Abstract
Background
Tracking the emergence, introduction and spread of SARS-CoV-2 variants of concern are essential for informing public health strategies. In 2021, Cambodia faced two major epidemic waves of SARS-CoV-2 triggered by the successive rise of the Alpha and Delta variants.
Methods
Phylodynamic analysis of 1,163 complete SARS-CoV-2 genomes from Cambodia, along with global sequences, were conducted between February and September 2021 to infer viral introductions, molecular epidemiology and population dynamics. The relationship between epidemic trends and control strategies were evaluated. Bayesian phylogeographic reconstruction was employed to estimate and contrast the spatiotemporal dynamics of the Alpha and Delta variants over time.
Results
Here we reveal that the Alpha variant displays rapid lineage diversification, accompanied by the acquisition of a spike E484K mutation that coincides with the national implementation of mass COVID-19 vaccination. Despite nationwide control strategies and increased vaccination coverage, the Alpha variant was quickly displaced by Delta variants that exhibits a higher effective reproductive number. Phylogeographic inference indicates that the Alpha variant was introduced through south-central region of Cambodia, with strong diffusion rates from the capital of Phnom Penh to other provinces, while the Delta variant likely entered the country via the northern border provinces.
Conclusions
Continual genomic surveillance and sequencing efforts, in combination with public health strategies, play a vital role in effectively tracking and responding to the emergence, evolution and dissemination of future emerging variants.
Subject terms: SARS-CoV-2, Viral epidemiology, Epidemiology, Epidemiology
Plain language summary
Tracking how SARS-CoV-2, the virus that causes COVID-19, changes over time is important for public health. In Cambodia, there were two major COVID-19 waves in 2021, driven by the Alpha and Delta variants. We analyzed 1,163 virus samples from Cambodia, plus samples from other places, to understand how these variants spread. We found that the Alpha variant quickly spread and changed as Cambodia started a mass vaccination campaign. Despite efforts to control it, the Delta variant, which spreads more easily, soon took over. The Alpha variant likely came into Cambodia from the south, while the Delta variant probably entered from the north. Monitoring these changes helps us respond better to future outbreaks.
Su et al. dissect SARS-CoV-2 variant patterns in Cambodia. They uncover divergent evolutionary trajectories and population dynamics, along with contrasting migration patterns, between Alpha and Delta variants in 2021.
Introduction
Cambodia is a lower-to-middle income, least developed country located in the Greater Mekong Subregion of Southeast Asia. The country is a hotspot of emerging and endemic infectious diseases and highly interconnected with other parts of Asia. In January 2020, before the Public Health Emergency of International Concern (PHEIC) declaration due to the outbreak of Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2), Cambodia utilized its existing surveillance and outbreak response capacity to commence testing of and response to suspected cases of coronavirus disease 2019 (COVID-19). The first case of COVID-19 in Cambodia was detected on 27 January 2020 in a traveler from Wuhan1.
Genomic surveillance and sequencing have been a critical factor in the global response to COVID-19, being essential for tracking the evolution of variants, monitoring mutations and tracing the transmission of SARS-CoV-2 variants from regional outbreaks to global predominance2–5. In 2020, sequencing in Cambodia was primarily used for monitoring imported cases; however, it became critical for outbreak response by aiding the determination of the probable origin and transmission during limited community outbreaks6. Global sequencing efforts played a critical role in confirming that these clusters stemmed from importation and were not a result of undetected community transmission. Since the emergence of COVID-19, SARS-CoV-2 has undergone rapid diversification, giving rise to a succession of variant strains with enhanced transmissibility that became predominant across different regions.
One such Variant of Concern (VoC) B.1.1.7, also known as Alpha, first emerged in the United Kingdom (UK) in September 20207. The B.1.1.7 variant was characterized by numerous unique mutations, including six key mutations in the spike protein: the 69/70 deletion, the 144Y deletion, N501Y, A570D, D614G, and P681H8. It was estimated to be 30–80% more transmissible than Wuhan-lineage wild-type SARS-CoV-28–10. Another VoC that posed major challenges was B.1.617.2 also known as Delta, first detected in India in late 2020. The Delta variant contained several key mutations in the spike protein such as L452R, T478K, and P681R, which increased binding affinity to the human ACE2 receptor11–13. Additionally, mutations like T478K and P681R potentially helped the virus evade neutralizing antibodies from previous infection and vaccination14–16. As a result, the Delta variant was estimated to be around 40–60% more transmissible than Alpha14,17,18. Furthermore, Delta was associated with higher viral loads, hospitalization rates and secondary attack rates compared to Alpha and ancestral strains19–22. These features facilitated the rapid spread and takeover of Delta leading to replacement of previously dominant variants and explosive outbreaks globally17,20.
Here, we employ an efficient real-time PCR and whole-genome sequencing analytical pipeline to detect and obtain complete genomes of SARS-CoV-2 variants during two successive large-scale outbreaks in Cambodia in 2021. We use phylodynamic inference and phylogeographic reconstruction to investigate the evolutionary trajectories and spatiotemporal patterns of Alpha and Delta variants following their importation into Cambodia. We demonstrate the correlation between changes in virus populations and the implementation of vaccinations and other public health measures in Cambodia. Our findings uncover divergent evolutionary trajectories and population dynamics, along with contrasting migration patterns, between Alpha and Delta variants throughout the course of the COVID-19 pandemic in Cambodia. This work highlights the importance of ongoing genomic surveillance and the critical role of strategic public health measures and vaccinations in monitoring the genetic diversity and spread of emerging variants, as well as in mitigating larger disease outbreaks.
Methods
Ethics statement
All SARS-CoV-2 sequences and accompanying information (e.g., collection date, location) included in this analysis were obtained as part of the national outbreak response and routine first-line testing of suspected cases. All patient samples were de-identified prior to analysis, and no other individual-specific information was utilized. As the study involved de-identified data and samples collected as part of a public health response, explicit patient consent was not required. Additionally, formal ethics approval was not sought, as the work fell under the remit of national public health surveillance activities, where individual consent and ethics approval are typically not required for anonymized, aggregate data used for outbreak investigations.
All data used in this manuscript, including genetic sequences and daily case numbers, are publicly available and can be accessed through relevant public databases. For additional context related to outbreaks and vaccination efforts in Cambodia, relevant data can also be found in public media sources.
COVID-19 reporting, vaccination, and mobility data
Official COVID-19 notification and vaccination data for Cambodia was downloaded from Our World in Data23. Mobility data was downloaded from Google COVID-19 Community Mobility Reports (https://www.google.com/covid19/mobility). The data used in this study spans the period from 1 January to 30 September 2021. Due to changes in the Cambodian government’s testing strategy from 1 October 2021 onwards, geographic and demographic representation of samples was significantly reduced and virus genomes from this date onward were not included in this study.
Sample collection, metadata, RNA extraction, and initial testing of SARS-CoV-2
Between 1 January 2021 and 30 September 2021, paired nasopharyngeal and oropharyngeal swab samples were collected in the same tube from suspected local cases and overseas travelers as part of multisource surveillance in Cambodia. Total viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. The detection of COVID-19 cases was confirmed by RT-PCR detection methods throughout the study period24. Positive samples were then selected for whole genome sequencing based on genomic surveillance strategy or for cluster/outbreak assessment. Information on geographical location at the provincial level was collected from the sequencing database. No identifying information was used for any subjects in this study.
Genomic surveillance strategy and Variant of Concern testing
To analyze the incursion and dissemination of SARS-CoV-2 VoCs, Cambodia implemented a genomic surveillance strategy where samples were chosen based on availability of previous day’s testing and attempts were made to distribute testing and analysis across provinces and sample types. From the early pandemic until week 14 (April) of 2021, all variant determination relied on genomic sequencing6. However, starting from week 15 of 2021, Cambodia introduced the KogeneBiotech PowerChek™ SARS-CoV-2 S-gene Mutation Kit for routine genomic surveillance. This enabled a significant number of samples to be tested daily for VoCs in the country. Further confirmatory testing was conducted using individual SNP VoC-PCR tests from LGC Biosearch Technologies25. These tests confirmed the presence of several spike gene mutations that are specific to certain VoCs and/or mutations of concern detected by PowerChek. Sequencing of a subset (~15% per week) of confirmed VoC samples based on availability, viral load, and provincial distribution was conducted and identification of VoCs was determined by both Nextclade 2.30.026 and Pangolin 4.3.127.
Next-generation sequencing
Complete genomes of SARS-CoV-2 were obtained utilizing a highly multiplexed PCR tiled-amplicon approach28 using the ARTIC Network multiplex PCR primer set v3, and following the v4 protocols on Oxford Nanopore GridION/MinION technology29. Samples were multiplexed using Oxford Nanopore barcodes and run in batches on a single flow cell. Base calling was conducted with Guppy on MinKNOW software while being monitored on RAMPART in real-time. ONT reads were filtered by length (min 250 max 500) to remove chimeric reads. Consensus sequence building was carried out with the Artic v1.2.1 minion pipeline. Consensus sequences were quality checked manually using BioEdit30. Pango lineages and specific mutations were determined from consensus sequences using Nextclade 2.30.0 (https://clades.nextstrain.org/)26 and Pangolin 4.3.127 (https://pangolin.cog-uk.io/). All sequences of sufficient quality were submitted to GISAID database31 in a timely manner post sequencing to ensure data from Cambodia remained relevant for global comparisons.
