proteins of the sugar transporter family. We show here that SSY1 is required for transcriptional induction, in response to multiple amino acids, of the AGP1 gene encoding a low-affinity, broad-specificity amino acid permease. Total noninduction of the AGP1 gene in the ssy1Δ mutant is not due to impaired incorporation of inducing amino acids. Conversely, AGP1 is strongly induced by tryptophan in a mutant strain largely deficient in tryptophan uptake, but it remains unexpressed in a mutant that accumulates high levels of tryptophan endogenously. Induction of AGP1 requires Uga35p(Dal81p/DurLp), a transcription factor of the Cys6-Zn2 family previously shown to participate in several nitrogen induction pathways. Induction of AGP1 by amino acids also requires Grr1p, the F-box protein of the SCFGrr1 ubiquitin-protein ligase complex also required for transduction of the glucose signal generated by the Snf3p and Rgt2p glucose sensors. Systematic analysis of amino acid permease genes showed that Ssy1p is involved in transcriptional induction of at least five genes in addition to AGP1. Our results show that the amino acid permease homologue Ssy1p is a sensor of external amino acids, coupling availability of amino acids to transcriptional events. The essential role of Grr1p in this amino acid signaling pathway lends further support to the hypothesis that this protein participates in integrating nutrient availability with the cell cycle.
Yeast cells can selectively use the wide variety of nitrogenous compounds that they find in their rich natural environment. Some of these molecules can be directly used as ready-made metabolites. Many of them can also be catabolized to sustain the synthesis of glutamate and glutamine, the predominant nitrogen donors in biosynthetic reactions (20, 87). The synthesis of many enzymes and permeases involved in nitrogen metabolism and the activity of some of these proteins are tightly regulated according to the nitrogen source(s) available in the medium (20, 35, 45, 59, 87). It is generally assumed that these regulations are triggered solely by variations in the intracellular concentrations of specific metabolites. For instance, many enzymes involved in nitrogen anabolism are inhibited and/or their synthesis is repressed upon accumulation of the end or intermediate products of biosynthetic pathways (35, 45). Similarly, expression of most genes encoding amino acid biosynthetic enzymes is stimulated severalfold in response to starvation for any one of several amino acids (35, 45). Nitrogen repression (NR) is yet another example of regulation apparently triggered upon variation of the concentration of intracellular effectors (59). For instance, repression in the presence of NH4+ of at least some NR-sensitive genes is relieved in cells partially starved for glutamine due to a thermosensitive mutation in the glutamine synthetase GLN1 gene (25, 87).
Whether yeast cells also possess regulatory systems responding specifically to the extracellular concentration of nitrogenous compounds has been studied very little to date. It seems reasonable, however, to speculate that such nitrogen sensors exist. For instance, two sensors of external glucose concentration (Snf3p and Rgt2p) have recently been discovered in yeast cells (67). These proteins are members of the sugar transporter superfamily (14, 15) and play a central role in the transcriptional regulation of the HXT genes encoding glucose transporters (55, 66, 67). Although Snf3p and Rgt2p show significant sequence similarity with hexose transporters, they seem unable to mediate glucose transport, or if they do, this activity is not sufficient to confer a measurable glucose uptake activity or to restore the ability to use glucose in a mutant lacking the six main glucose transporters (Hxt1, -2, -3, -4, -6, and -7) (55, 66, 73). Two other proteins of the sugar transport family, namely, the Rco3 regulator of conidiation in Neurospora crassa (58) and the Mst1 protein from the ectomycorrhiza Amita muscaria (64), may also serve as glucose sensors. Similarly, the uhpC gene of Escherichia coli encodes a protein highly similar in sequence to UhpT, a permease for several organophosphate compounds including glucose-6-phosphate (47). The UhpC protein seems unable to mediate uptake of glucose-6-phosphate and is involved, rather, in transcriptional induction of the uhpT permease gene in response to micromolar levels of external glucose-6-phosphate (48). Some cell surface proteins that effectively mediate transmembrane solute transport also seem to have a regulatory function. For instance, it was recently shown that Mep2p, a high-affinity NH4+ transporter (61), is essential to diploid-cell differentiation into a filamentous, pseudohyphal growth when the sole nitrogen source is NH4+ at a low concentration. This suggests that this transporter also acts as a sensor of low levels of extracellular NH4+ (56). These studies raise the possibility that yet other transporters or transporter homologs act as sensors of external compounds in addition to (or instead of) mediating their uptake across the plasma membrane. In fact, transmembrane solute transporters, by their location, diversity, and ability to recognize a wide variety of exogenous compounds with different specificities and affinities, seem ideally “qualified” to serve as sensors of external nutrients.
It was recently reported that the amino acid permease homologue encoded by the YDR160w/SSY1 gene is required for transcriptional induction by leucine of the amino acid permease genes, BAP2, TAT1, and BAP3, and of the peptide transporter, PTR2. Induction of BAP2 by l- or d-leucine also occurs in a strain largely deficient in l- and d-leucine transport, suggesting that Ssy1p mediates transcriptional induction of permease genes in response to external leucine (24). We report here the results of an independent approach indicating that this amino acid permease homologue acts as a sensor of multiple external amino acids. This sensor is required for transcriptional induction of at least six amino acid permease genes. We show that one of these genes, AGP1, encodes a wide-specificity amino acid permease induced by all amino acids except proline. Together with the general amino acid permease (Gap1p), this permease plays a major role in amino acid utilization. We show that induction of AGP1 occurs in response to extracellular rather than intracellular amino acids. This induction requires the Cys6-Zn2 transcription factor encoded by the UGA35(DAL81/DURL) gene. It also requires the F-box protein Grr1p involved in cell cycle regulation and in Snf3p- and Rgt2p-mediated glucose sensing.
MATERIALS AND METHODS
Strains, growth conditions, and methods.
The Saccharomyces cerevisiae strains used in this study are all isogenic with the wild-type Σ1278b (12) except for the mutations mentioned (Table 1). Cells were grown in a minimal buffered (pH 6.1) medium with 3% glucose as the carbon source (41). To this medium, urea (5 mM), proline (5 mM), (NH4)2SO4 (10 mM), amino acids (1 to 10 mM), or combinations of these compounds were added as a source(s) of nitrogen. Assays for resistance to toxic amino acid analogues were carried out on plates with (NH4)2SO4 (10 mM) as the sole nitrogen source. Analogue concentrations were as follows: 20 μg/ml, β-(2-thienyl)-dl-alanine; 20 μg/ml, p-fluoro-dl-phenylalanine; 20 μg/ml, d,l-ethionine; 500 μg/ml, 6-fluoro-tryptophan; and 1 mg/ml, hydroxy-tryptophan. All procedures for manipulating DNA were standard ones (6, 74). The E. coli strain used was JM109.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| 23344c | MATα ura3 | LPCGLa |
| 23346c | MATa ura3 | LPCGL |
| 32501a | MATα ura3 | This study |
| 32501b | MATα gap1Δ::kanMX2 ura3 | This study |
| 32501c | MATa ssy1Δ::kanMX2 ura3 | This study |
| 32501d | MATa gap1Δ::kanMX2 ssy1Δ::kanMX2 ura3 | This study |
| 30629c | MATa gap1Δ::kanMX2 ura3 | This study |
| 30633c | MATa gap1Δ::kanMX2 agp1Δ::kanMX2 ura3 | This study |
| 32502b | MATα gap1Δ::kanMX2 agp1Δ::kanMX2 ssy1Δ::kanMX2 ura3 | This study |
| 30622a | MATα gap1-92 agp1-1 ura3 | 36a |
| 30701a | MATa aro80Δ::kanMX2 ura3 | 36a |
| 26247b | MATa trp2fbr | 82 |
| RE1 | MATa trp2fbr ura3Δ::kanMX2 | This study |
| JA115 | MATα grr1Δ::kanMX2 ura3 | This study |
| CD17 | MATα uga35Δ::TYR1 tyr1 ura3 | 21 |
LPCGL, Laboratoire de Physiologie Cellulaire et de Génétique des Levures.
Construction of ssy1Δ, agp1Δ, gap1Δ, and grr1Δ deletion strains.
