Abstract
Phosphorylation provides an important mechanism by which transcription factor activity is regulated. Estrogen receptor α (ERα) is phosphorylated on multiple sites, and stimulation of a number of growth factor receptors and/or protein kinases leads to ligand-independent and/or synergistic increase in transcriptional activation by ERα in the presence of estrogen. Here we show that ERα is phosphorylated by protein kinase A (PKA) on serine-236 within the DNA binding domain. Mutation of serine-236 to glutamic acid prevents DNA binding by inhibiting dimerization by ERα, whereas mutation to alanine has little effect on DNA binding or dimerization. Furthermore, PKA overexpression or activation of endogenous PKA inhibits dimerization in the absence of ligand. This inhibition is overcome by the addition of 17β-estradiol or the partial agonist 4-hydroxy tamoxifen. Interestingly, treatment with the complete antagonist ICI 182,780 does not overcome the inhibitory effect of PKA activation. Our results indicate that in the absence of ligand ERα forms dimers through interaction between DNA binding domains and that dimerization mediated by the ligand binding domain only occurs upon ligand binding but that the complete antagonist ICI 182,780 prevents dimerization through the ligand-binding domain. Heterodimer formation between ERα and ERβ is similarly affected by PKA phosphorylation of serine 236 of ERα. However, 4-hydroxytamoxifen is unable to overcome inhibition of dimerization by PKA. Thus, phosphorylation of ERα in the DNA binding domain provides a mechanism by which dimerization and thereby DNA binding by the estrogen receptor is regulated.
Estrogen receptor α (ERα) mediates the effects of estrogens on cell growth and differentiation. ERα is a member of a superfamily of transcription factors that includes receptors for steroid (androgen, ecdysone, glucocorticoid, mineralocorticoid, estrogen, and progesterone) and thyroid hormones; vitamin D3; retinoic acid; and peroxisome proliferator-, farnesoid-, and arachidonic acid-activated receptors, as well as “orphan” receptors for which no ligands have as yet been identified. These receptors are characterized by highly conserved DNA and ligand-binding domains (LBDs) and regulate transcription by binding to cis-acting enhancer elements in promoters of responsive genes as monomers or as homo- or heterodimers (12, 13, 55, 56, 83). Comparison of the amino acid sequences of ERα from different species shows that the sequences can be divided into six regions, A to F, on the basis of differing amino acid sequence homologies (49). This division can be extended to all other members of the nuclear receptor superfamily. Region C encodes two zinc fingers comprising the DNA-binding domain (DBD). A region N-terminal to the DBD (regions A/B) contains a transcription activation function (AF-1) which can act in a ligand-independent manner when isolated from the LBD. The LBD (region E) contains a ligand-dependent transcription activation function (AF-2). AF-1 and AF-2 activate transcription independently and synergistically and act in a promoter- and cell-specific manner (14, 50–52, 79, 84). Antiestrogens such as tamoxifen and ICI 164,384 antagonize the effects of estrogens by competing with estrogen for binding to ERα. Tamoxifen or its derivative 4-hydroxytamoxifen is a partial antagonist; it inhibits transcriptional activation by AF-2 but enables transcription through AF-1 (14, 58, 59). ICI 164,384, on the other hand, is a complete antagonist which inhibits transcriptional activation by both AF-1 and AF-2 (57, 60). No known antiestrogens prevent DNA binding by hERα, although ICI 164,384 treatment reduces the half-life of ERα (24, 29, 71) and results in the progressive loss of ERα from the nucleus (23). Furthermore, in vivo or in vitro treatment with ICI 164,384 results in a loss in the ability of hERα to bind DNA in vitro at elevated temperatures through a mechanism which is at present unclear (60).
Phosphorylation is a common covalent modification of proteins which provides an important mechanism by which the activity of transcription factors is regulated. Cell surface receptors for polypeptide hormones, cytokines, etc., stimulate signal transduction pathways, leading to phosphorylation and/or dephosphorylation of substrate proteins, including transcription factors (40, 43, 46). The steroid receptors for androgen, glucocorticoids and progesterone, the vitamin D3 receptor, c- and v-erbA, the retinoic acid receptors, and NGFI-B (Nur77) have all been shown to be phosphoproteins. Phosphorylation of these receptors is often ligand induced, although constitutive phosphorylation sites are frequently present in vivo (10, 12, 82, 83), and modulation of the activity of some of these receptors by phosphorylation has been demonstrated. The c-erbA-encoded thyroid hormone receptor and its homologue, v-erbA, found in the genome of the avian erythroblastosis virus, are phosphorylated by protein kinase A (PKA) and PKC (32). Mutation of the PKA and PKC phosphorylation sites abolishes the ability of v-erbA to inhibit erythroid differentiation (31). Modulation of transcriptional activation of the vitamin D3 (36, 37) and progesterone receptors (e.g., see references 11 and 77) by phosphorylation has been demonstrated, whereas phosphorylation of the glucocorticoid receptor appears to be important for nuclear translocation and for cell cycle regulation of its activity (for a review and additional references, see references 25, 38, and 39). The involvement of cell cycle-dependent kinases in regulating nuclear receptor function has recently been described for the progesterone receptor, which is phosphorylated by cdk2 (88). Retinoic acid receptor α is phosphorylated by cdk7, the kinase activity associated with the basal transcription factor TFIIH, the consequence of which is increased transcriptional activation (72).
The possible importance of phosphorylation for ERα function was indicated by the finding that dopamine can activate ERα in the absence of ligand (69). Other groups have since shown that epidermal growth factor, heregulin, insulin, and the insulin-like growth factor TGFα, as well as cyclic AMP (cAMP) and phorbol esters, can also activate ERα (9, 19, 41, 54, 61, 64, and 68). Phosphorylation of ERα on serine 104 and/or serine 106, serines 118 and 167, and on tyrosine 537 has been demonstrated by using deletional or point mutation analyses (1, 20, 44, 53) or by purification and high-pressure liquid chromatography analysis (3–6). Mutation of serine 118 to alanine reduces its transcriptional activation ability (1, 53). Phosphorylation of human ERα at serine 118 is mediated by the RAS/MAPK pathway (19, 47); activation of the MAPK pathway enables ligand-independent transactivation by human ERα (19). Mutation of tyrosine 537 (tyrosine 541 of mouse ERα) enables, variously, ligand-independent transcriptional activation (85, 86) or altered sensitivity to estrogen, and it affects DNA binding and dimerization (7, 8, 20). Phosphorylation of ERα by casein kinase II and pp90rsk1 on serine 167 in vitro has also been demonstrated (3, 45), while tyrosine 537 is phosphorylatable by members of the Src family of tyrosine kinases in vitro (6).