Phylodynamic analysis
A total of 1163 complete genomes of SARS-CoV-2 virus from COVID-19 cases in Cambodia from February 2021 to September 2021 were available for analysis. To infer the evolutionary relationships of Cambodian genomes, we used the Nextstrain SARS-CoV-2 phylogenetic tree generation pipeline26 to reconstruct a phylogeny using 5964 genomes from Cambodia and worldwide. Virus lineage classification of SARS-CoV-2 was performed with Pangolin v.4.3.1 and Nextstrain v.2.30.0. A Nextstrain time-resolved tree was visualized using Auspice v.2.0.
To infer the past population dynamics of SARS-CoV-2 Alpha and Delta variants in Cambodia, we employed independent Bayesian coalescent-based analyses in BEAST v1.10.432. The Alpha dataset was sub-sampled proportionally from an inferred maximum likelihood tree using SMOT software33. We used linear regression analysis of root-to-tip distance in TempEst34 to check the temporal signal and remove the sequence outliers. The final datasets for the Alpha and Delta variants comprised 500 and 295 Cambodian genomes, respectively. For each variant, the changes in effective population size were inferred using the strict clock model under the HKY85 nucleotide substitution model with gamma distributed rate heterogeneity, and the Gaussian Markov Random Field (GMRF) Bayesian skyride coalescent model35 in BEAST. Four independent runs of 100 million generations, with every 10,000 generations were sampled. The effective sampling size (ESS) values of all parameters were reached above 200 and convergence of Markov Chain Monte Carlo (MCMC) runs were checked using Tracer v.1.7.236 after excluding appropriate burn-in values to ensure the ESS values have been reached for all parameters. The resulting tree and log files were summarized using LogCombiner v.1.10.4. The mean and 95% highest posterior density (HPD) interval of relative genetic diversity over time was calculated from both files using the GMRF skyride reconstruction with Tracer v.1.7.2 and visualized using in-house scripts.
The changes in effective reproductive number (denoted as Re) of the Alpha and Delta variants of SARS-CoV-2 virus in Cambodia were estimated using BEAST 2 v.2.6.637. The estimates of Re represent the average number of secondary infections caused by an infected individual in a population, which is a useful indicator for assessing the growth of the epidemic or the effectiveness of interventions38. This parameterization is analogous to dividing the “births” of new infectious individuals by the termination or “death” of their infectious status. An Re > 1 at any point of time signifies a growing epidemic, while an Re < 1 suggests recession39. To estimate the Re of SARS-CoV-2 Alpha and Delta variants in Cambodia, we employed a birth-death skyline serial (BDSKY) model as implemented in BEAST 2. For each variant, an optimized relaxed clock model with the HKY85 nucleotide substitution model was used40. The clock rate prior was set to 0.0006 sites/substitutions/year and the kappa prior set to 5.5 was applied. The reproductive number was set to 10 dimensions across the study period. Four independent runs of 300 million generations was run with a Bactrian operator schedule using BEAST 2, with sampling every 10,000 generations. Convergence of the runs were checked in Tracer and the appropriate burn-in values were removed. The resulting log files were summarized using LogCombiner v.1.10.4. The mean and 95% highest posterior density (HPD) interval of Re over time was calculated and visualized using the R package bdskytools39. Additionally, we conducted independent Bayesian Skyride and Re analyses based on Cambodian Delta Group 1 (n = 80) and 2 sequences (n = 215) separately, and their resulting plots are visualized.
Correlation analyses
To examine the relationship between daily case numbers and other parameters (effective population size, mobility data for residential and workplace/retail), pairwise Pearson correlation coefficients (r) were calculated using the R package corrplot v0.9241. Correlation analyses were conducted separately for Alpha and Delta cases in Cambodia. Significance tests were performed, with P-values < 0.05 indicating significance. Correlation coefficients greater than 0.7 were considered a very strong correlation, 0.5–0.7 a strong correlation, 0.3–0.5 as moderate correlation, and <0.3 as weak correlation.
Discrete phylogeographic analysis
To assess the spatiotemporal dynamics of SARS-CoV-2 in Cambodia, geographic locations were coded as discrete location states. The Alpha and Delta datasets consisted of 545 and 384 sequences, respectively. We coded Cambodia into nine location states, namely: Battambang, Kampong (including Kampong Cham, Kampong Chhnang, Kampong Speu, Kampong Thom), Kandal, Meanchey (Banteay Meanchey and Oddar Meanchey), Phnom Penh, PSRKM (including Preah Vihear, Stung Treng, Ratnanakiri, Kratie and Mondulkiri), Southeast Cambodia (including Tbong Kmoum, Prey Veng, Svay Rieng and Takeo), Siem Reap and Southwest Cambodia (including 4 provinces: Pursat, Koh Kong, Preah Sihanouk and Kampot). During the COVID-19 pandemic, there are two potential routes of SARS-CoV-2 introduction into Cambodia, either by international air travel from foreign countries into the capital city of Phnom Penh, or by land transport from Southeast Asian regions into the northern land borders of Cambodia. Two additional location states were coded to represent Southeast Asia and others (rest of world). For each variant, asymmetric non-zero migration rates between paired location states were inferred using the Bayesian Stochastic Search Variable Selection (BSSVS) method42 as implemented in BEAST. Four MCMC chains were run for 100 million states, sampling every 10,000 states. Ancestral trunk proportions for each location were also inferred through time using the 10,000 output trees using the program PACT v.0.9.543. To visualize the spatiotemporal history and assess the statistical significance of migration rates between location states, Bayes factor and posterior probabilities were inferred using SpreaD3 version 0.9.644. To understand the impact of sampling bias on the transmission patterns of SARS-CoV-2 within Cambodia, we randomly subsampled the datasets for the Alpha (n = 355) and Delta (n = 250) variants separately. Phylogeographic analyses were conducted using the same criteria as mentioned above. Circular migration diagrams and estimated diffusion rates between geographic locations were visualized using in-house scripts.
Results
Timeline of Alpha and Delta outbreaks
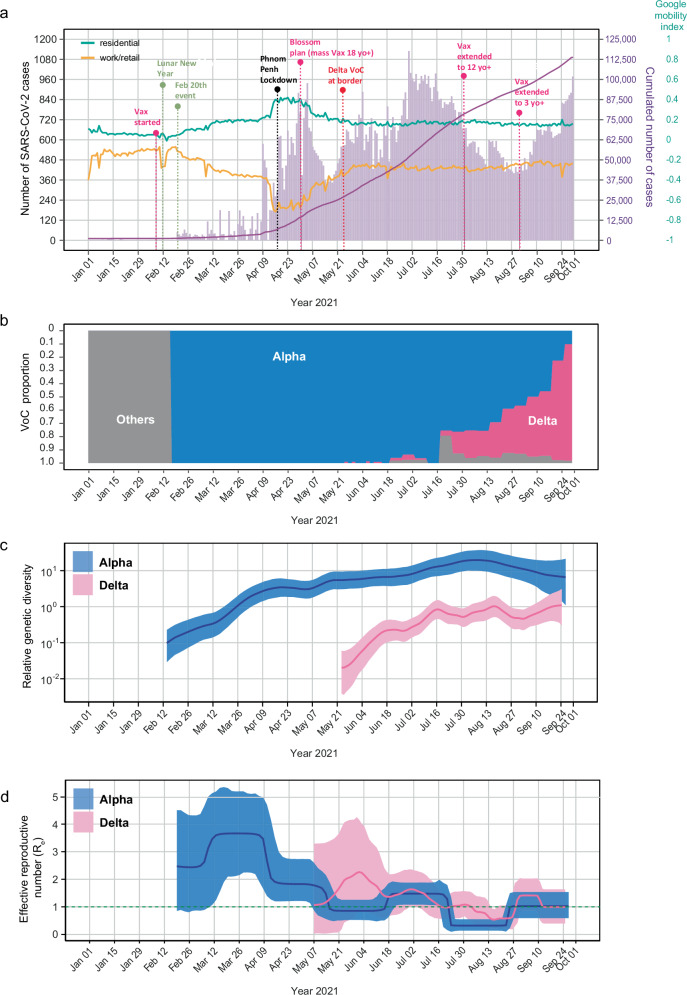
In January 2021, Cambodia detected the Alpha variant in several travelers entering the country, but these cases were effectively contained through ongoing control measures. However, during the Lunar New Year on 12 February 2021, a superspreader event occurred in Phnom Penh, and on 20 February 2021, the Cambodian Ministry of Health announced the country’s first major community outbreak. This marked the onset of the first wave of COVID-19 with a substantial surge in cases caused by Alpha variant, leading to the closure of schools. By early March 2021, over 10,000 Phnom Penh residents were placed under quarantine amidst escalating cases. As the number of cases continued to rise, a strict curfew was imposed in Phnom Penh for two weeks and non-essential businesses were closed from 1 April 2021. A subsequent 14-day lockdown (Fig. 1a) was enacted in the capital from 14 April 2021 to 28 April 2021. During this period, the Alpha variant was predominant in Cambodia, accounting for more than 65% of SARS-CoV-2 cases by July 2021 (Fig. 1b).