The ssy1Δ, agp1Δ, grr1Δ, and gap1Δ null mutations were constructed by the PCR-based gene deletion method (86). The DNA segments used to introduce these mutations were generated by using the kanMX2 gene from plasmid pFA6a-kanMX2 as a template and the following PCR primers: ssy1Δ:kanMX2, 5′-CTCTAGGGGAAAAAAGGAAACAGGCGTGTGATAAGAGGCCGCGGCCGCCAGCTGAAGCTTCGTACGC-3′ and 5′-CAGTTACCCGCACAATCTAGTGCGTAAAGCAGTGTCAATAGCGGCCGCATAGGCCACTAGTGGATCTG-3′; agp1Δ:kanMX2, 5′-CCAGAAGGCAACGACCCTTTTCCAATAAGGTCCGTTCCGCGGCCGC GCATAGGCCACTAGTGGATCTG-3′ and 5′-TCGTCGTCGAAGTCTCTATACGAACTGAAAGACTTGGCGGCCGCCAGCTGAAGCTTCGTACGA-3′; gap1Δ:kanMX2, 5′-CTATCAGGCAGCCTCACTAATCTACCCATTGACCTCATGCGCGGCCGCCAGCTGAAGCTTCGTACGC-3′ and 5′-GAAGCTCACACAGAT TAG T T T TCATCTCGCT G TC TACTAAGCGGCCGCATAGGCCACTAGTGGATCTG-3′; and grr1Δ:kanMX2, 5′-ATGGATCAGGATAACAACAACCACAATGACAGCAATAGGCTGCACCCATTCGTACGCTGCAGGTCGAC-3′ and 5′-GGGCGTTCCTGATGCTTCATCCATTTGAGAATCAATGGCAGTGTCAGGCGCATAGGCCACTAGTGGATCTG-3′. The yeast strain 23344c was transformed with the PCR fragments by the lithium method (39) as described previously (30). Transformants were selected on complete medium containing 200 μg of G418 (Geneticin; Gibco BRL) per ml.
Plasmids.
The YCpARO9-lacZ plasmid has been described (36). The YCpAGP1-lacZ plasmid was constructed by inserting into the BamHI- and HindIII-cleaved YCpAJ152 plasmid (4) a 996-bp DNA fragment flanked by HindIII and BamHI restriction sites and spanning the five first codons of AGP1 plus 979 bp of upstream sequences. This DNA fragment was obtained by PCR with, as a template, plasmid p16.2 bearing the AGP1 gene cloned from strain Σ1278b (36a) and the following PCR primers: 5′-CCGAAGCTTCCTCAACCTACCATGGCAAAC-3′ and 5′-CGCGGATCCGACTTCGACGACGACATTGT-3′. The accuracy of the PCR-amplified fragment was checked by sequencing.
Enzyme and permease assays.
All permease and enzyme assays were performed on cells that reached the state of balanced growth. Incorporation of 14C-labeled amino acids (Amersham) was measured as previously described (33). β-Galactosidase activities were measured as described earlier (4) and are expressed in nanomoles of o-nitrophenol formed per minute per milligram of protein. Protein concentrations were measured with the Folin reagent (57) and, as the standard, bovine serum albumin.
Measurements of intracellular tryptophan concentration.
Cells having reached the state of balanced growth were collected by filtration (Millipore 0.45-μm-pore-size filters) and washed four times with ice water. Cells were immediately resuspended in 5 ml of 5% trichloroacetic acid and incubated at 0°C for 10 min with several inversions of the tubes. The extracts were harvested after filtration (Millipore 0.45-μm-pore-size filters) and stored at −20°C. Tryptophan concentrations were determined by high-pressure liquid chromatography with electrochemical detection as previously described (29).
RNA analysis by RT-PCR.
Total yeast cell RNA was prepared by using the RNeasy Mini-Kit (Qiagen) as recommended by the manufacturer. RNA preparations were treated with DNase (Boehringer) for 1 h at 37°C and washed with RNeasy mini spin columns as described by the manufacturer (Qiagen). A PCR test was performed on each RNA preparation to make sure it was DNA-free. Reverse transcriptase (RT)-PCRs were performed by using the Titan One Tube RT-PCR Kit (Boehringer Mannheim) and a Tecne (Cambridge) thermocycler. The samples were first incubated at 55°C for 30 min (for reverse transcription) and then as follows for thermocycling: 94°C for 2 min (1 time); then 94°C for 30 s, 52°C for 30 s, and 68°C for 75 s, plus 5 s for each cycle starting at cycle 11 (25 times); and then 68°C for 7 min (1 time). The PCR primers were chosen so that they had similar melting temperatures and generated PCR fragments of similar sizes. Their sequences were as follows: ACT1, 5′-GACTCCTACGTTGGTGATGA-3′ and 5′-CTGGAGGAGCAATGATCTTG-3′; GAP1, 5′-ATCGGTACTGGTCTGCTGGT-3′ and 5′-TCTACGGATTCACTGGCAGC-3′; AGP1, 5′-TCTTACGTCGGCTATCTCAC-3′ and 5′-GATGCAACAGCAATGACATA-3′; GNP1, 5′-TGGTCACTGCATCCATGACT-3′ and 5′-GAGGCACAGAATGCAATGAC-3′; BAP2, 5′-TCGAGACGTACTTCATGATC-3′ and 5′-TCAGTCTTGGACCAGCATAC-3′; TAT1, 5′-GTCACTTAGTCATGATCAGT-3′ and 5′-ATGTGATGCAACAGCAATGA-3′; TAT2, 5′-ACCGTACAGTACTGGAACTC-3′ and 5′-CTGATATGTGACAGGTTGAT-3′; BAP3, 5′-ATCGGTTACGTTATGGTGTC-3′ and 5′-GCTGCCAAGACATATGGTGA-3′; YNL270c, 5′-GACAGAAGCAGTGCCTCTAG-3′ and 5′-CACCTCTGGTCACGTTAGAC-3′; YBR132c, 5′-CATTACTGTGTCTACAGCGG-3′ and 5′-AGTGTAAGCGTTACCAGCAG-3′; YPL274c, 5′-TGTCAGTAGGTTCATAGATG-3′ and 5′-GTCCATGTAGGAACATACCG-3′; YLL061w, 5′-TATCAAGATGACCGCATTCA-3′ and 5′-ATCACTAGCGTCCGGACCTG-3′; YFL055w, 5′-ATAGCGATGCACTGCCTGCA-3′ and 5′-TGCAGCTCCAACGCTCACAT-3′; MUP1, 5′-TCTGAATGTCAAGATTGGTC-3′ and 5′-GTAAGGAGCAATAATCAGGT-3′; and MUP3, 5′-ATAACCATCCATCGATACCA-3′ and 5′-CACGGATGATTCGTGGTCCA-3′.
RESULTS
Ydr160p, an amino acid permease homologue, displays unusual structural features.
Complete sequencing of the yeast genome has led to the discovery of many new genes encoding putative transmembrane solute transporters (3). The largest class of yeast transport proteins is the sugar transporter family, which includes 7 functionally characterized hexose transporters (Hxt1, -2, -3, -4, -6, and -7 and Gal2), 10 homologous proteins of still unknown function, and 17 more distantly related proteins, some of which have been functionally characterized (15, 53). It is now accepted that two proteins of this family, Snf3p and Rgt2p, ensure a regulatory rather than a catabolic function, i.e., they act as sensors of external glucose concentration (55, 67). These two proteins display features that clearly distinguish them from the Hxt transporters: they are expressed to much lower levels and their C-terminal cytosolic tail is much longer (14, 19, 67). These properties are obvious when the codon bias index (CBI [13]) values of the genes coding for Snf3p, Rgt2p, and the Hxt proteins are plotted against the amino acid chain lengths of these proteins (Fig. 1). We have applied the same plot representation to all other families of yeast transport proteins extracted by computational analysis (3). Analysis of the output data concerning the 18 proteins of the amino acid permease family revealed that one member of this family, namely, the YDR160w gene product (42), stands out from the others because of its unusually low CBI (0.013) and much larger size (852 amino acids) (Fig. 1). Like the other proteins of this family, Ydr160p consists of a central hydrophobic core of 12 predicted transmembrane (TM) domains flanked by N-terminal and C-terminal hydrophilic regions. The hydrophilic N terminus of Ydr160p (281 residues), however, is unusually large compared to those of the other members of the amino acid permease family (Fig. 2). Furthermore, several regions connecting TM domains and predicted to be extracellular are larger in Ydr160p than in classical amino acid permeases (Fig. 2). Finally, Ydr160p is more distantly related in sequence to the other members of the amino acid permease family (Fig. 2). The unusual structural features of Ydr160p are reminiscent of those distinguishing the Snf3p and Rgt2p glucose sensors from the other proteins of the sugar transporter family (14, 19, 67). On the basis of these criteria, we hypothesized that Ydr160p might perform a function different from that of classical amino acid permeases. For instance, just as Snf3p and Rgt2p play a determining role in regulating glucose transport, Ydr160p might be involved in regulating amino acid transport.
FIG. 1.
The yeast Ydr160p protein of the amino acid permease family displays unusual features resembling those of the Snf3p and Rgt2p glucose sensors. The CBI values (13) of the genes coding for the 17 Hxt proteins of the hexose transporter family, the glucose sensors Snf3p and Rgt2p (left panel), and the 18 proteins of the amino acid permease family (right panel) are plotted against the number of amino acid residues present in the proteins.
FIG. 2.
The Ydr160-Ssy1p protein displays unusual structural features compared to other members of the amino acid permease family. The amino acid sequences of the amino acid permeases Gap1p (43), Hip1p (79), Agp1p-Wap1p (65), and Ydr160p-Sys1p (42) were aligned by using the PILEUP program (23). Identical and conserved residues are indicated in black boxes. The transmembrane segments predicted by using the TMAP algorithm (72) are underlined.
Ydr160p-Ssy1p is required for induction of amino acid permeases.