Several studies have shown that upregulation of PKA activity results in ligand-independent activation of ERα (see references 9 and 19), as well as increased phosphorylation (21, 42, 53). We decided to investigate the mechanisms underlying PKA regulation of ERα activity. In vitro phosphorylation of human ERα (hERα) by PKA indicated the presence of at least two phosphorylation sites. Examination of the hERα amino acid sequence showed that there are two potential PKA phosphorylation sites, one within the DBD (serine 236) and a second at the N-terminal boundary of region E (serine 305) (53). Mutation of serine 236 gave reduced phosphorylation by PKA in vitro and in vivo. We further show that mutation of serine 236 to glutamic acid, but not to alanine, dramatically reduced hERα dimerization in the absence of ligand or in the presence of ICI 182,780 but had little effect on dimerization in the presence of estrogen or 4-hydroxytamoxifen. Stimulation of PKA activity also led to the inhibition of ERα dimerization in the absence or in the presence of ICI 182,780 but not in the presence of estrogen or 4-hydroxytamoxifen.
MATERIALS AND METHODS
Recombinants.
The expression vector pSG5 and constructs expressing human ERα (HEG0), deletion mutants expressing N- or C-terminal portions of hERα (HE15 and HEG19), GAL-VP16, and hERβ containing the FLAG epitope at the N terminus (hERβ1) have been described previously (33, 63, 78, 79), as has pSG5-PKA (73). Site-directed mutagenesis of serine 236 to alanine, glutamic acid, and threonine was performed for HEG0, HE15, and HEG19 by using oligonucleotides with the sequences 5′-AAAAACAGGAGGAAGGCCTGCCAGGCGTGTCGGCTC-3′, 5′-AAAAACAGGAGGAAGGAGTGCCAGGCGTGTCGGCTC-3′, and 5′-AAAAACAGGAGGAAGACCTGCCAGGCGTGTCGGCTC-3′, respectively, where changes from the wild-type hERα sequence are underlined. Positive clones were identified by loss of StuI and NaeI sites and confirmed by sequencing. The ERE-driven chloramphenicol acetyltransferase (CAT) gene reporter plasmids have all previously been described (14, 79, 84).
In vitro synthesis of hERα and hERβ polypeptides.
pSG5, HEG0, mutants in which the serine 236 has been altered, and hERβ1 were linearized with SalI. RNA was synthesised with 10 μg of linearized DNA template by using T7 RNA polymerase according to the manufacturer’s protocol (Promega). The RNA synthesized was quantified by spectroscopy at A260/280. Then 5 μg of the synthesized RNA was translated in the presence of 35S-labeled methionine (Amersham International) with rabbit reticulocyte lysate in a final volume of 50 μl according to manufacturer’s protocols (Promega). The synthesized proteins were analyzed by fractionation by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE); the gels were then dried and autoradiographed.
Cell transfection, in vivo labeling, and extraction.
COS-1 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS). Cells were plated in 9-cm petri dishes 16 to 24 h prior to transfection by the calcium phosphate coprecipitation technique with 5 μg of each expression plasmid, made up to a total of 20 μg of DNA with human placental DNA as the carrier. The precipitate was removed 16 h after transfection by a washing in DMEM supplemented with 5% FCS. The cells were harvested after a further 48 h. For experiments in which E2 or antiestrogens were added, the cells were cultured in DMEM without phenol red but supplemented with 5% dextran-coated charcoal-treated FCS (15). Ligands (E2, 10 nM; OHT, 100 nM; ICI, 100 nM), prepared in ethanol, were added 2 h before harvesting. The PKA activators 8-bromo-cAMP (8-Br-cAMP; 100 nM) and the PKA inhibitor H89 (150 nM final) were added at the same time as E2 or the antiestrogens.
For harvesting, transfected cells were washed with chilled phosphate-buffered saline (PBS), harvested in PBS, pelleted, and resuspended in HS buffer (400 mM KCl; 20 mM Tris-HCl, pH 7.5; 2 mM dithiothreitol (DTT); 20% glycerol [vol/vol]; 1 mM phenylmethylsulfonyl fluoride [PMSF]; protease inhibitor cocktail [2.5 μg each of leupeptin, pepstatin, chymostatin, antipain, and aprotinin per ml]). Whole-cell extracts (WCE) were prepared by three cycles of freezing (−80°C) and thawing (0°C), and centrifugation was done at 10,000 × g for 20 min at 4°C. The supernatants were stored at −80°C. The protein concentrations of the WCE were determined with the Bio-Rad protein assay kit (Bio-Rad).
For in vivo labeling with [35S]methionine transient transfections were carried out as described above. Cells were starved for 20 min in DMEM without phenol red or methionine but containing 5% dialyzed and charcoal-stripped FCS, followed by the addition of 150 μCi of 35S-labeled methionine and ligands. Cells were harvested after 1 h in high salt buffer.
For in vivo 32P-labeling of transfected cells, the medium was replaced with phosphate-free DMEM supplemented with 5% dialyzed FCS 64 h after transfection. After 4 h in the absence of phosphate, 1 mCi of [32P]orthophosphate (Amersham International) was added. The cells were labeled for 1 h, harvested, and processed as described above. 8-Br-cAMP (100 nM) was added at the same time as the [32P]orthophosphate.
Immunoprecipitations.