Fig. 1. Genomic epidemiology and population dynamics of confirmed SARS-CoV-2 cases in Cambodia from February to September 2021.
a Epidemic waves of confirmed COVID-19 cases and key control measures and events. The left y-axis represents the total number of positive cases (purple bars), while the right y-axis displays total cumulative number of cases (solid dark purple line) and local human mobility index (solid green and orange horizontal lines indicating residential and work/retail mobility, respectively). Dotted vertical lines denote key festive events and public health interventions. Abbreviations: VoC, variant of concern; Vax, COVID-19 vaccinations; yo+, years old above; b Proportion of the Alpha and Delta case numbers in Cambodia was indicated by shaded areas (blue and pink, respectively). Others denotes pre-VoC cases. c, d Relative genetic diversity and the effective reproductive number (Re) of the Alpha and Delta lineages based on Cambodian sequences. Solid lines represent median values, with the Alpha and Delta variants denoted by blue and pink lines, respectively. The shaded areas represent the corresponding 95% highest posterior density (HPD) intervals. The dotted green line represents an Re of 1. An Re greater than 1 signifies epidemic growth, while an Re less than 1 indicates recession.
Amidst the dominance of the Alpha variant, the Delta variant was first identified in Cambodia on 24 May 2021. Delta was initially detected in the northern border provinces, primarily among migrant workers returning from Thailand. In June 2021, curfews were imposed in cities close to border including Siem Reap and Poipet, and a lockdown was enforced in three areas of Oddar Meanchey Province. On 18 July 2021, Cambodia suspended border crossings with Vietnam for a month. Subsequently, on 29 July 2021, an additional 14-day lockdown was implemented in various provinces, including Banteay Meanchey, Oddar Meanchey, Battambang, Pailin, Pursat, Koh Kong, Preah Vihear and Siem Reap, to combat community transmission of the Delta variant. Despite these measures, the Delta variant spread widely across Cambodia, becoming the predominant strain towards the end of September 2021 (Fig. 1b).
COVID-19 vaccination
In tandem with non-pharmacological interventions, Cambodia initiated a COVID-19 vaccination program on 10 February 2021, with priority for healthcare workers and officials. In response to the COVID-19 outbreak driven by the Alpha variant, Cambodia launched a mass vaccination drive in April 2021, named the “Blossom Plan,” initially focusing on adults aged 18 and above receiving two vaccine doses in Phnom Penh before extending it nationwide (Fig. 1a). By July 2021, more than 76% of the initial target population of 10 million had received at least one vaccine dose. The program was further expanded on 1 August to include vaccination for all individuals aged 12 and above, and by September 2021 vaccination encompassed individuals aged 3 and above (Fig. 1a). From August 2021 third booster doses were introduced to protect frontline workers. By the end of September 2021, Phnom Penh reached nearly 99% of its adult population fully vaccinated and the country progressed to achieve full vaccination coverage of over 70% of its total population.
Genomic epidemiology of COVID-19 epidemic waves
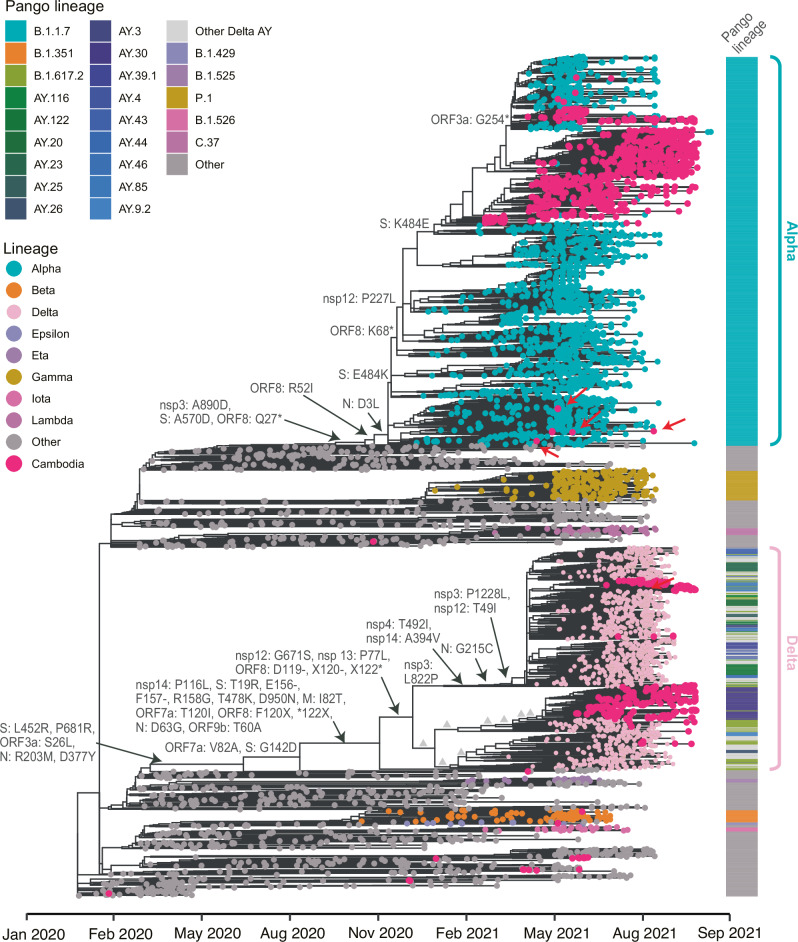
From January 2021 to September 2021, we sequenced a total of 1163 genomes from SARS-CoV-2 positive patients across Cambodia. During this period Alpha B.1.1.7 (n = 759) and Delta B.1.617.2 variants (n = 337) were prevalent, aligning with the global predominance of these variants. Evolutionary analysis indicated that the majority of Alpha and Delta sequences formed large monophyletic clusters within Cambodia (indicated by red tip circles in Fig. 2), alongside single independent cases detected in the country. From June 2021, Delta viruses spread locally in Cambodia and two large clusters were formed, with the AY.30 (n = 210) and AY.85 (n = 61) lineages being the most predominant (Fig. 2). The Cambodian AY.30 cluster was closely related to other AY.30 sequences from Thailand. The AY.85 cluster from Cambodia were related to AY.20 viruses from Mexico, AY.114 from USA, AY.116 from Africa, and B.1.617.2 from multiple regions including Australia, France, England, Romania, the Middle East, and the Netherlands. A minority of other SARS-CoV-2 variants were concurrently observed in Cambodia, including Epsilon B.1.427/B.1.429 (Fig. 2), albeit only in a few incoming cases and there was no evidence of their widespread community transmission.
Fig. 2. Phylogenetic analyses of global and Cambodian SARS-CoV-2 genome sequences, 2021.
A maximum likelihood phylogeny was reconstructed using Nextstrain based on 5964 full genomes comprising Cambodian and subsampled global sequences. Colored tip circles represent different virus lineages or clades, with light blue and pink tips circles denoting the Alpha and Delta variants, respectively. Deep pink tip circles represent sequences obtained from SARS-CoV-2 positive patients in Cambodia. Red arrows indicate single transmission events of Alpha cases introduced into Cambodia. Amino acid mutations are displayed at the major nodes. Grey triangles indicate additional mutations in the Delta lineage that are specified in Fig. 4. The annotation block provides colored Pango lineages for each respective tip sequence.
Our phylogenetic inference indicates that a limited number of Cambodian Alpha cases stemmed from single transmission events (marked by red arrows in Fig. 2) from other parts of the world, indicating multiple introductions of Alpha variants, consistent with epidemiological findings (Fig. 2, Supplementary Fig. 1). Subsequently, rapid dissemination of the Alpha variants throughout the country led to a substantial increase in cases, forming large monophyletic clusters between 5 April and 27 September 2021 (Figs. 2 and 3a). Several individual Alpha cases from Thailand grouped within Cambodian clusters. Notably, the Cambodian Alpha cluster was closely related to a large monophyletic cluster comprised primarily of Thailand sequences and some Alpha cases from Cambodia (Supplementary Fig. 1). Our results suggest bi-directional transmission of Alpha viruses between Cambodia and Thailand. We also observed that Delta cases from Thailand were grouped within Cambodian Delta lineages from 24 May–24 September 2021 (Supplementary Fig. 1).