As a first step in the functional analysis of the YDR160w gene, we isolated a yeast strain with a complete deletion of the corresponding coding region (see Materials and Methods). The deletion mutant was viable on both rich and minimal glucose medium. The ydr160Δ mutant displayed no clear growth defect on any of the amino acids that can be used as the sole nitrogen source (data not shown). We then compared the sensitivities of the wild-type and ydr160Δ strains to several toxic amino acid analogues. These experiments showed that lack of the YDR160w gene confers resistance to the phenylalanine analogues β-(2-thienyl)-dl-alanine and p-fluoro-dl-phenylalanine, to the methionine analogue d,l-ethionine, and to the tryptophan analogues 6-fluoro-tryptophan and hydroxy-tryptophan (data not shown). Resistance to several toxic amino acid analogues is a property shared by apf mutants deficient in the uptake of multiple amino acids (32). The YDR160w gene proved to be nonallelic with six previously isolated apf complementation groups and was initially called APF7 for amino acid permeability factor 7. In the course of preparation of this study, it was reported that the SSY1 gene originally identified on a genetic basis (46) is identical to YDR160w (24). Hence, the SSY1 nomenclature will hereafter be adopted.
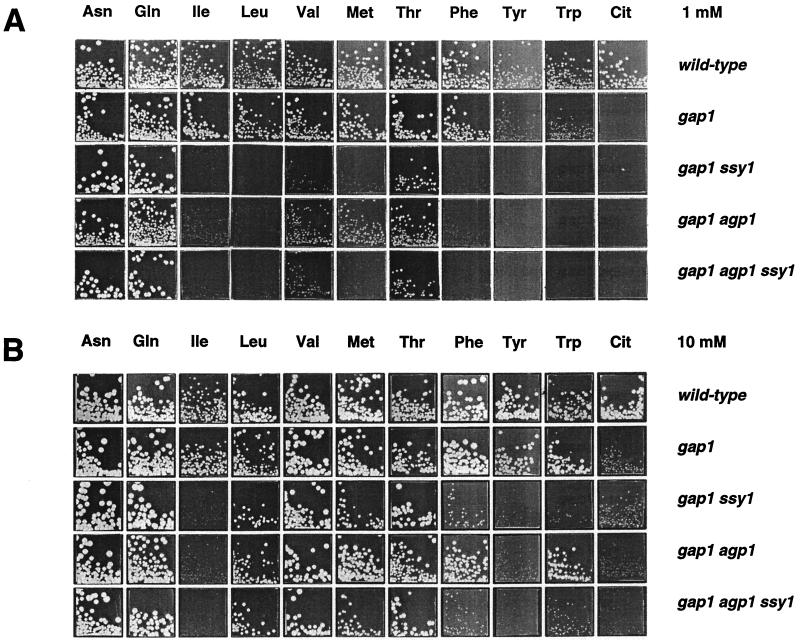
The activity of the general amino acid permease (Gap1p) was unaffected in the ssy1Δ mutant growing on urea or proline as the sole nitrogen source, i.e., under conditions where Gap1p is normally most active (not shown). This observation prompted us to examine the effect of deleting SSY1 in a gap1Δ mutant. We first compared growth of the gap1Δ and gap1Δ ssy1Δ strains on low (1 mM) and high (10 mM) concentrations of several amino acids, each used as the sole nitrogen source (Fig. 3). Deletion of the SSY1 gene in the gap1Δ strain dramatically reduced growth on low concentrations of isoleucine, leucine, valine, methionine, phenylalanine, tyrosine, tryptophan and, to a lesser extent, threonine (Fig. 3). Growth of the SSY1-deleted strain returned to normal on leucine, valine, and threonine when the amino acid concentration was increased to 10 mM. At this higher concentration, a clear growth defect due to lack of Ssy1p was still visible on isoleucine, phenylalanine, tyrosine, tryptophan and, to a lesser extent, methionine.
FIG. 3.
Deletion of the SSY1 or the AGP1 gene affects the utilization of several amino acids in cells lacking the general amino acid permease (Gap1p). Cells were spread on minimal medium with the indicated amino acid at the final concentration of 1 mM (A) or 10 mM (B) as the sole nitrogen source. The strains were 23344c (ura3), 32501b (gap1Δ ura3), 32501d (gap1Δ ssy1Δ ura3), 30633c (gap1Δ agp1Δ ura3), and 32502b (gap1Δ ssy1Δ agp1Δ ura3).
The effects of deleting SSY1 in the gap1Δ strain largely overlap with those produced in the same genetic background by the wap1Δ/agp1Δ mutation (Fig. 3). WAP1, a gene originally discovered during systematic sequencing of chromosome III (YCL025c), encodes a member of the amino acid permease family (65). The WAP1 gene (WAP1 for wide-specificity amino acid permease 1) was also isolated in our laboratory as part of a study focusing on induction by aromatic amino acids of the ARO9 gene encoding aromatic aminotransferase II. In that study, among mutants displaying reduced induction of an ARO9-lacZ fusion gene, one turned out to bear two mutations: one in the GAP1 gene and another in the WAP1 gene. In this gap1–92 wap-1-1 mutant strain, induction of ARO9-lacZ is reduced severalfold compared to the wild type, an effect most likely due to partial inducer exclusion (36a). As this study was being prepared, a functional and expression analysis of the YCL025c/WAP1 gene was reported and the gene was named AGP1 (for asparagine and glutamine permease) (76). Hence, the AGP1 nomenclature will hereafter be used here.
Deletion of the AGP1 gene in the wild-type strain did not affect amino acid utilization (data not shown), but in the gap1Δ strain it produced phenotypes similar to those of the gap1Δ ssy1Δ mutant, except on valine (1 mM), methionine (1 to 10 mM), phenylalanine (10 mM), and tryptophan (10 mM), where growth of the gap1Δ ssy1Δ strain was more strongly affected than that of the gap1Δ agp1Δ strain (Fig. 3). These results are consistent with SSY1 being needed for the function of the Agp1p permease, but since the growth deficiency caused by the ssy1Δ mutation is broader than that caused by the agp1Δ mutation, at least one additional amino acid permease is likely affected by the ssy1Δ deletion. The fact that the ssy1Δ strain is resistant to five toxic amino acid analogues and that the agp1Δ strain is sensitive to them (not shown) is also consistent with Ssy1p affecting other amino acid permeases in addition to Agp1p.
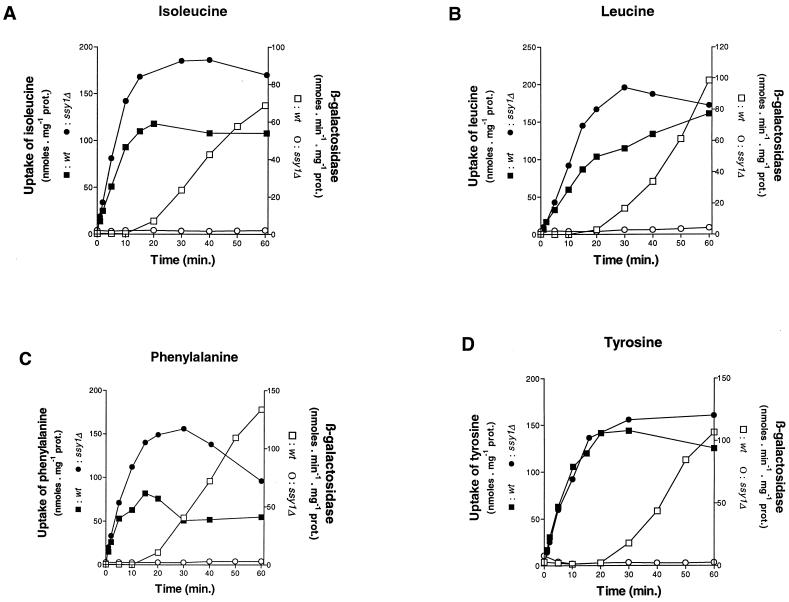
These assumptions were confirmed by amino acid uptake assays with cells growing on urea as the sole nitrogen source. Representative data obtained with the amino acids leucine, isoleucine, phenylalanine, and tyrosine are shown in Fig. 4. All four amino acids were immediately incorporated into gap1Δ cells at a relatively low rate, but uptake rapidly accelerated, a behavior typically observed in the case of permeases induced in the presence of their own substrates (4). In the gap1Δ agp1Δ double mutant, the inducible uptake of each amino acid was largely, though not entirely, suppressed (Fig. 4). Thus, the inducible leucine, isoleucine, phenylalanine, and tyrosine uptake activities displayed by gap1Δ cells growing on minimal urea medium are largely attributable to the product of the AGP1 gene. The fact that gap1Δ agp1Δ cells still display residual inducible uptake activity suggests that each amino acid is incorporated by at least one additional inducible permease. In the gap1Δ ssy1Δ mutant, finally, all four amino acids were incorporated at a low and apparently constant rate, indicating that Agp1p and the additional permease(s) normally induced in response to leucine, isoleucine, phenylalanine, and tyrosine are inactive in the ssy1Δ strain. These results are consistent with the growth test data and show that Ssy1p is required for the activity of at least two inducible amino acid permeases, one being Agp1p.