Immunoprecipitations of in vitro translation products or in vivo 35S- or 32P-labeled cell extracts were carried out with 500 μg of protein from WCEs or 50 μl of in vitro translation products; these were added to 1 ml of buffer A (1% Triton X-100; 400 mM NaCl; 1 mM DTT; 40 mM Tris-acetate, pH 7.5; 0.18 mg of PMSF per ml; 1.25 μg each of leupeptin, aprotinin, pepstatin, antitrypsin and chymostatin per ml). The samples were rotated at 4°C for 45 min with 2 mg of protein A-Sepharose, followed by incubation with 2 μg of B10, F3 (2), or M2 (IBI) monoclonal antibodies and incubation at 4°C for 30 min, when 2 μg of rabbit anti-mouse immunoglobulin G (IgG; Sigma, UK) was added and a further incubation at 4°C for 30 min was carried out. Then 2 mg of protein A-Sepharose was added, and the samples were rotated at 4°C for 45 min, followed by four washes with buffer A containing 0.2% SDS. The retained complexes were resolved by SDS-PAGE. The gels were either dried for autoradiography or transferred to nitrocellulose, and immunoblotting was performed prior to autoradiography. For in vitro kinase assays, the immunoprecipitates were processed as described below.
In vitro protein kinase assays.
Immunoprecipitates were washed twice with PKA buffer (10 mM Tris-HCl, pH 7.2; 6.25 mM MgCl2) or with PKC buffer (10 mM Tris-HCl, pH 7.2; 6.25 mM MgCl2; 0.625 mM CaCl2). PKA reactions were carried out in a total volume of 50 μl of PKA buffer containing 25 mM [γ-32P]ATP (specific activity, 1.5 Ci/mmol) and 2 μg/ml of the catalytic subunit of PKA (Promega) at 30°C for 30 min. PKC reactions were performed according to manufacturer’s protocols (Promega). Two washes with 1 ml of the PKA or PKC buffer were carried out after labeling, and the samples were resolved by SDS-PAGE; the gels were then dried and autoradiographed.
Phosphopeptide mapping by two-dimensional separation on thin-layer cellulose plates was performed essentially as described previously (16) with the Hunter thin-layer electrophoresis system (HTLE-7000; CBS Scientific). In brief, in vitro phosphorylated immunoprecipitates were separated by SDS-PAGE; the gels were then dried and autoradiographed. The labeled bands were excised and eluted from gel slices by incubation in 50 mM ammonium bicarbonate with 5% β-mercaptoethanol and 0.1% SDS for 16 to 20 h, followed by precipitation with trichloroacetic acid, oxidation with performic acid, and digestion with 10 μg of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (Sigma) for 8 h at 37°C and a further 8-h digestion with a second 10 μg of trypsin. After three lyophilization steps, the pelleted peptides were resolved in the first dimension by electrophoresis in pH 1.9 buffer (formic acid-glacial acetic acid-water [1:3.1:35.9]) at 1.5 kV for 25 min and in the second dimension by chromatography in pH 7.42 buffer (n-butanol–pyridine–glacial acetic acid–water [5:3.3:1:4]). Phosphopeptides were visualized by autoradiography.
Immunoblot analysis.
WCEs were fractionated by SDS–10% PAGE, and immunodetection was performed with monoclonal antibodies B10 or F3 directed against regions B and F, respectively, to detect hERα (2), except that immunodetection was carried out by incubation with alkaline phosphatase-labeled goat anti-rabbit IgG and revelation with BCIP/NTB (Promega). For detection of hERβ1, blots were incubated with M2 monoclonal antibody (IBI) at 1 μg/ml for 2 h at room temperature, followed by three 15-min washes with 0.05% Tween 20-PBS. The immunoblots were then incubated with alkaline phosphatase-labeled goat anti-mouse IgG for 2 h at room temperature; this was followed by further washing and revelation with BCIP/NTB.
Gel retardation and shift assay.
Gel shift assays (15 μl) contained 1 to 10 μg of WCE, 2.0 μg of poly(dI-dC), and 20,000 cpm of 5′-32P-end-labeled double-stranded estrogen response element (ERE; 5′-TCGAGCAAAGTCAGGTCACAGTGACCTGATCAAT-3′) in 80 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.5 mM DTT, 5% glycerol (vol/vol), 0.5 mM PMSF, and protease inhibitor cocktail (1.25 μg each of leupeptin, pepstatin, chymostatin, antipain, and aprotinin per ml). The mixtures were preincubated at 4°C for 15 min, followed by addition of ERE and incubation at 25°C for 15 min. Receptor-DNA complexes were separated on 5% polyacrylamide gels (30% acrylamide and 1% bisacrylamide, containing 0.5× TBE). Gels were electrophoresed in 0.5× TBE at 150 V and dried before autoradiography.
β-Galactosidase and CAT assays.
COS-1 cells were maintained as described above, split into 9-cm plates in DMEM without phenol red, supplemented with 5% dextran-coated charcoal-stripped FCS, and transfected by the calcium phosphate coprecipitation technique (78). The cells were transfected with 2 μg of the CAT reporter gene, along with 0.5 μg of the β-galactosidase reference plasmid pCH110 (Pharmacia) and 0.5 μg of pSG5, HEG0, HEG0236A, or HEG0236E expression plasmids, together with 1 μg of pSG5-PKA and 10 ng of GAL-VP16 where appropriate. Bluescribe vector M13+ DNA (BSM+; Stratagene) was used as a carrier DNA to make a total of 20 μg of DNA. Ligands (E2, 10 nM; OHT, 100 nM; ICI, 100 nM), prepared in ethanol, were added 30 min after the addition of the precipitates. The precipitates were removed 16 h after transfection by a washing in DMEM without phenol red but supplemented with 5% dextran-coated charcoal-stripped FCS, and fresh ligands were added. Cells were harvested after a further 24 h in 100 μl of 50 mM Tris-HCl (pH 7.5), and extracts were prepared by freeze-thaw three cycles and centrifugation at 10,000 × g for 20 min at 4°C. β-Galactosidase and CAT assays were performed as described previously (1).
RESULTS
Serine 236 of hERα is phosphorylated by PKA.