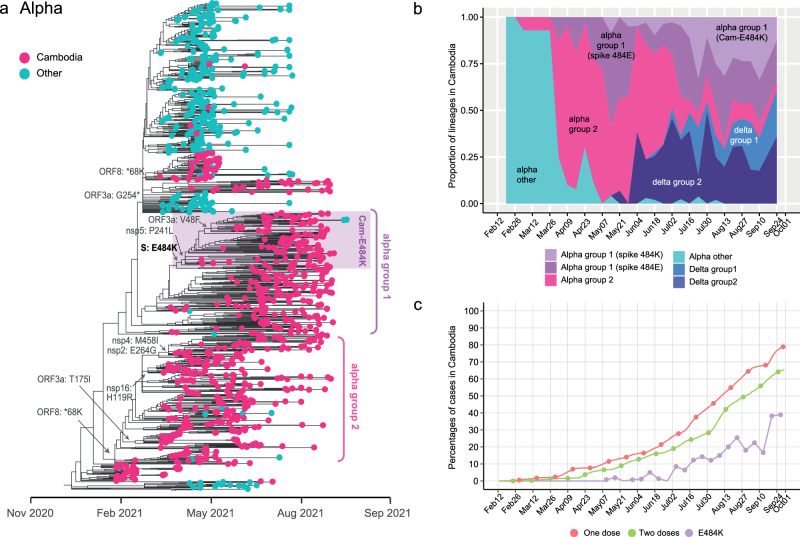
Fig. 3. Evolutionary relationships of the Alpha variant in Cambodia.
a Maximum likelihood phylogeny of SARS-CoV-2 viruses inferred from Nextstrain tree in Fig. 2. Light blue tip circles represent subsampled global Alpha sequences. Deep pink tip circles denote Cambodian sequences obtained from SARS-CoV-2 positive patients. Cambodian Alpha sequences that have acquired the spike E484K mutation are highlighted in the shaded purple area. Amino acid mutations are mapped at the major nodes. b Proportion of sequences representing different groups of Alpha and Delta variants within Cambodia. c Vaccination coverage over time among the Cambodian population (with one dose represented by the red line and two doses by the green line). The purple line displays the proportion of sequenced Alpha viruses with the E484K mutation.
All Cambodian Alpha viruses exhibited a two-amino-acid-deletion at positions 69–70 in the spike protein. This 69–70 deletion, also found in Alpha B.1.1.7 as well as other variants such as B.1.375, B.1.258 and P.1, has been associated with enhanced infectivity and syncytia formation45. An additional N501Y spike mutation was present in all Cambodian Alpha variants. This mutation, also found in early globally circulating Alpha strains from the United Kingdom, Brazil and South Africa, is considered a major determinant contributing to enhanced infection and transmission of SARS-CoV-2 virus46. Within Cambodia, two main clusters of the Alpha variant emerged during circulation: labeled as Alpha Group 1 and Group 2 viruses (Fig. 3a). The Group 1 viruses underwent genetic diversification and acquired an E484K amino acid mutation within the receptor-binding domain of the spike protein, which showed an increase in proportion of E484K cases (approximately 33%) in July 2021 (Fig. 3a, b). These Cambodian E484K viruses then acquired two additional mutations in ORF1a (nsp5: P241L) and in ORF3a (V48F). The nsp5 P241L (ORF1a: P3504L) mutation was identified in another variant of P.1 (Gamma)47. In contrast, Alpha Group 2 viruses did not possess the E484K spike mutation, but had mutations in the ORF1a (nsp2: E264G, nsp4: M458I), ORF1b (nsp16: H119R) and ORF8 (*68 K) regions. A gradual decline in the prevalence of 484E viruses occurred from July 2021 followed by an increase in Alpha viruses bearing the E484K mutation (Fig. 3b), coinciding with the implementation of mass vaccination in Cambodia from May 2021 onwards (Fig. 3c). Of note, the spike E484K mutation emerged early in the spread of global Alpha cases around November 2020, followed by a reversal to 484E around January 2021, prior to the introduction of the Alpha variant into Cambodia (Fig. 1). Later within the Cambodian Group 1 cluster, the 484K mutation became predominant, suggesting this mutation may play a crucial role in the persistence and circulation of the virus. However, by the end of September 2021, there was a substantial reduction in the prevalence of Alpha variant as they were replaced by Delta variant (Figs. 1b and 3b).
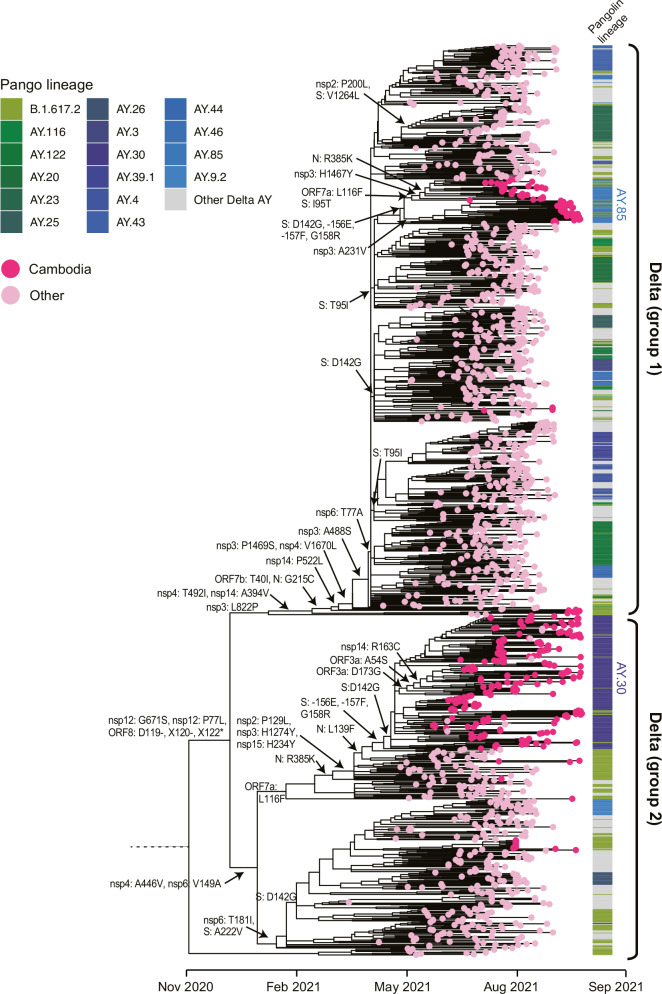
The global Delta virus lineage had key amino acid mutations at L452R, T478K and P681R in the spike protein (Fig. 2). Additionally, most globally circulating Delta viruses acquired two amino acid deletions at residues 156 and 157, as well as R158G in the N-terminal domain (NTD) of the spike protein, which are associated with immune evasion and reduced sensitivity to antibodies48. Based on phylogenetic inferences, the global Delta viruses evolved into two distinct monophyletic groups, referred to as Delta Group 1 and Group 2 (Fig. 4). The Delta Group 1 viruses exhibited sequential amino acid substitutions in ORF1a (L1640P, T3255I, P2046L, P2887S, A1306S, T3646A), ORF1b (A1918V, T40I) and N (G215C) regions. The Delta Group 2 viruses acquired mutations in ORF1a (A3209V, V3718A, P309L), ORF1b (H2285Y), spike (A222V, -156E, -157F, G158R), ORF7a (L116F) and N (R385K, L139F) regions. Of note, reversions of spike mutations occurred at amino acid position 158 (R158G) and 142 (G142D) (Figs. 2 and 4). Most Cambodian Delta viruses lacked two amino acid deletions at the 156 and 157 residues in the spike protein. Also, some of the AY.30 lineage clades from Cambodia had sequential mutations in the ORF3a (D173G), ORF3a (A54S) and nsp14 (R163C) (Fig. 4). Similarly, sequential mutations were observed in AY.84 lineage from Cambodia, including ORF7a (L116F), spike (I95T), nsp3 (H1467Y and A231Y) and N (R385K). Furthermore, the Delta viruses in Cambodia displayed notable local transmission, as larger nationwide clusters were evident from the phylogenetic tree (Fig. 4).
Fig. 4. Evolutionary relationships of the Delta variant in Cambodia.
Maximum likelihood phylogeny of SARS-CoV-2 viruses inferred from Nextstrain tree in Fig. 2. Pink tip circles represent subsampled global Delta sequences. Deep pink tip circles denote sequences generated from SARS-CoV-2 positive patients detected in Cambodia. Amino acid mutations are shown at the major nodes. The annotation block provides colored Pango lineages for each respective tip sequence.
Comparative population dynamics and growth of Cambodian Alpha and Delta variants
To compare the population dynamics of the Alpha and Delta variants of SARS-CoV-2 in Cambodia, relative genetic diversity of each variant was quantified over time. Skyride plots revealed that Alpha viruses exhibited a gradual increase in diversity from early 2021, peaking in August 2021, followed by a decline in diversity shortly afterwards (Fig. 1c). In contrast, later Delta strains displayed greater levels of genetic diversity around mid-July 2021 and mid-August 2021 compared to earlier Delta strains. These peaks coincided with the increased genetic diversification of Delta variants in Cambodia.
Further estimation of the effective reproductive number (Re) of the Alpha and Delta variants in Cambodia using the birth-death skyline serial (BDSKY) model inferred changes in epidemic growth, steady growth or recession over time. The Cambodian Alpha variant (represented by the blue line in Fig. 1d) initially experienced a rapid exponential increase in Re, shortly after introduction into the country with a peak estimated mean Re of approximately 3.7 (95% HPD interval: 2.1–5.2) in March 2021, reflecting the efficient transmission of Alpha variants. However, by April 2021, there was a two-fold decrease and the mean Re dropped to around 1.8 (95% HPD interval: 1.2–2.7). The Cambodian Alpha variant further declined in growth to Re ≈ 1 by June 2021 (Fig. 1d, Supplementary Data 1). This rapid fall in epidemic growth coincided with lockdowns restricting movement, and with implementation of mass vaccination programs (Fig. 1a, d). Although there was a slight increase in mean Re to around 1.5 from mid-June to mid-July 2021, this value remained markedly lower compared to the early stages of Alpha transmission in Cambodia. By August 2021, the mean Re of the Alpha variant dropped back to 0.4, soon after extending vaccination to individuals aged 12–17 years old, and its growth remained at 1 or below throughout September 2021 (Fig. 1d).