FIG. 4.
Deletion of the SSY1 or AGP1 gene alters incorporation of amino acids. The time course of 14C-labeled leucine, isoleucine, phenylalanine, and tyrosine (initial concentration, 0.1 mM) accumulation measured in cells growing on minimal medium with urea as the sole nitrogen source is shown. The strains were 30629c (gap1Δ ura3) (■), 30633c (gap1Δ agp1Δ ura3) (□) and 32501d (gap1Δ ssy1Δ ura3) (○).
Ssy1p is required for transcriptional induction of the AGP1 gene in response to multiple amino acids.
A DNA fragment composed of the first codons of the AGP1 gene preceded by its promoter region was fused in frame with the lacZ reporter gene in a low-copy-number plasmid. Wild-type cells transformed with this AGP1-lacZ-bearing plasmid were grown on minimal medium containing urea, urea-leucine, urea-isoleucine, urea-phenylalanine, or urea-tyrosine as the sole nitrogen source(s). β-Galactosidase assays in extracts of steady-state growing cells (Table 2) clearly showed that AGP1 is not expressed on urea medium but that its transcription is markedly induced in the presence of each of the four amino acids (lines 1 to 5). In the ssy1Δ strain, in contrast, the AGP1-lacZ gene remained uninduced (lines 1 to 5). We then tested the influence of other amino acids on expression of the AGP1-lacZ gene. Remarkably, many other amino acids induced transcription of the AGP1-lacZ gene, and in all cases induction was abolished in the ssy1Δ strain (lines 6 to 23). The level of induction in the wild type, however, varied strongly according to the amino acid tested. The highest induction levels were produced by leucine, isoleucine, phenylalanine, tyrosine, tryptophan, threonine, and methionine, i.e., amino acids on which growth of the gap1Δ strain was most affected after deletion of the SSY1 gene. Intermediate levels of induction were obtained with valine, citrulline, cysteine, alanine, and serine; and still lower levels were obtained with lysine, histidine, glutamate, glutamine, glycine, aspartate, and asparagine. 4-Aminobutyrate (GABA), arginine, and ornithine were very poor inducers, and the presence of proline or certain other nitrogenous compounds that can serve as nitrogen sources (allantoate, allantoin, cytosine, and adenine) had no influence on AGP1-lacZ expression. In conclusion, transcription of the AGP1 gene is induced by many amino acids (a notable exception being proline), and this induction requires a functional SSY1 product.
TABLE 2.
The Ssy1 protein is required for transcriptional induction of the AGP1 gene in response to multiple amino acidsa
| Line no. | Nitrogen sources |
AGP1-lacZ β-galactosidase activity (nmol · min−1 · mg of protein−1)
|
|
|---|---|---|---|
| Wild type | ssy1Δ | ||
| 1 | Urea | ≤2 | ≤2 |
| 2 | Urea + Leu | 713 | ≤2 |
| 3 | Urea + IIe | 1,069 | ≤2 |
| 4 | Urea + Phe | 943 | ≤2 |
| 5 | Urea + Tyr | 903 | ≤2 |
| 6 | Urea + Trp | 1,246 | ≤2 |
| 7 | Urea + Thr | 871 | ≤2 |
| 8 | Urea + Met | 825 | ≤2 |
| 9 | Urea + Val | 421 | ≤2 |
| 10 | Urea + citrulline | 294 | ≤2 |
| 11 | Urea + Cys | 241 | ≤2 |
| 12 | Urea + Ala | 198 | ≤2 |
| 13 | Urea + Ser | 177 | ≤2 |
| 14 | Urea + Lys | 84 | ≤2 |
| 15 | Urea + His | 75 | ≤2 |
| 16 | Urea + Glt | 54 | ≤2 |
| 17 | Urea + Gln | 49 | ≤2 |
| 18 | Urea + Gly | 46 | ≤2 |
| 19 | Urea + Asp | 33 | ≤2 |
| 20 | Urea + Asn | 27 | ≤2 |
| 21 | Urea + GABA | 7 | ≤2 |
| 22 | Urea + Arg | 5 | ≤2 |
| 23 | Urea + ornithine | 4 | ≤2 |
| 24 | Urea + Pro | ≤2 | ≤2 |
| 25 | Urea + allantoin | ≤2 | ≤2 |
| 26 | Urea + adenine | ≤2 | ≤2 |
| 27 | Urea + cytosine | ≤2 | ≤2 |
| 28 | Urea + allantoate | ≤2 | ≤2 |
Strains 23344c (ura3) and 30501c (ssy1Δ ura3) transformed with the CEN-based YCpAGP1-lacZ plasmid were grown on a minimal medium with the indicated compounds as the sole nitrogen source(s), each added at 5 mM final concentration. The reported β-galactosidase activities are means of two to three independent experiments. Variations were less than 20%.
AGP1 encodes a broad-specificity, low-affinity amino acid permease.
That the AGP1 gene is induced to various degrees by multiple amino acids suggests that the amino acid permease encoded by this gene has a broad substrate specificity. In amino acid uptake experiments (Fig. 4), induction of AGP1 clearly led to markedly increased uptake of leucine, isoleucine, phenylalanine, and tyrosine. These uptake assays in fact show that, apart from the general amino acid permease, Agp1p is the main entry pathway for these four amino acids (used at 0.1 mM concentrations) in cells growing under conditions of nitrogen derepression. To further explore the substrate specificity range of the Agp1p permease, we compared in the gap1Δ and gap1Δ agp1Δ strains the initial uptake rates of several amino acids present at a 0.1 mM concentration (Table 3). To induce AGP1 expression, we grew the strains on minimal urea medium supplemented with citrulline (0.5%). We found that citrulline used at this concentration does not significantly interfere with Agp1p-mediated uptake of amino acids (inhibition was ≤10%), i.e., it behaves like a gratuitous inducer of AGP1 expression. In the gap1Δ strain grown under these conditions, Agp1p proved responsible for a significant portion of the initial uptake of many amino acids, including leucine (96%), isoleucine (86%), tyrosine (83%), valine (82%), phenylalanine (78%), threonine (77%), methionine (68%), glutamine (64%), serine (62%), alanine (60%), histidine (54%), glycine (49%), and asparagine (25%). It contributed negligibly to the uptake of tryptophan, proline, arginine, aspartate, glutamate, and lysine (0 to 15%). Hence, most amino acids inducing high-level expression of AGP1 appear to be substrates of Agp1p. The fact that deletion of AGP1 in a gap1Δ mutant alters growth on only some of the Agp1p substrates is likely due to the existence of other permeases that can compensate for the lack of Agp1p. For instance, a permease able to transport glutamine (Gnp1p) has been described (90). Although we could not detect any significant contribution of Agp1p to the uptake of tryptophan, proline, arginine, lysine, glutamate, aspartate, or citrulline used at concentrations of up to 0.1 mM, at least some of these amino acids are likely incorporated via Agp1p when present at a higher concentration. For instance, Agp1p is required for a gap1 mutant to grow on tryptophan at a 1 mM concentration (Fig. 3). Agp1p is a relatively low-affinity permease, as judged by the kinetic parameters of Agp1p-mediated leucine uptake (Km = 0.16 mM; Vmax = 100 nmol · min−1 · mg of protein−1), isoleucine (Km = 0.6 mM; Vmax = 175 nmol · min−1 · mg of protein−1) and phenylalanine (Km = 0.6 mM; Vmax = 60 nmol · min−1 · mg of protein−1).
TABLE 3.
Agp1 is a broad-specificity amino acid permeasea
| Line no. | Amino acid | Uptake activity (nmol · min−1 · mg of protein−1)
|
|
|---|---|---|---|
| gap1Δ | gap1Δ agp1Δ | ||
| 1 | Leucine | 39 | 1 |
| 2 | Isoleucine | 40 | 5 |
| 3 | Valine | 22 | 4 |
| 4 | Threonine | 79 | 18 |
| 5 | Phenylalanine | 16 | 3 |
| 6 | Tyrosine | 5 | 1 |
| 7 | Serine | 130 | 50 |
| 8 | Methionine | 160 | 51 |
| 9 | Alanine | 54 | 20 |
| 10 | Glutamine | 103 | 38 |
| 11 | Histidine | 30 | 14 |
| 12 | Asparagine | 93 | 69 |
| 13 | Glycine | 19 | 10 |
| 14 | Aspartate | 122 | 111 |
| 15 | Glutamate | 41 | 41 |
| 16 | Arginine | 87 | 77 |
| 14 | Proline | 14 | 12 |
| 17 | Tryptophan | 2 | 2 |
| 18 | Lysine | 12 | 11 |
Strains 30629c (gap1Δ ura3) and 30633c (gap1Δ agp1Δ ura3) were grown on minimal urea (1%) medium supplemented with citrulline (0.5%) to induce the synthesis of Agp1. 14C-labeled amino acids were added at a 100 μM final concentration, and their initial uptake rates were determined in 2- or 4-min kinetic experiments.
Nonexpression of the AGP1 gene in the ssy1Δ mutant is not due to inducer exclusion.