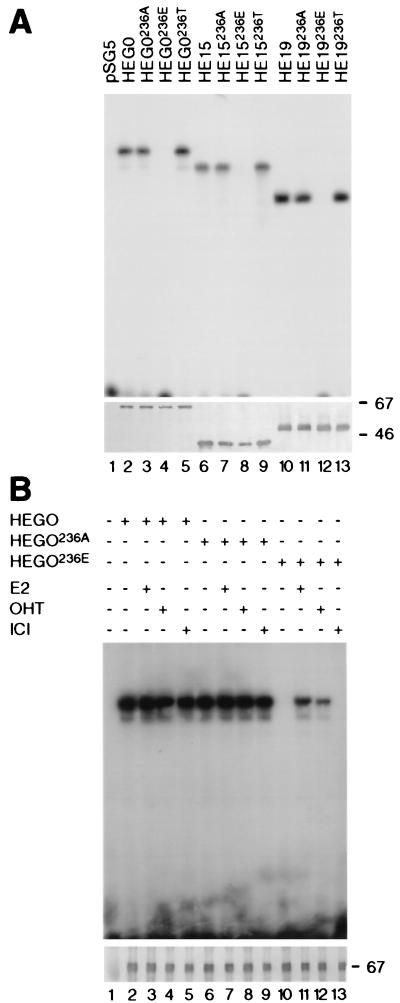
Previous studies have indicated that phosphorylation of hERα is increased by the activation of PKA (see references 21 and 53). In order to directly determine whether PKA can phosphorylate hERα in vitro, we transfected COS-1 cells with an expression vector (pSG5) containing the open reading frame encoding hERα (HEG0). At 48 h after transfection the cells were harvested, WCEs were prepared, and 100 μg of the WCEs were immunoprecipitated with monoclonal antibody F3. Immunoprecipitates were incubated with the purified catalytic subunit of PKA in the presence of [γ-32P]ATP, followed by resolution by SDS-PAGE and autoradiography. Incubation of the immunoprecipitates with PKA resulted in phosphorylation of HEG0 (Fig. 2A, lane 2).
FIG. 2.

Serine 236 is phosphorylated by PKA. (A) WCEs of COS-1 cells transiently transfected with HEG0, HEG19, or HE15 were immunoprecipitated with F3 (lanes 1 and 2) and B10 (lane 3) monoclonal antibodies, phosphorylated with PKA, and analyzed by SDS-PAGE and autoradiography. The solid triangle marks the position of an irrelevant 68-kDa polypeptide observed with the F3 monoclonal antibody and previously described (2). Another triangle at 40 kDa marks the presumed position of the catalytic subunit of PKA used for in vitro phosphorylation. (B) Phosphorylation of HE15 and HE15236A by PKA and PKC. Immunoprecipitates were resolved by SDS-PAGE, transferred to nitrocellulose, immunoprobed with B10, and revealed by the alkaline phosphatase method to determine the levels of HE15 and HE15236A (lower panel). The nitrocellulose membrane was autoradiographed for the phosphorylation signal (upper panel). The filled triangle shows the position of the catalytic subunit of PKA. The positions of molecular size markers (in kilodaltons) are shown. (C and D) Two-dimensional phosphopeptide maps of HE15 and HE15236A, respectively, following in vitro phosphorylation by PKA as in panel B. (D) The empty circles highlight the spots present in the HE15 map but missing from PKA phosphorylation of HE15236A. (E) COS-1 cells transiently transfected with pSG5 (lane 1), HEG0 (lanes 2, 3, and 6), or HEG0236A (lanes 4, 5, and 7) in the absence (lanes 1, 2, 4, 6, and 7) or the presence (lanes 3 and 5) of pSG5-PKA were labeled with [32P]orthophosphate. 8-Br-cAMP (100 nM) was added as appropriate (lanes 6 and 7). Immunoprecipitations were resolved by SDS-PAGE and autoradiographed (upper panel). Immunoblotting with monoclonal antibody B10 was used to control for hERα protein levels (lower panel).
hERα deletion mutants lacking either the N-terminal A/B region (HEG19) or the LBD and region F (HE15) (Fig. 1A) were transfected into COS-1 cells, and WCEs were immunoprecipitated with monoclonal antibodies F3 or B10, respectively, and phosphorylated with the catalytic subunit of PKA. Both HEG19 and HE15 were phosphorylated by PKA in vitro (Fig. 2A, lanes 1 and 3), indicating either that PKA phosphorylates hERα within or near the DBD and/or that hERα is phosphorylated by PKA at sites in the A/B region and in the LBD.
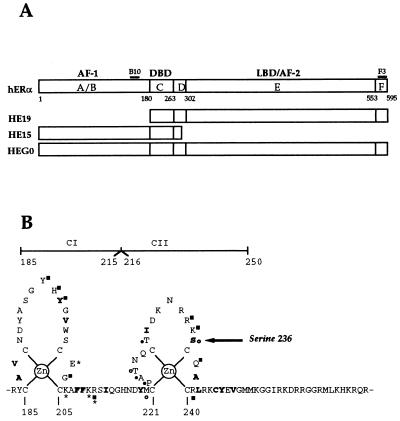
FIG. 1.
Schematic representation of hERα. (A) Regions A to F, as initially described by Krust et al. (49) are depicted, the numbers below referring to amino acid positions of the boundaries for the different regions. Also shown are the positions of the epitopes for monoclonal antibodies B10 and F3. The portion of hERα coding region contained within HEG0, HE15, and HEG19 are also shown. (B) The amino acids comprising the core DBD and their involvement in DNA binding, as determined crystallographically, is represented. The amino acids marked in boldface participate in the hydrophobic core. Asterisks mark the residues which interact with base pairs; those making phosphate contacts (squares) or participating in the dimer interface (circles) are also indicated. Closed and open circles and squares denote direct interactions and interactions via ordered water molecules, respectively (75). The position of serine 236 is marked by an arrow.
Examination of the amino acid sequence of hERα showed that two serine residues, serine 236 (NRRKSC) in the DBD (Fig. 1B) and serine 305 (SKKNSL) in the LBD could be potential PKA phosphorylation sites (53). Only one of these sites (serine 236) is present in both HE15 and HEG19 and was mutated to the nonphosphorylatable residue alanine. Immunoprecipitates were incubated with PKA as described above, resolved by SDS-PAGE, and transferred to nitrocellulose. Immunoblotting with B10 was performed to compare levels of HE15 and HE15236A protein, and autoradiography of the nitrocellulose membrane showed levels of phosphorylation. HE15236A was phosphorylated to a lower level than HE15, indicating that serine 236 is a substrate for PKA in vitro (Fig. 2B). HE15 was also phosphorylatable by PKC in vitro, albeit to a much lower degree. HE15 and HE15236A were phosphorylated to a similar extent by PKC (Fig. 2B, lanes 5 and 6, and data not shown). No signal was obtained when PKA or PKC were omitted from the kinase assay (data not shown).