In contrast, the initial exponential growth of the Cambodian Delta variant was relatively lower compared to the Alpha variant. We observed a gradual increase in growth, which peaked with an estimated mean Re of around 2.3 (95% HPD interval: 0.9–4.1) in May–June 2021 (Fig. 1d, Supplementary Data 1). This period also coincided with a substantial reduction in Re to below 1 for the Alpha variant. The Re estimates for the Delta variant started declining and reached a minimal value (mean Re = 0.5) in mid-August, indicating a recession of the epidemic during that month. The observed reduction in growth could potentially be attributed to the higher rates of COVID-19 vaccine uptake in Cambodia, combined with government mandates like border and travel restrictions, as well as strict isolation of positive cases. In late August 2021, the Re of the Cambodian Delta rose to around 1.5 in late August 2021, greater that of the Alpha (Fig. 1d), indicating a higher level of transmissibility of the Delta variant and coinciding with its increasing proportion to nearly 50% (Fig. 1b). Additionally, we conducted analyses to independently assess the population dynamics of the two Cambodian Delta groups. Our results indicate that the variations in relative genetic diversity and Re based on Delta Group 2 (n = 215) broadly mirrored those observed in the combined dataset of Cambodian Delta sequences (Supplementary Fig. 2). Delta Group 1 did not show the same trends as the combined analysis, likely due to the smaller number (n = 80) of Cambodian sequences in this clade.
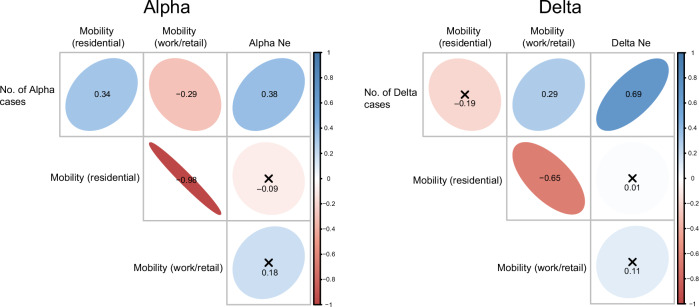
We further examined the relationship between the total number of daily cases and virus population dynamics. Our results show a positive correlation between the number of Alpha cases and relative genetic diversity (r = 0.38), with even stronger positive correlations observed between the number of Delta cases and its relative genetic diversity (r = 0.69, Fig. 5). These correlations suggest that genomic surveillance in Cambodia was sufficient to accurately capture virus dynamics. We also found that the number of Alpha cases showed a positive association with residential mobility (r = 0.34, Fig. 5), possibly indicating localized transmission during the 14-day stay-at home order and restriction of non-essential movement that was imposed in Phnom Penh in April 2021, when Alpha was the only circulating variant. Conversely, the number of Delta cases showed a positive correlation with work or retail mobility (r = 0.29), likely due to the relaxation of lockdowns during the period that Delta was becoming predominant in Cambodia (Fig. 5).
Fig. 5. Correlation matrix between SARS-CoV-2 case numbers and mobility data for Alpha and Delta variants in Cambodia.
Positive associations are represented by blue ellipsoids, while negative correlations are denoted by red ellipsoids. The intensity of color and ellipse size correspond to the strength of the correlation coefficients. Numbers in the center represent Pearson correlation coefficients. Statistical significance was determined using a P-value < 0.05, with non-significant correlations indicated by a cross symbol. Ne represents relative genetic diversity.
Spatial transmission dynamics of the Alpha and Delta variants in Cambodia
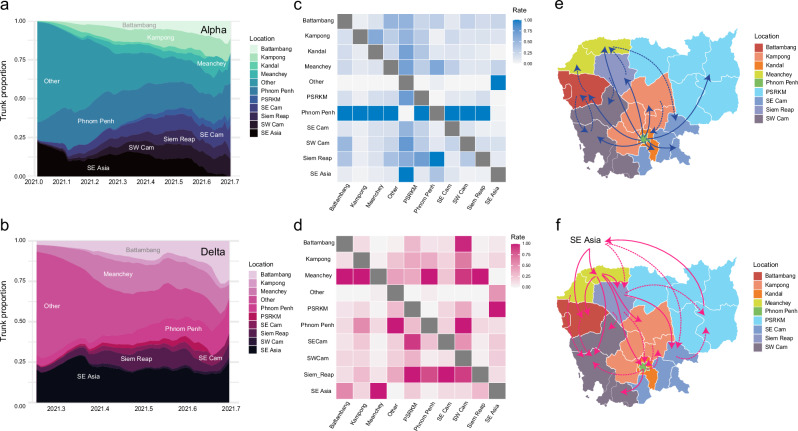
To unravel the spatial diffusion dynamics of the Alpha and Delta variants in Cambodia, phylogeographic analyses using Bayesian stochastic search variation selection (BBSVS) model estimated the rates of virus migration between discrete geographical states (Fig. 6). The trunk reward proportion of the Alpha variant was estimated for each ancestral location through time (Fig. 6a, Supplementary Data 2). Phylogeographic inference suggests that Phnom Penh was the initial source of Alpha variant within Cambodia in early 2021. From March 2021 onwards, a rapid dissemination of the Alpha variant occurred to all other regions of Cambodia surrounding the capital, particularly Kampong, Meanchey, Southeast Cambodia, Siem Reap and Southwest Cambodia (Supplementary Fig. 3). Notably, Phnom Penh consistently accounted for the majority of trunk proportion throughout this period, indicating that the capital was the primary source of continuous Alpha circulation. This observation was supported by the estimated diffusion rates between location states. Specifically, Alpha displayed greater diffusion rates (ranging from 0.79 to 3.56, Fig. 6c, Table 1) with significant support (Bayes Factor >100) from Phnom Penh to the rest of Cambodia. Strongly supported routes were observed from Phnom Penh to Battambang, Kampong, Kandal, Meanchey, SE Cam, SW Cam, and Siem Reap (Fig. 6e). In addition, moderate migration rates were inferred between some provinces, such as from Battambang to Meanchey, from Battambang to SW Cam and from Kampong to Kandal (Fig. 6c, e).
Fig. 6. Spatiotemporal diffusion of the Alpha and Delta variants in Cambodia using discrete phylogeographic reconstruction.
a, b Trunk reward proportion was estimated for each geographic location over time for the Alpha and Delta variant, respectively. c, d Asymmetric non-zero migration rates between pairwise location states were inferred using the Bayesian stochastic search variable selection (BSSVS) model. e, f Migration pathways with statistical support were inferred using SPREAD 3. Solid lines represent significant migration routes with strong support (Bayes factor of ≥100), and dotted lines represent transmission routes with moderate support (BF between 3 and 99). Other rest of the world, PSRKM include five provinces (Preah Vihear, Stung Treng, Ratanakiri, Kratie and Modulkiri), SE Cam Southeast Cambodia, SW Cam Southwest Cambodia, SE Asia Southeast Asia.
Table 1.
Asymmetric migration rates of Alpha B.1.1.7 variant of SARS-CoV-2 virus between pairwise geographic locations
| Location | Battambang | Kampong | Kandal | Meanchey | Other | PSRKM | Phnom Penh | SE Cam | SW Cam | Siem Reap | SE Asia |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Battambang | – | 0.3 | 0.27 | 0.64a | 0.23 | 0.25 | 0.44 | 0.31 | 0.66a | 0.49 | 0.7 |
| Kampong | 0.3 | – | 0.83a | 0.43 | 0.23 | 0.32 | 0.33 | 0.53 | 0.37 | 0.41 | 0.52 |
| Kandal | 0.33 | 0.36 | – | 0.32 | 0.27 | 0.35 | 0.36 | 0.36 | 0.33 | 0.31 | 0.32 |
| Meanchey | 0.37 | 0.45 | 0.26 | – | 0.27 | 0.31 | 0.57 | 0.72a | 0.34 | 0.37 | 0.73 |
| Other | 0.08 | 0.08 | 0.09 | 0.08 | – | 0.08 | 0.34b | 0.09 | 0.09 | 0.18 | 1.87b |
| PSRKM | 0.31 | 0.29 | 0.31 | 0.3 | 0.28 | – | 0.31 | 0.29 | 0.3 | 0.48 | 0.31 |
| Phnom Penh | 1.36b | 3.56b | 1.32b | 1.18b | 0.12 | 0.79b | – | 2.13b | 2.16b | 1.74b | 0.34 |
| SE Cam | 0.26 | 0.29 | 0.29 | 0.27 | 0.23 | 0.27 | 0.34 | – | 0.4 | 0.26 | 0.38 |
| SW Cam | 0.72a | 0.29 | 0.29 | 0.36 | 0.28 | 0.31 | 0.36 | 0.61 | – | 0.47 | 0.57a |
| Siem Reap | 0.58 | 0.32 | 0.27 | 0.54a | 0.25 | 0.58 | 0.73a | 0.28 | 0.82a | – | 0.45 |
| SE Asia | 0.18 | 0.17 | 0.18 | 0.19 | 0.34 | 0.16 | 0.48 | 0.34b | 0.17 | 0.25 | – |
Other rest of world, PSRKM represents 5 provinces (Preah Vihear, Stung Treng, Ratanakiri, Kratie and Mondulkiri), SE Cam Southeast Cambodia, SW Cam Southwest Cambodia, SE Asia Southeast Asia.
aDenotes moderate support with Bayes Factor of 3–99.