As the Ssy1p protein is a member of the amino acid permease family, one might argue that noninduction of AGP1 in the ssy1Δ strain is due to insufficient incorporation of the inducing amino acids. To test this possibility, we grew the wild type and the ssy1Δ strain, both harboring the AGP1-lacZ fusion gene, on minimal urea medium. After addition of 14C-labeled leucine, isoleucine, tyrosine, or phenylalanine, culture samples were withdrawn at intervals and used in parallel experiments to assay β-galactosidase activity and the incorporation of 14C-labeled amino acids (Fig. 5). AGP1-lacZ was induced in response to each amino acid tested, and this induction was abolished in the ssy1Δ strain. The ssy1Δ strain incorporated all four amino acids at a high rate, so noninduction of the AGP1-lacZ gene in the ssy1Δ strain is not due to inducer exclusion. In fact, the ssy1Δ strain even accumulated greater amounts of 14C-labeled amino acids than did the wild type, an effect not further investigated here. Whatever might cause this effect, it is clear that noninduction of the AGP1 gene in the ssy1Δ mutant is not due to reduced uptake of amino acid inducers. Rather, Ssy1p behaves like a regulatory factor essential to transcriptional induction of the AGP1 gene in response to multiple amino acids.
FIG. 5.
Noninduction of the AGP1 gene in the ssy1Δ mutant is not due to inducer exclusion. The time course of accumulation of 14C-labeled amino acids (initial concentration, 0.1 mM) (solid symbols) and of the increase in β-galactosidase activity (open symbols) in strains 32501a (ura3) (squares) and 32501c (ssy1Δ ura3) (circles) transformed with the YCpAGP1-lacZ plasmid and initially growing on minimal medium with urea as the sole nitrogen source is shown.
Ssy1p mediates induction of the AGP1 gene in response to external amino acids.
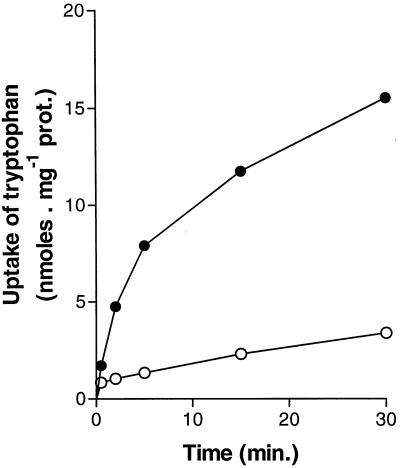
In a next set of experiments we addressed the question of whether Ssy1p-dependent expression of the AGP1 gene is induced in response to intracellular or extracellular amino acids. For comparison, we extended this analysis to the ARO9 gene inducible by aromatic amino acids. Unlike AGP1, induction of ARO9 by tryptophan is normal in the ssy1Δ mutant but is abrogated in cells deleted of ARO80, a gene encoding a transcription factor of the Cys6-Zn2 family. Conversely, induction of AGP1 by aromatic amino acids is normal in the aro80Δ mutant (36a). Thus, the presence of tryptophan leads to transcriptional induction of both the AGP1 and ARO9 genes, but the regulatory pathway involved in induction is apparently different for each gene. Expression of AGP1-lacZ and ARO9-lacZ was assayed in a trp2fbr mutant, in which anthranilate synthase (Trp2p) is resistant to feedback inhibition by tryptophan (51). In keeping with previous reports (51, 82), this mutant growing on urea medium was found to accumulate endogenously synthesized tryptophan to levels that were at least 70-fold higher than in the wild-type (Table 4, lines 1 and 2). Expression of AGP1-lacZ and ARO9-lacZ was also assayed in the gap1–92 agp1-1 strain, in which tryptophan is incorporated at a much lower rate than in the wild type (Fig. 6). In the gap1–92 agp1-1 strain growing on urea medium, the intracellular tryptophan pool is as low as in the wild type; by 90 min after the addition of tryptophan (5 mM), it was about the same as in the trp2fbr mutant growing on urea medium (Table 4, line 3). The results of β-galactosidase assays show that ARO9-lacZ is induced in cells that accumulate intracellular tryptophan. In contrast, AGP1 remains insensitive to the intracellular accumulation of tryptophan (lines 1 and 2) and is specifically induced when tryptophan is added to the culture medium (line 3). This result clearly shows that AGP1 is induced in response to extracellular rather than intracellular tryptophan. The role of Ssy1p is likely to detect the external amino acid and to activate a signal transduction pathway leading ultimately to transcriptional induction of the AGP1 gene.
TABLE 4.
Transcription of AGP1 is induced in response to external amino acidsa
| Line | Strain | Nitrogen source | Intracellular tryptophan (nmol · mg−1 [dry wt]) | β-Galactosidase activity (nmol · min−1 · mg of protein−1)
|
|
|---|---|---|---|---|---|
| AGP1-lacZ | ARO9-lacZ | ||||
| 1 | Wild type | Urea | 0.15 | ≤2 | ≤2 |
| 2 | trp2fbr | Urea | 10 | ≤2 | 93 |
| 3 | gap1-92 agp1-1 | Urea | 0.09 | ≤2 | ≤2 |
| Urea plus Trp (90 min) | 12 | 560 | 80 | ||
Strains 23346c (ura3), RE1 (trp2fbr ura3), and 30622a (gap1-92 agp1-1 ura3) harboring the CEN-based plasmids YCpAGP1-lacZ or YCpARO9-lacZ were grown on minimal medium with urea (5 mM) as the sole nitrogen source. Tryptophan was added at a 5 mM final concentration where indicated. The reported β-galactosidase activities are means of two independent experiments. The values of intracellular tryptophan concentrations are means of three experiments.
FIG. 6.
The gap1-92 agp1-1 strain is largely defective in tryptophan transport. The time course of 14C-labeled tryptophan accumulation (initial external concentration, 5 mM) in strains 23344c (ura3) (•) and 30622a (gap1-92 agp1-1 ura3) (○) initially growing on minimal medium with urea (5 mM) as the sole nitrogen source is shown.
Induction of AGP1 requires Grr1p.
The Snf3p and Rgt2p proteins of the sugar transporter family act as sensors of external glucose concentration, sensors that can generate intracellular signals leading to glucose-regulated transcription (55, 67). The Grr1p protein plays a central role in the transduction of this glucose signal. This is shown, for instance, by the fact that grr1 mutations relieve repression of many glucose-repressible genes (8, 28) and prevent induction by glucose of several HXT genes encoding glucose transporters (68). Grr1p is also involved in regulating the cell cycle, as it is required for degradation of the G1 cyclins Cln1p and Cln2p (11). Grr1p is in fact a component of a ubiquitin-protein ligase complex (SCFGrr1) (52) possibly involved in coupling nutrient availability to gene expression and cell cycle regulation (11, 54). In addition to impaired glucose signaling, grr1 mutants display a number of other defects, including reduced uptake of aromatic amino acids (28) and leucine (69). These observations prompted us to test the role of Grr1p in Ssy1p-mediated induction of the AGP1 gene. A mutant strain with a complete deletion of GRR1 was isolated and used to monitor expression of the AGP1-lacZ gene. Induction of AGP1-lacZ by amino acids was abolished in the grr1Δ strain (Table 5). Hence, Grr1p is essential to the transduction of signals generated not only by the Snf3p and Rgt2p glucose sensors, but also by the Ssy1p amino acid sensor.
TABLE 5.
Grr1p and Uga35p are essential to the transcriptional induction of the AGP1 genea
| Line no. | Nitrogen sources |
AGP1-lacZ β-galactosidase activity (nmol · min−1 · mg of protein−1)
|
||
|---|---|---|---|---|
| Wild type | grr1Δ | uga35Δ | ||
| 1 | Urea | ≤2 | ≤2 | |
| 2 | Urea + Leu | 638 | ≤2 | |
| 3 | Urea + IIe | 895 | ≤2 | |
| 4 | Urea + Phe | 960 | ≤2 | |
| 5 | Urea + Tyr | 724 | ≤2 | |
| 6 | Urea + Met | 744 | ≤2 | |
| 7 | Urea + citrulline | 284 | ≤2 | |
| 8 | Proline | ≤2 | ≤2 | |
| 9 | Proline + Leu | 839 | 18 | |
| 10 | Proline + IIe | 1,227 | 8 | |
| 11 | Proline + Phe | 1,343 | 10 | |
| 12 | Proline + Tyr | 791 | 5 | |
| 13 | Proline + Met | 950 | 11 | |
| 14 | Proline + citrulline | 235 | ≤2 | |
Strains 23344c (ura3), JA115 (grr1Δ ura3) and CD17 (uga35Δ ura3) transformed by the CEN-based YCpAGP1-lacZ plasmid were grown on minimal medium with the indicated compounds as the sole nitrogen source(s), each added at a 5 mM concentration. The uga35Δ was grown on proline instead of urea medium because of its inability to use urea as the sole nitrogen source. The reported β-galactosidase activities are means of two to three independent experiments. Variations were less than 20%.
Induction of AGP1 requires Uga35p, a transcription factor of the Cys6-Zn2 family.