Two-dimensional phosphopeptide mapping of in vitro-phosphorylated immunoprecipitates showed the absence of three spots (dotted) from HE15236A (Fig. 2D) compared with HE15 (Fig. 2C), indicating that serine 236 is phosphorylated by PKA in vitro. The two strong spots in both the HE15 and HE15236A phosphopeptide maps suggest the presence of at least one other major PKA site resides within amino acids 1 to 281 of hERα (HE15).
We next examined the ability of PKA to alter the phosphorylated state of wild-type hERα (HEG0) in vivo. COS-1 cells transiently transfected with HEG0 were labelled with [32P]orthophosphate and subjected to immunoprecipitation, SDS-PAGE, and autoradiography. Immunoblotting with monoclonal antibody B10 was used to control for equivalent levels of ER protein (Fig. 2E). A single band at 67 kDa in HEG0- but not in pSG5-transfected cells shows that hERα is phosphorylated in untreated cells (Fig. 2E, lanes 1 and 2). Cotransfection of pSG5-PKA resulted in a 5.6-fold increase in the intensity of the phosphorylated band (lane 3). 8-Br-cAMP treatment similarly increased hERα phosphorylation 3.65-fold (lane 6). HEG0236A was phosphorylated to a lower extent than was HEG0 (lane 4), and cotransfection of pSG5-PKA (lane 5) or the addition of 8-Br-cAMP (lane 7) resulted in lower increases (2.7- and 1.44-fold, respectively) in phosphorylation intensities compared to HEG0; 2.0- and 2.5-fold-lower signals were obtained for HEG0236A than for HEG0 with pSG5-PKA and 8-Br-cAMP, respectively.
Comparison of in vitro DNA binding by wild-type and mutant hERα.
Since serine 236 lies within the second zinc finger (CII) of the DBD (Fig. 1B) we wished to investigate the effect of its mutation on DNA binding by hERα. The mutants in which serine 236 was replaced by alanine in HEG0, HE15, and HEG19 were used to investigate DNA binding when phosphorylation at this position is prevented. Mutants in which serine 236 of HEG0, HE15, and HEG19 were replaced by glutamic acid were also created in order to examine the effect of a “constitutive” negative charge at this position. As PKA is a serine-threonine kinase, serine 236 was also mutated to threonine.
HEG0, HE15, and the serine 236 mutants were overexpressed in COS-1 cells by transient transfection; cells were then harvested, and extracts were prepared in a high salt buffer. Gel shifts were performed by preincubation of the extracts in gel shift buffer (containing 80 mM KCl) at 4°C for 15 min, followed by the addition of radiolabeled ERE and incubation at 25°C for 15 min, before the receptor-ERE complexes were analyzed by PAGE as previously described (59, 63). A specific retarded complex was observed for HEG0, and faster-migrating complexes were seen for HE15 and HEG19 (Fig. 3A, lanes 2, 6, and 10). Mutation of serine 236 to alanine or threonine had no obvious effect on complex formation (Fig. 3A, lanes 3, 5, 7, 9, 11, and 13). Interestingly, replacement of serine 236 with glutamic acid drastically reduced the amount of complex seen (Fig. 3A, lanes 4, 8, and 12). Immunoblotting showed that the reduced DNA binding by HEG0236E, HE15236E, and HEG19236E was not due to the absence of protein or to its degradation during incubation (Fig. 3A, lower panel). These results suggest that the phosphorylation of serine 236 may adversely affect DNA binding by hERα.
FIG. 3.
Comparison of the in vitro DNA binding by wild-type hERα and mutants of serine 236. (A) COS-1 cells were transiently transfected with pSG5 (lane 1), HEG0 (lane 2), mutants of HEG0 (lanes 3 to 5), HE15 and its mutants (lanes 6 to 10), or HEG19 and its mutants (lanes 10 to 13) in the presence of 10−7 M E2. Lysates were prepared in HS buffer, and gel shifts were performed as described in Materials and Methods. (B) COS-1 cells were transiently transfected with pSG5 (lane 1), HEG0 (lanes 2 to 5), HEG0236A (lanes 6 to 9), or HEG0236E (lanes 10 to 13). Carrier (ethanol) or the ligands E2, OHT, or ICI (prepared in ethanol) were added to a final concentration of 10−7 M 2 h before harvesting. Western blot analysis of the extracts was performed with monoclonal antibodies B10 (panel A, lanes 1 to 9; panel B) or F3 (panel A, lanes 10 to 13) to control for the levels of receptor proteins.
In order to investigate the effect of ligand on DNA binding by the wild-type and mutant ERα, COS-1 cells grown in DMEM lacking phenol red and containing 5% charcoal-stripped FCS were transfected with HEG0, HEG0236A, or HEG0236E, and ligands were added 1 h prior to harvesting. No differences in DNA binding were observed between HEG0 and HEG0236A in the absence of ligand or in the presence of 17β-estradiol (E2), the partial agonist 4-hydroxytamoxifen (OHT), or the complete antagonist ICI 182,780 (ICI) (Fig. 3B lanes 2 to 9). As described above (Fig. 3A), little binding was evident for HEG0236E in the absence of ligand (Fig. 3B, lane 10). In the presence of E2 and OHT, however, some DNA binding was seen (lanes 11 and 12), although no DNA binding was observed with ICI (lane 13), suggesting that the inhibition of DNA binding can be prevented, at least partially, by some ligands. Again, immunoblotting was used to control for levels of protein and degradation (Fig. 3B, lower panel).