Bold numbers and bdenote strong support with Bayes Factor ≥100.
In marked contrast, Delta variant experienced source-sink dynamics in Cambodia. Meanchey, located in northern Cambodia, was the primary source of Delta variant during May–June 2021 (Fig. 6b, Supplementary Fig. 3, Supplementary Data 3), as they were likely introduced through land transport. Consequently, Phnom Penh and other regions of Cambodia became the sink as Delta spread nationally. Significant diffusion rates (1.14–1.70 with strong Bayes Factor support of >100, Table 2, Fig. 6d) indicate Delta introduction from Southeast Asia into adjacent provinces of Battambang and Meanchey, which was likely driven by movement of individuals from Cambodia’s northern land border. Subsequently, viral transmission was observed from Meanchey, with greater migration rates (ranging from 1.52 to 2.27 with strong Bayes Factor support >100, Table 2) from Meanchey into most Cambodian provinces, including Battambang, Kampong, Phnom Penh, SW Cam and Siem Reap (Fig. 6d, f). Furthermore, local spread of Delta variants was observed among neighboring regions, particularly from Phnom Penh to Kampong, from Phnom Penh to SW Cam, and from Siem Reap to PSRKM (5 provinces: Preah Vihear, Stung Treng, Ratanakiri, Kratie and Mondulkiri). Taken together, our results revealed differential transmission dynamics of the Alpha and Delta variants in Cambodia; the Alpha viruses exhibited higher migration rates from Phnom Penh into other regions of Cambodia, while the Delta displayed stronger rates of migration from the northern regions of Cambodia into the rest of country.
Table 2.
Asymmetric migration rates of Delta B.1.617.2 variant of SARS-CoV-2 virus between pairwise geographic locations
| Location | Battambang | Kampong | Meanchey | Other | PSRKM | Phnom Penh | SE Cam | SW Cam | Siem Reap | SE Asia |
|---|---|---|---|---|---|---|---|---|---|---|
| Battambang | – | 0.29 | 0.29 | 0.22 | 0.31 | 0.44 | 0.32 | 0.72a | 0.28 | 0.26 |
| Kampong | 0.32 | – | 0.33 | 0.33 | 0.36 | 0.87a | 0.47 | 0.51 | 0.35 | 0.29 |
| Meanchey | 1.96b | 1.52b | – | 0.42 | 0.36 | 1.84b | 0.6 | 0.69a | 2.27b | 0.24 |
| Other | 0.11 | 0.11 | 0.35 | – | 0.09 | 0.37a | 0.1 | 0.1 | 0.24 | 0.4b |
| PSRKM | 0.28 | 0.32 | 0.3 | 0.35 | – | 0.29 | 0.44 | 0.34 | 0.32 | 0.57b |
| Phnom Penh | 0.36 | 0.88a | 0.49 | 0.7a | 0.29 | – | 0.48 | 0.74a | 0.55 | 0.27 |
| SE Cam | 0.29 | 0.44 | 0.31 | 0.31 | 0.73a | 0.71 | – | 0.43 | 0.34 | 0.31 |
| SW Cam | 0.41 | 0.33 | 0.33 | 0.31 | 0.33 | 0.37 | 0.31 | – | 0.31 | 0.3 |
| Siem Reap | 0.33 | 0.53 | 0.42 | 0.4 | 0.79b | 1.6a | 2.14b | 0.68a | – | 0.27 |
| SE Asia | 1.14b | 0.35 | 1.7b | 0.21 | 0.24 | 0.35 | 0.38a | 0.18 | 0.86a | – |
Other rest of world, PSRKM represents 5 provinces (Preah Vihear, Stung Treng, Ratanakiri, Kratie and Mondulkiri), SE Cam Southeast Cambodia, SW Cam Southwest Cambodia, SE Asia Southeast Asia.
aDenotes moderate support with Bayes Factor of 3–99.
bold numbers and bdenote strong support with Bayes Factor ≥100.
To evaluate the effect of sampling bias, we conducted additional analyses with smaller subsampled datasets of Alpha and Delta, respectively (Supplementary Fig. 4). Circular migration plots based on both large and small datasets consistently identified Phnom Penh in the south as the primary source of Alpha transmission, while Meanchey in the north was the main source for Delta transmission (Supplementary Fig. 4). However, there were some disparities in transmission rate estimates. The larger dataset indicated higher dispersal rates of Alpha from Phnom Penh to eight other regions in Cambodia, whereas the smaller dataset suggested higher rates to only three regions (Supplementary Fig. 4). Similarly, for Delta transmission, while the larger dataset showed higher dispersal rates from Meanchey to seven other regions, the smaller dataset indicated higher rates to five other regions. These findings underscore the necessity of exercising caution when interpreting phylogeographic inferences.
Discussion
During the early COVID-19 pandemic, Cambodia leveraged its existing infectious disease surveillance infrastructure to effectively curb the transmission of the SARS-CoV-2 virus. Through rigorous quarantine and testing measures, Cambodia successfully controlled the spread of the virus for 2020, with only a limited number of reported cases6. However, the subsequent emergence and global dissemination of VoCs with enhanced transmissibility posed new challenges, particularly in a resource-limited country like Cambodia. Genomic surveillance and sequencing played a critical role in tracking the introductions, mutations, and transmission patterns of VoCs, forming a key component of public health strategies. In this study, we utilized targeted next-generation sequencing and phylodynamic approaches to analyze genomic data of SARS-CoV-2. Our findings offer valuable insights into understanding the evolution, transmissibility and spatial dynamics of the Alpha versus the Delta variants that were responsible for the first two waves of large-scale community outbreaks in Cambodia from February to September 2021.
The Alpha variant likely entered Cambodia through initial introduction into the capital Phnom Penh, sparking widespread community transmission nationally. All sequenced Cambodian Alpha genomes harbored canonical mutations like the 69/70 deletion and N501Y substitution in the spike protein, that are known to confer increased transmissibility9,49. The spike 69/70 deletion facilitates faster syncytium formation through improved fusogenicity49 and the N501Y substitution in the receptor binding domain (RBD) enhances the binding affinity of the spike protein for the ACE2 receptor46. Upon establishing and circulating within Cambodia, the Alpha variant diversified into local sublineages, notably acquiring the E484K mutation in the RBD. This E484K mutation has also been observed in other globally circulating variants including Delta (B.1.617.2), Beta (B.1.351) and Gamma (P.1) viruses. Studies have shown that the E484K mutation can reduce, but not abolish, the neutralizing activity of post-infection and post-vaccination sera50, while also conferring resistance to certain monoclonal antibodies51–53.
We show that the Alpha variant displayed a higher level of genetic diversity and more variation in effective reproductive number than the Delta variant. The epidemic experienced greater expansion with the onset of Alpha transmission, followed by a notable reduction by April 2021. In comparison, the Delta epidemic peaked in early June 2021, coinciding with a decline in the Alpha epidemic. Interestingly, the epidemic growth of the Alpha variant exhibited a slight rebound in epidemic growth from mid-June to mid-July 202. Soon after, both variants showed a recessive epidemic (Re<1) from mid-July to mid-August, possibly due to the launch of Cambodia’s mass vaccination campaign in April 2021, which effectively reduced virus transmission.
Our Alpha and Delta Re estimates corresponded to published estimates (0.28–2.11, and 0.48–2.68, respectively54) made using the same Birth-Death Skyline (BDSKY) model. From June to September 2021, the Delta variant rapidly displaced the Alpha variant in Cambodia, following the global epidemic trend20,55,56. In particular, the epidemic growth of the Delta variant showed a 50% increase in the effective reproductive number (Re) compared to the Alpha variant in late August 2021. Our estimates corroborate previous findings indicating that the Delta also demonstrated a higher Re than Alpha across 64 countries57. While the COVID-19 vaccination has been effective in curtailing transmission of the Alpha variant, the Delta variant has acquired three key amino acid mutations, L452R, T478K and P681R in the spike protein58. The L452R mutation in the RBD was demonstrated to reduce antibody recognition and virus neutralization14,59. The T478K mutation, also in the RBD, could enhance the binding affinity to human ACE2 receptor60,61. Additionally, the P681R and D950N substitutions at the S1/S2 cleavage site can promote cleavage rate and facilitate faster transmission13. Like globally circulating Delta variant, all Cambodian Delta viruses similarly possessed these three mutations. Despite the increasing uptake of COVID-19 vaccinations in the adult populations, the second wave of infections in 2021 was still driven by the Delta variant. This highlights the impact of newer variants on vaccine efficacy and the vulnerability of younger age populations that remain unvaccinated. Furthermore, some studies have suggested that COVID-19 vaccines can exert selective pressure that leads to the emergence of immune evasion mutations in different SARS-CoV-2 variants, thereby enabling the virus to evade the vaccine-induced immunity62–64. However, COVID-19 vaccination remains protective against Delta-related severe illness and mortality65,66.