The UGA35(DAL81/DURL) gene encodes a transcription factor of the Cys6-Zn2 family, which is required for full induction of several nitrogen utilization genes (16, 21). These include the GABA-inducible genes involved in GABA utilization (UGA1, UGA2, and UGA4) (84, 85) and the allophanate-inducible genes involved in urea (DUR1-2 and DUR3), allantoate (DAL7), and arginine (CAR2) utilization (34, 40, 80). Induction of these genes requires a second transcription factor which, unlike Uga35p, is inducer specific (85). In the case of the GABA-inducible genes, the second factor is Uga3p (2); it is DurMp(Dal82p) in the case of allophanate-inducible genes (5, 40). In previous experiments, we observed that the uga35 mutant displays resistance to β-2-thienylalanine (unpublished data). Although this resistance was not as pronounced as for the ssy1Δ mutant, we tested the role of Uga35p in the induction of AGP1 (Table 5). The results clearly show that the induction of AGP1-lacZ is dramatically reduced in a uga35Δ mutant. It is unaltered, however, in uga3 and durM mutants (data not shown). These results reinforce the previous conclusion that Uga35p is a pleiotropic factor involved in several transcriptional induction pathways (85). Uga35p does not seem, however, to specifically mediate Ssy1p-dependent induction, since it is also essential to allophanate-induced transcription (40, 85). Furthermore, Uga35p is required for induction of the UGA4 gene by GABA (84), a regulation unaltered in the ssy1Δ mutant (data not shown).
Ssy1p is required for induced transcription of at least five additional amino acid permease genes.
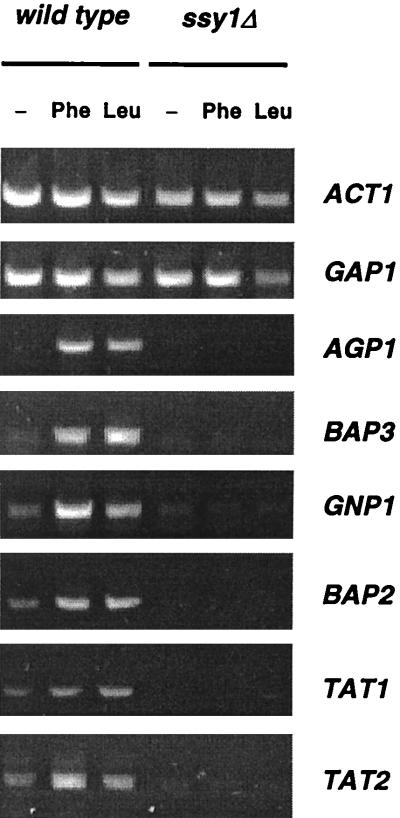
To find additional genes that might display Ssy1p-dependent induction by amino acids, we purified total RNA from wild-type and ssy1Δ strains grown in the presence or absence of amino acids, treated it with DNase, and used it in RT-PCR experiments (see Materials and Methods). This approach enabled us to rapidly estimate the relative amounts of transcripts of several genes of the amino acid permease family. These included the inducible BAP2 gene proposed to encode a branched-chain amino acid permease (31), the BAP3(PAP1) gene encoding a close homologue of Bap2p (60), and several genes encoding probable amino acid permeases of unknown substrate specificity, namely, ALP1 (78), YFL055w (63), YPL274w (17), YBR132c (27), and YLL061w (44). Some previously characterized genes, such as MUP1 and MUP3 (38), TAT1 and TAT2 (9, 75), and GNP1 (90), were also included in this analysis because the reported experiments on expression of these genes were carried out in complex medium or in minimal medium containing amino acids to compensate for amino acid auxotrophies, i.e., under conditions where the expression of Ssy1p-responsive genes should appear constitutive. In the RT-PCR experiments (Fig. 7), the amplified signal corresponding to the AGP1 gene was undetectable in urea-grown cells but was very strong when phenylalanine or leucine was added to the cultures 90 min before they were harvested. As expected, the signal remained undetectable in the ssy1Δ mutant after amino acid addition. The BAP3, TAT1, TAT2, BAP2, and GNP1 genes also displayed increased expression upon addition of leucine or phenylalanine, whereas their expression was barely detectable in the ssy1Δ mutant. Expression of ALP1, YFL055w, YPL274w, YBR132c, YLL061w, MUP1, and MUP3 did not appear significantly different in wild-type and ssy1Δ cells (data not shown). As expected, the signal corresponding to the GAP1 gene was roughly constant under all of the tested conditions. Although the results of these RT-PCR experiments must be confirmed by more-quantitative methods, they clearly show that several genes encoding amino acid permeases are under the positive control of the Ssy1p amino acid sensor.
FIG. 7.
The Ssy1p amino acid sensor affects expression of multiple amino acid permease genes. The results of RT-PCR analysis of the total RNA from strains 23344c (ura3) and 32501c (ssy1Δ ura3), with oligonucleotide primers specific to the actin gene (ACT1) and to several genes encoding amino acid permeases (see Materials and Methods), are shown. The cells were grown on minimal urea medium. At time zero, phenylalanine (5 mM) (Phe) or leucine (5 mM) (Leu) was added to part of the cultures, and the cells were collected after 90 min.
DISCUSSION
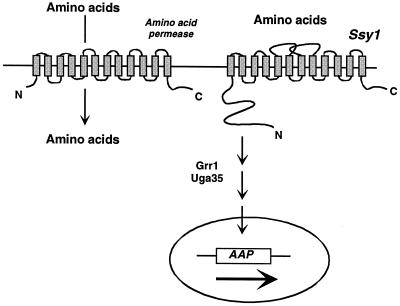
In this study, we report the characterization in S. cerevisiae of Agp1p, a new broad-specificity amino acid permease. We show that transcription of the corresponding gene is induced in response to extracellular amino acids in a manner dependent on (i) an amino acid permease homologue acting as a sensor (Ssy1p), (ii) an F-box protein involved in glucose signaling and cell cycle control (Grr1p), and (iii) a transcription factor of the Cys6-Zn2 family involved in other nitrogen induction pathways (Uga35p) (Fig. 8). Furthermore, we provide evidence that this new nutritional signaling pathway influences the transcription of at least five other amino acid permease genes.
FIG. 8.
Schematic presentation of the role of the permease-like amino acid sensor Ssy1p in the transcriptional regulation of amino acid permease genes in S. cerevisiae.
Agp1p, a broad-specificity amino acid permease in S. cerevisiae.
Agp1p has the properties of a wide-specificity amino acid permease. Synthesis of this permease is associated with more-rapid uptake of many amino acids, including leucine, isoleucine, valine, threonine, phenylalanine, tyrosine, serine, methionine, alanine, glutamine, histidine, asparagine, and glycine (Table 3). Furthermore, growth tests suggest that Agp1p can also import tryptophan if present at a sufficiently high concentration. Agp1p appears as a relatively low-affinity permease. For instance, the Km values for isoleucine, leucine, and tyrosine transport are in the 10−4 molar range and those for the transport of other amino acids, such as tryptophan, are probably much higher. Growth tests illustrate the importance of Agp1p in amino acid utilization: together with Gap1p, Agp1p is the main amino acid permease ensuring growth on isoleucine, leucine, phenylalanine, tyrosine, and tryptophan as the sole nitrogen source and also contributes significantly to the utilization of valine, methionine, and threonine (1 mM). The absence of any clear growth defect of the gap1 agp1 strain when grown on higher concentrations of some of these amino acids (10 mM) or on other substrates of Agp1p such as glutamine, asparagine, serine, and alanine is likely due to the activity of other amino acid permeases which can compensate for the lack of Agp1p. Interestingly, our growth tests failed to show any contribution of Bap2p, defined as the major branched-chain amino acid permease (31), in the utilization of leucine and isoleucine (1 mM) as the sole nitrogen source (Fig. 3). Similarly, Tat1p, defined as a major tyrosine permease in yeast cells (75), is unable to ensure tyrosine utilization at a 1 to 10 mM final concentration (Fig. 3). These observations raise questions as to the actual physiological function of these permeases.
As this paper was being written, it was reported by Schreve et al. (76) that the primary substrates of Agp1p are asparagine (Km = 0.29 mM) and glutamine (Km = 0.79 mM), and the permease was named Agp1p to reflect this substrate specificity. It was further suggested that Agp1p mediates the uptake of other amino acids but that the affinity of the permease for these amino acids is lower than for asparagine and glutamine (76). These data are hardly consistent with the Km values of Agp1p for leucine (0.16 mM), isoleucine (0.6 mM), and phenylalanine (0.6 mM) transport. Rather, the affinity of Agp1p appears to be in the same range for many amino acids. Schreve et al. also reported that the pattern of AGP1 expression according to the nitrogen source is very similar to that of the GAP1 gene: the reason why the induction by amino acids was not revealed in these experiments is probably because the leucine was systematically added to the growth medium in order to compensate for a leu2 auxotrophy.