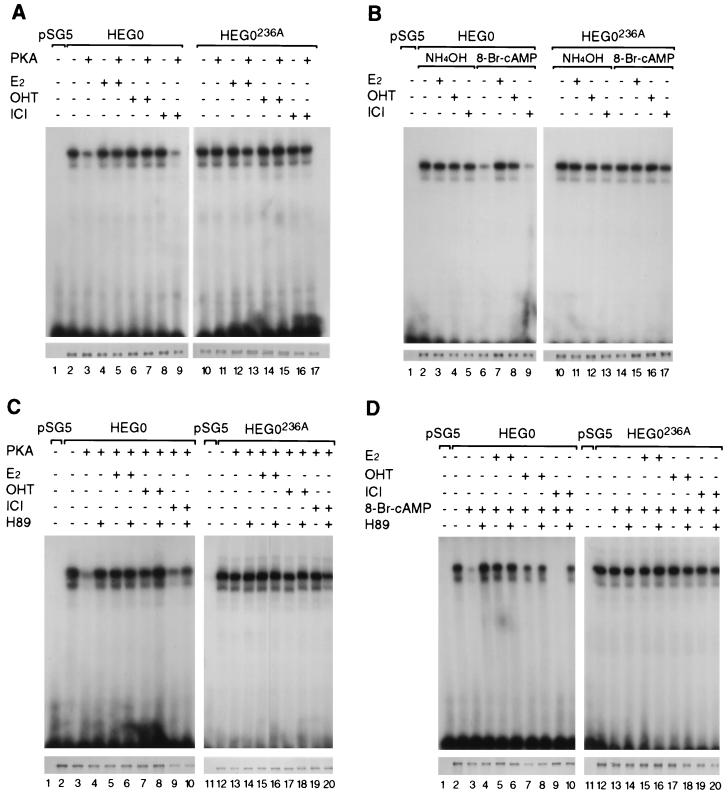
In order to demonstrate that the apparent inhibition of DNA binding is due to phosphorylation of serine 236 and to evaluate its ligand dependence, cells were transfected with HEG0 or HEG0236A, as well as the expression vector pSG5 containing the open reading frame encoding the catalytic subunit of PKA (pSG5-PKA) (73). Gel shifts performed under standard conditions demonstrated that the overexpression of PKA had little effect on DNA binding by HEG0 in the presence of E2 or OHT (Fig. 4A, lanes 4 to 7). In the absence of ligand (Fig. 4A, lanes 2 and 3) or in the presence of ICI (Fig. 4A, lanes 8 and 9), however, in vitro DNA binding was dramatically reduced when HEG0 was cotransfected with pSG5-PKA. No reduction in DNA binding was observed when HEG0236A was cotransfected with pSG5-PKA in the presence or absence of any of the ligands (Fig. 4A, lanes 10 to 17).
FIG. 4.
DNA binding by hERα is inhibited by phosphorylation of serine 236. (A to D) COS-1 cells were transiently transfected with pSG5, HEG0, or HEG0236A. E2, OHT, and ICI were added 2 h prior to harvesting. (A and C) An expression plasmid encoding the catalytic subunit of PKA (63) was transfected, along with the pSG5, HEG0, or HEG0236A. (B and D) Carrier (NH4OH) or 8-Br-cAMP was added at the same time as E2, OHT, or ICI. (C and D) The PKA inhibitor H89 was also added at the same time as E2, OHT, or ICI. Immunoblotting with B10 was performed to control for levels of receptor protein (A to D, lower panels).
Activation of endogenous PKA by 8-Br-cAMP resulted in a similar loss of DNA binding by HEG0 in the absence of ligand (Fig. 4B, compare lanes 2 and 6) or in the presence of ICI (Fig. 4B, compare lanes 5 and 9) but not in the presence of E2 or OHT (Fig. 4B, lanes 3, 4, 7, and 8). As seen when PKA was overexpressed, no differences in DNA binding were evident for HEG0236A in the absence or presence of 8-Br-cAMP (Fig. 4B, lanes 10 to 17). Similar results were obtained when forskolin was used (data not shown). Overexpression of the catalytic subunit of PKA or activation of PKA with 8-Br-cAMP did not result in altered protein levels in transfections, nor did they increase the degradation of wild-type or mutant ERα (Fig. 4A and B, lower panels).
H89 is a specific inhibitor of PKA (35). Treatment with H89 prevented the loss of DNA binding by HEG0, a result observed in the absence of ligand when PKA was overexpressed (Fig. 4C, compare lanes 2 and 3 with lane 4). The loss of DNA binding observed in the presence of ICI was also prevented when H89 was added (Fig. 4C, compare lanes 8 and 9 with lane 10). The reduction in DNA binding induced by 8-Br-cAMP was similarly prevented by H89 (Fig. 4D). There was no effect of H89 on the DNA binding by HEG0 in the presence of E2 or OHT (Fig. 4C and 4D, lanes 5 to 8) or on the DNA binding by HEG0236A in the absence of ligand or in the presence of E2, OHT, or ICI (Fig. 4C and 4D, lanes 11 to 20).
Replacement of serine 236 by glutamic acid inhibits dimerization by hERα.
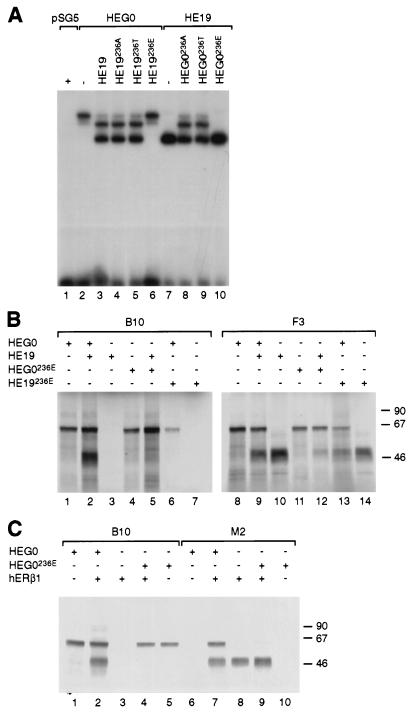
ERα binds DNA as a homodimer or as a heterodimer with ERβ (63). Inability of the receptor to dimerize would result in the loss of DNA binding. In order to investigate the effect of the mutation of serine 236 on dimerization, we analyzed the dimeric status of hERα bound in vitro to an ERE by using extracts of COS-1 cells transfected with HEG0 or HEG19 and cotransfected with the mutants of serine 236. The HEG19-ERE complex migrates faster than the complex obtained with HEG0 (Fig. 3A; Fig. 5A compare lanes 2 and 7). A weak lower band observed for HEG0 probably corresponds to a degradation product (see also Fig. 3 and 4) and has also previously been observed (60). When coexpressed in COS-1 cells, an additional complex migrating at a position intermediate between those observed for HEG0 and HEG19 is observed, corresponding to the binding of HEG0-HEG19 heterodimers (Fig. 5A, lane 3). HEG0 and either HEG19236A or HEG19236T formed similar amounts of heterodimer as HEG0 and HEG19 (Fig. 5A, lanes 3 to 5). Heterodimer formation was similar when HEG0236A or HEG0236T were cotransfected with HEG19 (Fig. 5A, lanes 8 to 9). When HEG0 was cotransfected with HEG19236E, however, a complex corresponding to HEG19236E was not observed, nor was a heterodimeric complex seen while the HEG0 homodimeric complex was present at levels similar to those seen with HEG0 alone (Fig. 5A, compare lanes 2 and 6). The reciprocal event was observed when HEG0236E was cotransfected with HEG19 (Fig. 5A, compare lanes 7 and 10). The fact that the amount of HEG0 and HEG19 complexes observed when cotransfected with HEG19236E and HEG0236E, respectively, is similar to levels observed for HEG0 and HEG19 alone suggests that dimerization is impaired by mutation of serine 236 to glutamic acid.