The rapid displacement of a previously predominant variant by a newer, more adapted variant has been a hallmark of SARS-CoV-2 evolution. The rapid rate of evolution coupled with strong selective forces drive continuous genetic drift and the emergence of advantageous mutations67. Moreover, new SARS-CoV-2 variants have been observed to emerge from specific locations worldwide. In this study, we uncovered contrasting spatial dissemination patterns between Alpha and Delta variants in Cambodia, consistent with the epidemiological patterns reported. Notably, the spread of the Alpha variant from Phnom Penh into the rest of country coincided with the introduction of the virus from the rest of the world due to a quarantine breach in the capital. Conversely, the Delta variant was likely introduced into Cambodia through northern border provinces, possibly via returning migrant workers from Thailand in May 2021. The prevalent Delta cases in Cambodia were associated with AY.30 and AY.85 lineages, aligning with the dominant Delta lineages observed in Thailand68. Subsequently, the Delta variant continued to spread to densely populated areas in the southern provinces. Phylogeographic reconstruction has provided valuable insights into the geographic spread and epidemic dynamics of SARS-CoV-2 variants69–72. However, sampling bias due to a limited number of sequences available from different geographic locations may affect interpretation of phylogeography. In some instances, virus sequences may not be available from certain locations, which may impact the estimated number of migration pathways and estimated dispersal rates73,74. Interpretation of phylogeographic inference should therefore be approached with caution. To mitigate these challenges, alternative approaches74 are necessary for comparative analyses to reinforce the consistent identification of key migration events and enhance the robustness and reliability of findings.
Our phylodynamic analyses provide important insights into the evolutionary trajectories and transmission patterns of major SARS-CoV-2 variants in Southeast Asia. However, the limited sampling and geographic representation highlights the necessity for expanded genomic surveillance, given the scarcity of data from the region. Unlike the singular introduction in Cambodia, a recent evolutionary study from Vietnam75 analyzed 1303 genomes of both Alpha and Delta variants and suggested multiple introductions of the Alpha variant that sparked sporadic outbreaks. However, like Cambodia, the Delta variant quickly became prevalent and caused a major nationwide wave of infections. In Vietnam, the Delta outbreak traced back to a single AY.57 lineage introduction in March 2021 which then diversified into localized sub-clusters coinciding with domestic lockdowns75. As such, we observe the similarities and differences in variant transmission even between neighboring countries and the value of genomic epidemiology in deciphering SARS-CoV-2 transmission pathways and evolutionary patterns, thus informing effective public health strategies in Southeast Asia. Nevertheless, comprehensive understanding of these complex dynamics, particularly in under-sampled regions, requires expanded sequencing and integration with epidemiological data.
In summary, Cambodia, like other countries, faced the emergence and introduction of new SARS-CoV-2 variants. Our study emphasizes the effectiveness of incorporating genomic surveillance into routine monitoring to track and control regional COVID-19 epidemics driven by highly transmissible variants. We show that the successive epidemic waves of both Alpha and Delta variants were driven by virus importation, followed by subsequent community transmission within the country. Phnom Penh, in particular, is well-connected to rest of world, while transmission between provinces occurred due to business and trade activities. To mitigate large local outbreaks, the Cambodian government continues routine genomic sequencing to monitor current SARS-CoV-2 variants. Future research should prioritize understanding the epidemic behavior and onward transmission dynamics of Omicron and emerging variants in Cambodia. However, due to changes in testing requirements and public health intervention measures, complete genetic data from the entire country is limited from October 2021 onwards. Maintaining a vigilant approach through viral genomic surveillance and strengthening resources for strategic public health measures and vaccination campaigns are crucial in mitigating future waves of SARS-CoV-2 variants and other endemic and emerging viruses with pandemic potential.
Supplementary information
Acknowledgements
The authors would like to acknowledge the Cambodian Ministry of Health and Cambodian Communicable Disease Control Department; Rapid Response Team members; all of the Provincial Health Departments; and the doctors, nurses, and staff involved in the COVID-19 response in Cambodia. We thank all the technicians and staff at Institut Pasteur du Cambodge in the Virology and Epidemiology/Public Health Units involved in this work, especially all of the tirelessly dedicated COVID-19 teams. The investigators also thank all the authors and laboratories that contribute genetic sequence data via GISAID. The work at Institut Pasteur du Cambodge is supported by the WHO, the European Union, The Pasteur International Center for Research on Emerging Infectious Diseases National Institutes of Health, Department of Health and Human Services funded project No. 1U01AI151758-01, Wellcome Trust grant 222574/Z/21/Z, the British Embassy in Cambodia, and French Development Agency funded ECOnomic development, ECOsystem MOdifications, and emerging infectious diseases Risk Evaluation (ECOMORE, phase 2) COVID-19 top up project No. CZZ 2146 01A. E.A.K. was funded, in part, by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services Contract No. 75N93021C00015. YCFS and GJDS are supported by the Duke-NUS Signature Research Programme funded by the Ministry of Health, Singapore, and by contract 75N93021C00016 from the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, USA. H.A. is supported by the German Centre for International Migration and Development. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the funders. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization: Y.C.F.S., G.J.D.S., and E.A.K. Data curation: Y.C.F.S., M.A.Z., T.P.O., J.M., L.P., R.Z., S.R., V.H., S. Kol, R.L., K.L.C., L.K., L.H., P.R., V.I., V.K., S.P., B.S., V.S.H., S.Y., H.A., J.Y.S., C.T., N.B., K.V., M.C., S.S., S.L., V.D., G.J.D.S., and E.A.K. Formal analysis: Y.C.F.S., M.A.Z., T.P.O., J.M., L.P., R.Z., S.R., V.H., S. Kol, R.L., K.L.C., L.K., L.H., V.S.H., S.Y., H.A., J.Y.S., C.T., N.B., K.V., M.C., S.S., V.D., G.J.D.S., and E.A.K. Funding acquisition: L.B., V.D., and E.A.K. Investigation: Y.C.F.S., M.A.Z., T.P.O., L.P., R.Z., S.R., V.H., S. Kol, R.L., K.L.C., L.K., L.H., S. Krang, P.R., M.H.K., V.I., V.K., S.P., B.S., V.S.H., S.Y., H.A., J.Y.S., C.T., N.B., K.V., M.C., S.S., Y.S., S.H., C.D., C.S., A.K., S.L., V.D., L.S., G.J.D.S., and E.A.K. Methodology: Y.C.F.S., M.A.Z., H.A., J.Y.S., K.V., W.F.Y., G.G.K.N., S.L., G.J.D.S., and E.A.K. Project administration: Y.C.F.S., V.S.H., S.L., L.B., A.S., V.D., L.S., G.J.D.S., and E.A.K. Resources: S. Krang, P.R., M.H.K., V.I., V.K., S.P., B.S., Y.S., S.H., C.D., C.S., V.D., L.S., G.J.D.S., and E.A.K. Software: K.V. Supervision: Y.C.F.S., V.S.H., H.A., C.T., N.B., Y.S., S.H., A.K., S.L., L.B., A.S., V.D., L.S., G.J.D.S., and E.A.K. Validation: Y.C.F.S., T.P.O., L.P., V.H., S. Kol, R.L., K.L.C., L.K., L.H., V.S.H., S.Y., H.A., J.Y.S., C.T., N.B., K.V., M.C., S.S., S.L., V.D., G.J.D.S., and E.A.K. Visualization: Y.C.F.S., M.A.Z., T.P.O., R.Z., G.J.D.S., and E.A.K. Writing - original draft: Y.C.F.S., G.J.D.S., and E.A.K. Writing - review & editing: Y.C.F.S., M.A.Z., T.P.O., J.M., L.P., R.Z., S.R., V.H., S. Kol, R.L., K.L.C., L.K., L.H., S. Krang, P.R., M.H.K., V.I., V.K., S.P., B.S., V.S.H., S.Y., H.A., J.Y.S., C.T., N.B., K.V., W.F.Y., G.G.K.N., M.C., S.S., Y.S., S.H., C.D., C.S., A.K., S.L., L.B., A.S., V.D., L.S., G.J.D.S., and E.A.K.
Peer review
Peer review information
Communications Medicine thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
The SARS-CoV-2 sequences from Cambodia have been deposited in the GISAID database as part of the COVID-19 response. All sequences analyzed in this study are available from 10.55876/gis8.240605ho76. The GISAID database is available at https://gisaid.org. Source data for the six figures in the manuscript are available in Supplementary Data 4–5.
Competing interests
Y.C.F.S. and G.J.D.S. report grants from National Institutes of Health. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yvonne C. F. Su, Email: yvonne.su@duke-nus.edu.sg
Erik A. Karlsson, Email: ekarlsson@pasteur-kh.org
Supplementary information
The online version contains supplementary material available at 10.1038/s43856-024-00685-7.
References
- 1.Manning, J. E. et al. Rapid metagenomic characterization of a case of imported COVID-19 in Cambodia. bioRxiv, https://www.biorxiv.org/content/10.1101/2020.03.02.968818v1 (2020).