Agp1p also mediates methionine uptake (Table 3). It was previously reported that methionine is transported in yeast cells by at least three different permeases: the high-affinity permease (Km = 0.013 mM) encoded by the MUP1 gene, a low-affinity permease (Km = 0.2 mM) whose gene (MUP2) remained uncharacterized, and a very-low-affinity permease (Km = 1 mM) encoded by the MUP3 gene (38). The low-affinity permease is probably Agp1p: its activity was measured under growth conditions leading to the induction of AGP1, it displays a broad specificity range, and methionine transport mediated by this permease is inhibited by leucine with an apparent Ki value (0.30 mM) (38) very close to the Km value of Agp1 for leucine.
AGP1 is induced by multiple amino acids: role of a permease-like sensor of external amino acids.
Transcription of the AGP1 gene is induced by many amino acids, a notable exception being proline (Table 2). Induction levels vary considerably according to the amino acid tested, and most inducing amino acids are also substrates of the Agp1p permease. Induction of AGP1 is abolished in mutants lacking Ssy1p, a protein of the amino acid permease family acting as a sensor of extracellular amino acids. The first evidence for a role of this protein in amino acid sensing was recently provided (24). It was found that induction by leucine (0.23 mM) of the BAP2, BAP3, and TAT1 genes encoding amino acid permeases (31, 60, 75) and of the PTR2 gene encoding a peptide transporter (71) is abolished in the ssy1 strain. Induction of BAP2 by l- or d-leucine still occurs in a mutant strain largely deficient in l- and d-leucine uptake, leading to the suggestion that Ssy1p acts as a sensor of external leucine (24). Using RT-PCR, we have confirmed that expression of BAP2, TAT1, and BAP3 is under the positive control of Ssy1p and found that the same is true of AGP1, GNP1 encoding a glutamine permease (90), and TAT2 encoding a tryptophan permease (75). These genes seem, in fact, to be induced by other amino acids besides leucine (Table 2; Fig. 7, and unpublished data); this means that Ssy1p is probably involved in the transcriptional induction of several permease genes in response to various amino acids. Our data on the AGP1 gene show that permease genes under the control of Ssy1p are transcriptionally induced in response to extracellular rather than intracellular amino acids. For instance, AGP1 remains unexpressed in a trp2fbr mutant endogenously accumulating tryptophan to relatively high levels. It is, however, markedly induced after the addition of tryptophan to a mutant strain largely defective in tryptophan uptake even though the intracellular tryptophan pool is about the same as in the trp2fbr strain failing to induce AGP1. We thus propose that the Ssy1p permease homologue detects external amino acids and activates, in turn, a transduction pathway leading ultimately to transcriptional activation of several permease genes.
The Ssy1p protein displays structural features that clearly distinguish it from Agp1p, Gap1p, and the other members of the amino acid permease family. In particular, its hydrophilic N terminus is much larger, as are several regions connecting TM domains and predicted to be extracellular (Fig. 2). These features are reminiscent of those displayed by the Snf3p and Rgt2p glucose sensors, which differ from the other members of the sugar transporter family by their unusually long cytoplasmic C-terminal domain (55, 66, 67). These domains have been shown to be essential to the role of Snf3p and Rgt2p as glucose sensors (66). Furthermore, grafting these domains onto the C terminus of Hxt1p confers to this glucose transporter the properties of a glucose sensor (66). The large N-terminal domain of Ssy1p likely plays an important role in generating the amino acid signal and could, for instance, mediate interaction with another protein. A candidate protein is the PTR3 gene product (10), a hydrophilic 76-kDa protein originally discovered as a factor essential to the induction of the PTR2 gene in response to amino acids (37). Mutants affected in this gene were subsequently and independently isolated as shr6 (49), ssy3 (46), and apf3 mutants (13a). Although the exact function of Ptr3p remains undetermined, the phenotypes of the ptr3/apf3/ssy3/shr6 and ssy1 mutants appear indistinguishable (13a, 46, 49).
The Ssy1p amino acid sensor might activate a signal transduction pathway upon binding of amino acids without translocating them across the plasma membrane. As such, Ssy1p would act as a receptor. Alternatively, Ssy1p-mediated transport of amino acids could be essential to the protein’s signaling function. This hypothesis, however, implies that the transport capacity of Ssy1p should be very low, since a gap1Δ agp1Δ SSY1+ strain is unable to grow on several amino acids as the sole nitrogen source (1 mM) (Fig. 3), even though Ssy1p effectively transmits signals in response to these amino acids.
Grr1p: an F-box protein involved in glucose signaling, amino acid signaling, and cell cycle control.
The Grr1p protein plays a central role in transducing signals generated by the Snf3p and Rgt2p glucose sensors (54) and in degrading G1 cyclins (11). This F-box protein is a component of a so-called SCF complex (for Skp1–Cullin–F-box) belonging to a novel class of ubiquitin-protein ligases (E3) that probably exist in organisms ranging from yeast to humans (26, 52). A typical SCF complex consists of (i) Cdc53p, a protein functioning as a scaffold subunit and belonging to a large, evolutionarily conserved family of proteins called cullins (62, 70, 88); (ii) an F-box protein, involved in selecting which potential targets are to be ubiquitinated (70, 77); and (iii) Skp1p, which forms a bridge between Cdc53p and one of several F-box proteins (Grr1p, Cdc4p, and Met30p) (7). SCF complexes function in combination with an E2 enzyme catalyzing transfer of ubiquitin to the target protein. For instance, an SCF complex containing Grr1p as the F-box protein promotes ubiquitination of G1 cyclins in conjunction with the E2 enzyme Cdc34p (22, 81, 89). On the other hand, transduction of the glucose signal generated by Snf3p leading to induction of the HXT1 gene requires Grr1p, Cdc53p, and Skp1p, but Cdc34p is not needed, suggesting that another E2 enzyme is involved (54).
In this study, we show that Grr1p is also essential to Ssy1p-mediated induction of the AGP1 gene, thus extending the role of this F-box protein to an additional regulatory pathway. It seems reasonable, therefore, to speculate that transduction of the signal generated by the Ssy1p sensor involves an SCF complex with Grr1p as the F-box–protein subunit. The complex could be involved, for instance, in ubiquitinating a downregulator of Ssy1p-regulated genes in response to external amino acids. Regarding possible targets of this putative SCFGrr1 complex, it is noteworthy that PTR2, a peptide transporter gene whose induction by amino acids is Ssy1p dependent (24), is under the negative control of Cup9p (50), a short-lived homeodomain protein (t1/2, ∼5 min) whose degradation involves the ubiquitin pathway (18). The Ptr1p-Ubr1p protein proposed to act as a ubiquitin-protein ligase (83) is essential both to Cup9p degradation (18) and to PTR2 induction (1, 37). Furthermore, the E2 enzymes encoded by the UBC2 and UBC4 genes appear essential to Cup9 degradation (18). We are currently conducting experiments to test the roles of Cup9p, Ptr1p-Ubr1p, Ubc2p, Ubc4p and of the SCF complex components Skp1p, Cdc53p, and Cdc34p in the expression of amino acid and peptide permease genes under Ssy1p control.
ACKNOWLEDGMENTS
We gratefully acknowledge the excellent technical assistance of Catherine Jauniaux. We are also grateful to Michel Hanocq, Jacques Dubois, Bart Scherens, and Mohamed El Bakkoury for their help in measuring intracellular tryptophan pools. We also thank Anne-Marie Marini for fruitful discussions and for critical reading of the manuscript.
This work was supported by the following contracts: The Commission of the European Communities (EUROFAN, BIO4-CT95-0080), The Fund for Medical Scientific Research (Belgium, FRSM 3.4602.94), The International Brachet Stiftung (grant GR97/9-02), and the Communauté Française de Belgique, Direction de la Recherche Scientifique, Actions de Recherche Concertées. I.I. is a recipient of a predoctoral fellowship from the Communauté Française de Belgique and from the Fondation Universitaire David et Alice Van Buuren (Belgium). J.-O.D. and F.B. are recipients of FRIA (Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture) predoctoral fellowships.
REFERENCES
- 1.Alagramam K, Naider F, Becker J M. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. doi: 10.1111/j.1365-2958.1995.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 2.André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990;220:269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- 3.André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 4.André B, Hein C, Grenson M, Jauniaux J C. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:17–25. doi: 10.1007/BF00282779. [DOI] [PubMed] [Google Scholar]
- 5.André B, Jauniaux J C. Nucleotide sequence of the DURM gene coding for a positive regulator of allophanate-inducible genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:7136. doi: 10.1093/nar/18.23.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 7.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 8.Bailey R B, Woodward A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- 9.Bajmoczi M, Sneve M, Eide D J, Drewes L R. TAT1 encodes a low-affinity histidine transporter in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;243:205–209. doi: 10.1006/bbrc.1998.8082. [DOI] [PubMed] [Google Scholar]
- 10.Barnes D, Lai W, Breslav M, Naider F, Becker J M. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1998;29:297–310. doi: 10.1046/j.1365-2958.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- 11.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 12.Béchet J, Grenson M, Wiame J M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 13.Bennetzen J L, Hall B D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982;257:3018–3025. [PubMed] [Google Scholar]
- 13a.Bernard, F., and B. André. Unpublished observations.