FIG. 5.
Loss of hERα DNA binding is due to inhibition of dimerization. (A) Gel shifts were performed with extracts prepared from COS-1 cells transfected with HEG0 alone (lane 2) or together with wild-type HEG19 or serine 236 mutants (lanes 3 to 6) and HEG19 alone (lane 7) or together with serine 236 mutants of HEG0 (lanes 8 to 10). (B) HEG0, HEG19, HEG0236E, and HEG19236E were synthesised in vitro either separately or together in the presence of [35S]methionine. The in vitro translation products were divided in two and immunoprecipitated with the B10 monoclonal antibody (lanes 1 to 7). Immunoprecipitation with the F3 (lanes 8 to 14) monoclonal antibody was used to show that HEG0, HEG19, and the 236E mutants are present in the appropriate in vitro translations. (C) HEG0, HEG0236E, and hERβ1 were synthesized in vitro either separately or together in the presence of [35S]methionine. The in vitro translation products were divided in two and immunoprecipitated with B10 (lanes 1 to 5) or M2 (lanes 6 to 10) monoclonal antibodies.
The effect of mutation of serine 236 to glutamic acid upon dimerization was ascertained directly by using an assay in which HEG0, HEG19, and their respective glutamic acid mutants were synthesised by in vitro transcription followed by in vitro translation with a rabbit reticulocyte lysate system in the presence of 35S-labeled methionine, followed by immunoprecipitation of one-half of the lysates with the monoclonal antibody B10, which recognizes an epitope in the A/B region and does not therefore, recognize HEG19 (or its mutants) (Fig. 1A). Immunoprecipitation of HEG0 gave a band at 67 kDa, whereas HEG19 was not immunoprecipitated by B10 (Fig. 5B, compare lanes 1 and 3). Immunoprecipitation of HEG0 and HEG19 cotranslated with B10 resulted in the detection of HEG0 and HEG19, showing that they can dimerize. Only one band, at 67 kDa, was observed when HEG0236E and HEG19 (Fig. 5B, lane 5) or HEG0 and HEG19236E (Fig. 5B, lane 6) were cotranslated and immunoprecipitated with B10, indicating that the mutation of serine 236 to glutamic acid impairs dimerization by hERα. Immunoprecipitation of the second half of all of the lysates with F3, which recognizes an epitope present in both HEG0 and HEG19 (Fig. 1A), was also performed to show that HEG19 and HEG19236E were not absent in lysates in which they were not coprecipitated with HEG0236E (Fig. 5B, lanes 8 to 14).
We and others have previously shown that hERα and hERβ can heterodimerize (22, 62, 63, 65). Given the almost complete amino acid sequence identity between hERα and β in the DBD and their ability to heterodimerize, we investigated whether hERα and β can heterodimerize in solution and whether heterodimerization between hERα and β is impaired by mutation of serine 236 of hERα to glutamic acid. HEG0, HEG0236E, and hERβ1 (pSG5 containing the hERβ open reading frame with the FLAG tag at the 5′ end, enabling immunodetection with the M2 monoclonal antibody [63]) were in vitro translated in the presence of 35S-labeled methionine, and immunoprecipitations were performed with B10 and M2 monoclonal antibodies as shown (Fig. 5C). Immunoprecipitation of HEG0 with hERβ1 by using B10 or M2 resulted in immunoprecipitation of both polypeptides (Fig. 5C, lanes 2 and 7), whereas when HEG0236E and hERβ1 were immunoprecipitated with B10 only HEG0236E was seen (Fig. 5C, lane 4). Immunoprecipitation with M2 resulted in the coprecipitation of HEG0 but not of HEG0236E with hERβ1 (Fig. 5C), indicating that serine 236 is important for the dimerization of hERα and -β, as well as for homodimerization of hERα.
Phosphorylation on serine 236 inhibits dimerization by hERα in the absence of ligand.
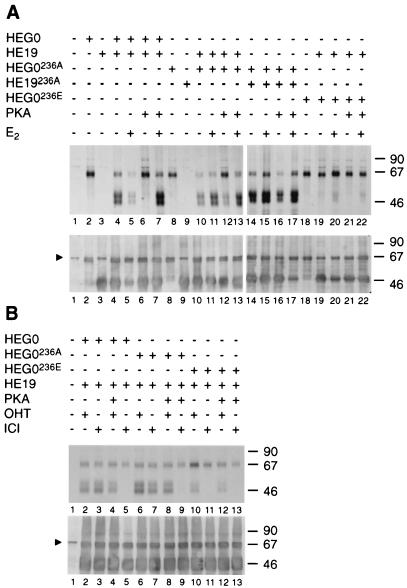
E2 and OHT can overcome the inhibitory effect on DNA binding of mutating serine 236 to glutamic acid or activation of PKA (Fig. 3 and 4). The gel shift and immunoprecipitation results described above show that the 236E mutants fail to dimerize (Fig. 5). In order to determine whether the inhibition of dimerization is overcome by ligand treatment, COS-1 cells were transiently transfected with HEG0 and HEG19 or their respective mutants, either separately or together. The cells were labelled with [35S]methionine, and WCEs were immunoprecipitated with B10. Immunoblotting of extracts with F3 established that HEG0 and/or HEG19 (and their mutants) were present in the appropriate extracts (Fig. 6B, lower panel). In order to investigate the effects of phosphorylation of serine 236, the cells were transfected with pSG5 or pSG5-PKA in addition to the ERα constructs.