- 2.Zeller, M. et al. Emergence of an early SARS-CoV-2 epidemic in the United States. Cell184, 4939–4952.e4915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodcroft, E. B. et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature595, 707–712 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Giandhari, J. et al. Early transmission of SARS-CoV-2 in South Africa: An epidemiological and phylogenetic report. Int. J. Infect. Dis.103, 234–241 (2021). [DOI] [PMC free article] [PubMed]
- 5.Nadeau, S. A. et al. Swiss public health measures associated with reduced SARS-CoV-2 transmission using genome data. Sci. Transl. Med. 15, eabn7979 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su, Y. C. F. et al. Genomic epidemiology of SARS-CoV-2 in Cambodia, January 2020 to February 2021. Virus Evol.9, veac121 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraemer, M. U. G. et al. Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence. Science373, 889–895 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supasa, P. et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell184, 2201–2211.e2207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, N.G. et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science372, eabg3055 (2021). [DOI] [PMC free article] [PubMed]
- 10.Walker, A.S. et al. Increased infections, but not viral burden, with a new SARS-CoV-2 variant. medRxiv, https://www.medrxiv.org/content/10.1101/2021.01.13.21249721v1 (2021).
- 11.Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature599, 114–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherian, S. et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms9, 1542 (2021). [DOI] [PMC free article] [PubMed]
- 13.Furusawa, Y. et al. In SARS-CoV-2 delta variants, Spike-P681R and D950N promote membrane fusion, Spike-P681R enhances spike cleavage, but neither substitution affects pathogenicity in hamsters. EBioMedicine91, 104561 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Edara, V. V. et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med.385, 664–666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzmina, A. et al. SARS CoV-2 Delta variant exhibits enhanced infectivity and a minor decrease in neutralization sensitivity to convalescent or post-vaccination sera. iScience24, 103467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCrone, J. T. et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature610, 154–160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, P. et al. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the Delta variant. Science374, eabl9551 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Wintersdorff, C. J. H. et al. Infections with the SARS-CoV-2 Delta variant exhibit fourfold increased viral loads in the upper airways compared to Alpha or non-variants of concern. Sci. Rep.12, 13922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolze, A. et al. SARS-CoV-2 variant Delta rapidly displaced variant Alpha in the United States and led to higher viral loads. Cell Rep. Med.3, 100564 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trobajo-Sanmartín, C. et al. Differences in Transmission between SARS-CoV-2 Alpha (B.1.1.7) and Delta (B.1.617.2) Variants. Microbiol. Spectr.10, e0000822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twohig, K. A. et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis.22, 35–42 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathieu, E. et al. Coronavirus (COVID-19) cases. Our World in Data, https://ourworldindata.org/covid-cases (2020).
- 24.Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill.25, 2000045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdulnoor, M. et al. Real-Time RT-PCR Allelic Discrimination Assay for Detection of N501Y Mutation in the Spike Protein of SARS-CoV-2 Associated with B.1.1.7 Variant of Concern. Microbiol. Spectr.10, e0068121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics34, 4121–4123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Toole, Á. et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol.7, veab064 (2021). [DOI] [PMC free article] [PubMed]
- 28.Quick, J. et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc.12, 1261–1276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itokawa, K., Sekizuka, T., Hashino, M., Tanaka, R. & Kuroda, M. A proposal of an alternative primer for the ARTIC Network’s multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing. bioRxiv, https://www.biorxiv.org/content/10.1101/2020.03.10.985150v1 (2020).
- 30.Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser.41, 95–98 (1999).
- 31.Elbe, S. & Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall.1, 33–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol.4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arendsee, Z., Baker, A. & Anderson, T. smot: a python package and CLI tool for contextual phylogenetic subsampling. J. Open Source Softw.7, 4193 (2022). [Google Scholar]
- 34.Rambaut, A., Lam, T. T., Max Carvalho, L. & Pybus, O. G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol.2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minin, V. N., Bloomquist, E. W. & Suchard, M. A. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol.25, 1459–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol.67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol.10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gostic, K. M. et al. Practical considerations for measuring the effective reproductive number, Rt. PLoS Comput. Biol.16, e1008409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadler, T., Kühnert, D., Bonhoeffer, S. & Drummond, A. J. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc. Natl Acad. Sci. USA110, 228–233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa, M., Kishino, H. & Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol.22, 160–174 (1985). [DOI] [PubMed] [Google Scholar]
- 41.Wei T. & Simko V. R package ‘corrplot’: Visualization of a Correlation Matrix. (version 0.92), https://github.com/taiyun/corrplot (2021).
- 42.Lemey, P., Rambaut, A., Drummond, A. J. & Suchard, M. A. Bayesian phylogeography finds its roots. PLoS Comput. Biol.5, e1000520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedford, T., Cobey, S., Beerli, P. & Pascual, M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog.6, e1000918–e1000918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielejec, F. et al. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol. Biol. Evol.33, 2167–2169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcantara, L. C. J. et al. Methods for fighting emerging pathogens. Nat. Methods19, 395–397 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Liu, Y. et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature602, 294–299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimerman, R. A. et al. Comparative Genomics and Characterization of SARS-CoV-2 P.1 (Gamma) Variant of Concern From Amazonas, Brazil. Front. Med.9, 806611 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng, B. et al. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep.40, 111220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng, B. et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep.35, 109292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jangra, S. et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe2, e283–e284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen, R. E. et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med.27, 717–726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhary, M. C. et al. Emergence of SARS-CoV-2 escape mutations during Bamlanivimab therapy in a phase II randomized clinical trial. Nat. Microbiol.7, 1906–1917 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreano, E. et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl Acad. Sci. USA118, e2103154118 (2021). [DOI] [PMC free article] [PubMed]
- 54.Jimenez-Silva, C. et al. Genomic epidemiology of SARS-CoV-2 variants during the first two years of the pandemic in Colombia. Commun. Med.3, 97 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen, P. A. et al. Delta Variants of SARS-CoV-2 Cause Significantly Increased Vaccine Breakthrough COVID-19 Cases in Houston, Texas. Am. J. Pathol.192, 320–331 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Earnest, R. et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med.3, 100583 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell, F. et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro. Surveill.26, 2100509 (2021). [DOI] [PMC free article] [PubMed]
- 58.Tian, D., Sun, Y., Zhou, J. & Ye, Q. The Global Epidemic of the SARS-CoV-2 Delta Variant, Key Spike Mutations and Immune Escape. Front Immunol.12, 751778 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tada, T. et al. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. iScience24, 103341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng, M. H. et al. Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions. iScience25, 103939 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan, M.I. et al. Impact of the Double Mutants on Spike Protein of SARS-CoV-2 B.1.617 Lineage on the Human ACE2 Receptor Binding: A Structural Insight. Viruses13, 2295 (2021). [DOI] [PMC free article] [PubMed]
- 62.McCallum, M. et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science374, 1621–1626 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Beltran, W. F. et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell184, 2372–2383.e2379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koyama, T. et al. Evasion of vaccine-induced humoral immunity by emerging sub-variants of SARS-CoV-2. Fut. Microbiol.17, 417–424 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez Bernal, J. et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med.385, 585–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamandjou Tchuem, C. R. et al. Vaccine effectiveness and duration of protection of COVID-19 mRNA vaccines against Delta and Omicron BA.1 symptomatic and severe COVID-19 outcomes in adults aged 50 years and over in France. Vaccine41, 2280–2288 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol.21, 361–379 (2023). [DOI] [PubMed] [Google Scholar]
- 68.Aiewsakun, P. et al. Spatiotemporal evolution of SARS-CoV-2 in the Bangkok metropolitan region, Thailand, 2020-2022: implications for future outbreak preparedness. Microb. Genom9, 001170 (2023). [DOI] [PMC free article] [PubMed]
- 69.Lemey, P. et al. Untangling introductions and persistence in COVID-19 resurgence in Europe. Nature595, 713–717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dellicour, S. et al. Dispersal dynamics of SARS-CoV-2 lineages during the first epidemic wave in New York City. PLoS Pathog.17, e1009571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tegally, H. et al. The evolving SARS-CoV-2 epidemic in Africa: Insights from rapidly expanding genomic surveillance. Science378, eabq5358 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tegally, H. et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med.28, 1785–1790 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu, P., Song, Y., Colijn, C. & MacPherson, A. The impact of sampling bias on viral phylogeographic reconstruction. PLOS Glob. Public Health2, e0000577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lemey, P. et al. Accommodating individual travel history and unsampled diversity in Bayesian phylogeographic inference of SARS-CoV-2. Nat. Commun.11, 5110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tam, N. T. et al. Spatiotemporal Evolution of SARS-CoV-2 Alpha and Delta Variants during Large Nationwide Outbreak of COVID-19, Vietnam, 2021. Emerg. Infect. Dis.29, 1002–1006 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su, Y.C.F. et al. Accession numbers for GISAID: Ackowledgment of data contributors, https://epicov.org/epi3/epi_set/240605ho?main=true (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SARS-CoV-2 sequences from Cambodia have been deposited in the GISAID database as part of the COVID-19 response. All sequences analyzed in this study are available from 10.55876/gis8.240605ho76. The GISAID database is available at https://gisaid.org. Source data for the six figures in the manuscript are available in Supplementary Data 4–5.