- 14.Bisson L F, Coons D M, Kruckeberg A L, Lewis D A. Yeast sugar transporters. Crit Rev Biochem Mol Biol. 1993;28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- 15.Boles E, Hollenberg C P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 16.Bricmont P A, Daugherty J R, Cooper T G. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1161–1166. doi: 10.1128/mcb.11.2.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussey H, Storms R K, Ahmed A, Albermann K, Allen E, Ansorge W, Araujo R, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XVI. Nature. 1997;387:103–105. [PubMed] [Google Scholar]
- 18.Byrd C, Turner G C, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celenza J L, Marshall-Carlson L, Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci USA. 1988;85:2130–2134. doi: 10.1073/pnas.85.7.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- 21.Coornaert D, Vissers S, André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991;97:163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- 22.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didion T, Regenberg B, Jorgensen M U, Kielland-Brandt M C, Andersen H A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 25.Dubois E, Vissers S, Grenson M, Wiame J M. Glutamine and ammonia in nitrogen catabolite repression of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1977;75:233–239. doi: 10.1016/0006-291x(77)91033-6. [DOI] [PubMed] [Google Scholar]
- 26.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 27.Feldmann H, Aigle M, Aljinovic G, André B, Baclet M C, Barthe C, Baur A, Becam A M, Biteau N, et al. Complete DNA sequence of yeast chromosome II. EMBO J. 1994;13:5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnier P, Grosclaude J M, Goudey-Perriere F, Gervat V, Gayral P, Jacquot C, Perriere C. Presence of norepinephrine and other biogenic amines in stonefish venom. J Chromatogr B. 1996;685:364–369. doi: 10.1016/s0378-4347(96)00203-4. [DOI] [PubMed] [Google Scholar]
- 30.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grauslund M, Didion T, Kielland-Brandt M C, Andersen H A. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1269:275–280. doi: 10.1016/0167-4889(95)00138-8. [DOI] [PubMed] [Google Scholar]
- 32.Grenson M. Amino acid transporters in yeast: structure, function and regulation. In: De Pont J J L L L M, editor. Molecular aspects of transport proteins. New York, N.Y: Elsevier Sscience; 1992. pp. 219–245. [Google Scholar]
- 33.Grenson M, Mousset M, Wiame J M, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. Biochim Biophys Acta. 1966;127:339–346. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- 34.Hennaut C. l-Ornithine transaminase synthesis in Saccharomyces cerevisiae. Induction by allophanate, intermediate and inducer of the urea degradative pathway, adds to arginine induction. Curr Genet. 1981;4:69–72. doi: 10.1007/BF00376788. [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch A G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. New York, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 319–414. [Google Scholar]
- 36.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- 36a.Iraqui, I., S. Vissers, B. André, and A. Urrestarazu. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 37.Island M D, Perry J R, Naider F, Becker J M. Isolation and characterization of S. cerevisiae mutants deficient in amino acid-inducible peptide transport. Curr Genet. 1991;20:457–463. doi: 10.1007/BF00334772. [DOI] [PubMed] [Google Scholar]
- 38.Isnard A D, Thomas D, Surdin-Kerjan Y. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J Mol Biol. 1996;262:473–484. doi: 10.1006/jmbi.1996.0529. [DOI] [PubMed] [Google Scholar]
- 39.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs E, Dubois E, Wiame J M. Regulation of urea amidolyase synthesis in Saccharomyces cerevisiae, RNA analysis, and cloning of the positive regulatory gene DURM. Curr Genet. 1985;9:333–339. doi: 10.1007/BF00421602. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs P, Jauniaux J C, Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980;139:691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- 42.Jacq C, Alt-Morbe J, André B, Arnold W, Bahr A, Ballesta J P, Bargues M, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature. 1997;387:75–78. [PubMed] [Google Scholar]
- 43.Jauniaux J C, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- 44.Johnston M, Hillier L, Riles L, Albermann K, André B, Ansorge W, Benes V, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature. 1997;387:87–90. [PMC free article] [PubMed] [Google Scholar]
- 45.Jones E W, Fink G R. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 181–299. [Google Scholar]
- 46.Jorgensen M U, Bruun M B, Didion T, Kielland-Brandt M C. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 47.Kadner R J, Island M D, Dahl J L, Webber C A. A transmembrane signalling complex controls transcription of the Uhp sugar phosphate transport system. Res Microbiol. 1994;145:381–387. doi: 10.1016/0923-2508(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 48.Kadner R J, Webber C A, Island M D. The family of organo-phosphate transport proteins includes a transmembrane regulatory protein. J Bioenerg Biomembr. 1993;25:637–645. doi: 10.1007/BF00770251. [DOI] [PubMed] [Google Scholar]
- 49.Klasson H, Ljungdahl P O. 15th Small Meeting on Yeast Transport and Energetics, Mexico. 1997. SHR5 and SHR6 participate in extracellular amino acid sensing; p. 12. [Google Scholar]
- 50.Knight S A, Tamai K T, Kosman D J, Thiele D J. Identification and analysis of a Saccharomyces cerevisiae copper homeostatis gene encoding a homeodomain protein. Mol Cell Biol. 1994;14:7792–7804. doi: 10.1128/mcb.14.12.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kradolfer P, Niederberger P, Hutter R. Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases. Arch Microbiol. 1982;133:242–248. doi: 10.1007/BF00415010. [DOI] [PubMed] [Google Scholar]
- 52.Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 53.Kruckeberg A L. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 54.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenz M C, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurements with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 58.Madi L, McBride S A, Bailey L A, Ebbole D J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics. 1997;146:499–508. doi: 10.1093/genetics/146.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 283–317. [Google Scholar]
- 60.Mai B, Lipp M. Cloning and chromosomal organization of a gene encoding a putative amino-acid permease from Saccharomyces cerevisiae. Gene. 1994;143:129–133. doi: 10.1016/0378-1119(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 61.Marini A M, Soussi-Boudekou S, Vissers S, André B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathias N, Johnson S L, Winey M, Adams A E, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S, Sasanuma M, et al. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- 64.Nehls U, Wiese J, Guttenberger M, Hampp R. Carbon allocation in ectomycorrhizas: identification and expression analysis of an Amanita muscaria monosaccharide transporter. Mol Plant-Microbe Interact. 1998;11:167–176. doi: 10.1094/MPMI.1998.11.3.167. [DOI] [PubMed] [Google Scholar]
- 65.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 66.Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozcan S, Dover J, Rosenwald A G, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozcan S, Schulte F, Freidel K, Weber A, Ciriacy M. Glucose uptake and metabolism in grr1/cat80 mutants of Saccharomyces cerevisiae. Eur J Biochem. 1994;224:605–611. doi: 10.1111/j.1432-1033.1994.00605.x. [DOI] [PubMed] [Google Scholar]
- 70.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry J R, Basrai M A, Steiner H Y, Naider F, Becker J M. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol Cell Biol. 1994;14:104–115. doi: 10.1128/mcb.14.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 73.Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- 74.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 75.Schmidt A, Hall M N, Koller A. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol. 1994;14:6597–6606. doi: 10.1128/mcb.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schreve J L, Sin J K, Garrett J M. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J Bacteriol. 1998;180:2556–2559. doi: 10.1128/jb.180.9.2556-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 78.Sychrova H, Chevallier M R. APL1, a yeast gene encoding a putative permease for basic amino acids. Yeast. 1994;10:653–657. doi: 10.1002/yea.320100509. [DOI] [PubMed] [Google Scholar]
- 79.Tanaka J, Fink G R. The histidine permease gene (HIP1) of Saccharomyces cerevisiae. Gene. 1985;38:205–214. doi: 10.1016/0378-1119(85)90219-7. [DOI] [PubMed] [Google Scholar]
- 80.Turoscy V, Cooper T G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982;151:1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vang N H. Régulation de la biosynthèse du tryptophane chez la levure Saccharomyces cerevisiae. Ph.D. thesis. Brussels, Belgium: Université Libre de Bruxelles; 1976. [Google Scholar]
- 83.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vissers S, André B, Muyldermans F, Grenson M. Positive and negative regulatory elements control the expression of the UGA4 gene coding for the inducible 4-aminobutyric-acid-specific permease in Saccharomyces cerevisiae. Eur J Biochem. 1989;181:357–361. doi: 10.1111/j.1432-1033.1989.tb14732.x. [DOI] [PubMed] [Google Scholar]
- 85.Vissers S, André B, Muyldermans F, Grenson M. Induction of the 4-aminobutyrate and urea catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur J Biochem. 1990;187:611–616. doi: 10.1111/j.1432-1033.1990.tb15344.x. [DOI] [PubMed] [Google Scholar]
- 86.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 87.Wiame J M, Grenson M, Arst H N J. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv Microbiol Physiol. 1985;26:1–87. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- 88.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 89.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu X, Garrett J, Schreve J, Michaeli T. GNP1, the high-affinity glutamine permease of S. cerevisiae. Curr Genet. 1996;30:107–114. doi: 10.1007/s002940050108. [DOI] [PubMed] [Google Scholar]