FIG. 6.
Inhibition of dimerization by PKA is prevented by 17β-estradiol and 4-hydroxytamoxifen but not by ICI 182,780. COS-1 cells were transiently transfected with HEG0, HEG19, HEG0236A, HEG19236A, HEG0236E, and HEG19236A and pSG5-PKA as shown. The cells were labelled for 1 h with [35S]methionine in the presence or absence of E2, OHT, or ICI (final concentrations, 10−7 M). The extracts were divided in two and immunoprecipitated with B10 (upper panels) or F3 (lower panels) as shown. Triangles indicate the position of a nonspecific band (2). The molecular size markers are shown (in kilodaltons) on the right.
Monoclonal antibody B10 immunoprecipitated HEG0, HEG0236A, and HEG0236E (Fig. 6A, lanes 2, 8, and 18, respectively) but not HEG19 or HEG19236A (lanes 3 and 9). As expected, immunoprecipitation of WCEs from cells transfected with HEG0 and HEG19 resulted in precipitation of HEG19 along with HEG0 in the presence or absence of E2 (Fig. 6A, lanes 4 and 5). When HEG0 and HEG19 were transfected along with pSG5-PKA, however, HEG19 was not brought down in the absence of ligand (Fig. 6A, lane 6). HEG19 was brought down when E2 was added in vivo (lane 7), indicating that phosphorylation of hERα by PKA inhibits dimerization in vivo in the absence but not in the presence of E2. In agreement with this, immunoprecipitation of WCEs containing HEG0236A, HEG19, and pSG5-PKA or containing HEG0236A, HEG19236A, and pSG5-PKA resulted in the coprecipitation of HEG19 and HEG19236A, respectively, in the presence or absence of E2 (Fig. 6A, lanes 10 to 17). Immunoprecipitation of WCEs from cells transfected with HEG0236E and HEG19 resulted in coprecipitation of HEG19 in the presence but not in the absence of E2 (Fig. 6A, lanes 19 and 20).
Similar experiments performed in the presence of OHT or ICI showed that the PKA-mediated inhibition of dimerization is prevented by OHT (Fig. 6B, lanes 2 and 4). OHT also enabled the dimerization between HEG0236E and HEG19 (lane 10). The addition of ICI, however, did not prevent inhibition of dimerization by PKA (Fig. 6B, lanes 3 and 5), nor did HEG0236E and HEG19 dimerize in the presence of ICI (lane 11).
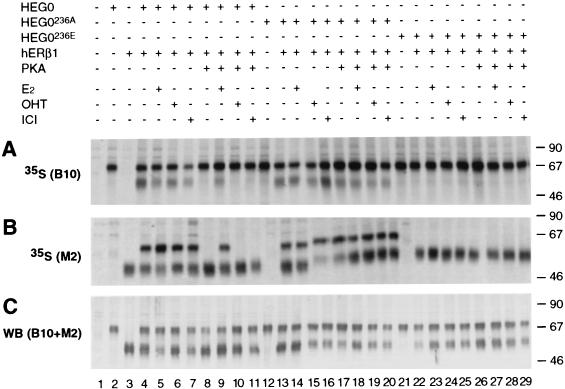
The importance of ligands for heterodimerization of hERα and -β was also investigated. COS-1 cells were transiently transfected with HEG0, HEG0236A, or HEG0236E in the presence or absence of hERβ1 and pSG5-PKA and labeled with [35S]methionine. The lysates were divided into three equal fractions, and immunoprecipitations were performed with B10 (Fig. 7A) or M2 (Fig. 7B) monoclonal antibodies. As expected, hERβ1 was coimmunoprecipitated with HEG0 in the presence or absence of ligand (Fig. 7A, lanes 4 to 7). Overexpression of the catalytic subunit of PKA (pSG5-PKA), however, inhibited dimer formation in the absence of ligand (Fig. 7A, lane 8), and addition of ICI did not prevent the inhibition (Fig. 7A, lane 11), as found for hERα. Addition of E2 enabled heterodimer formation (Fig. 7A, lane 9). However, whereas the addition of OHT allowed dimerization between HEG0 and HEG19, dimer formation between HEG0 and hERβ1 in the presence of OHT was undetectable when the catalytic subunit of PKA was overexpressed (Fig. 7A, lane 10). Dimer formation between HEG0236A and hERβ1 was ligand independent and was not inhibited by PKA (Fig. 7A, lanes 13 to 20). Furthermore, in contrast to the observations with hERα, no heterodimer formation between HEG0236E and hERβ1 was seen in the presence of E2 or OHT (Fig. 7A, lanes 22 to 29). Immunoprecipitations performed with M2 to immunoprecipitate hERβ1 gave results similar to those obtained for HEG0 with B10 (Fig. 7B). Immunoblotting of the remainder of these lysates with B10 and M2 served to control for the presence of HEG0 (or mutants of serine 236) and hERβ1, respectively, in the appropriate lysates (Fig. 7C).
FIG. 7.
Dimerization between hERα and -β is inhibited by PKA. COS-1 cell extracts were immunoprecipitated with B10 (A) or M2 (B) after transient transfection with HEG0, HEG0236A, HEG0236E, and/or hERβ1, as well as pSG5-PKA (as appropriate), and [35S]methionine labeling in the presence or absence of ligands as described in Fig. 6. (C) Immunoblotting with the B10 and M2 monoclonal antibodies, used together, served to control for the presence of HEG0, HEG0236A, HEG0236E, and hERβ1 in the appropriate cell lysates. The molecular size markers are shown (in kilodaltons) on the right.
Stimulation of hERα-mediated transcriptional activation by PKA is modulated in a promoter-specific manner.
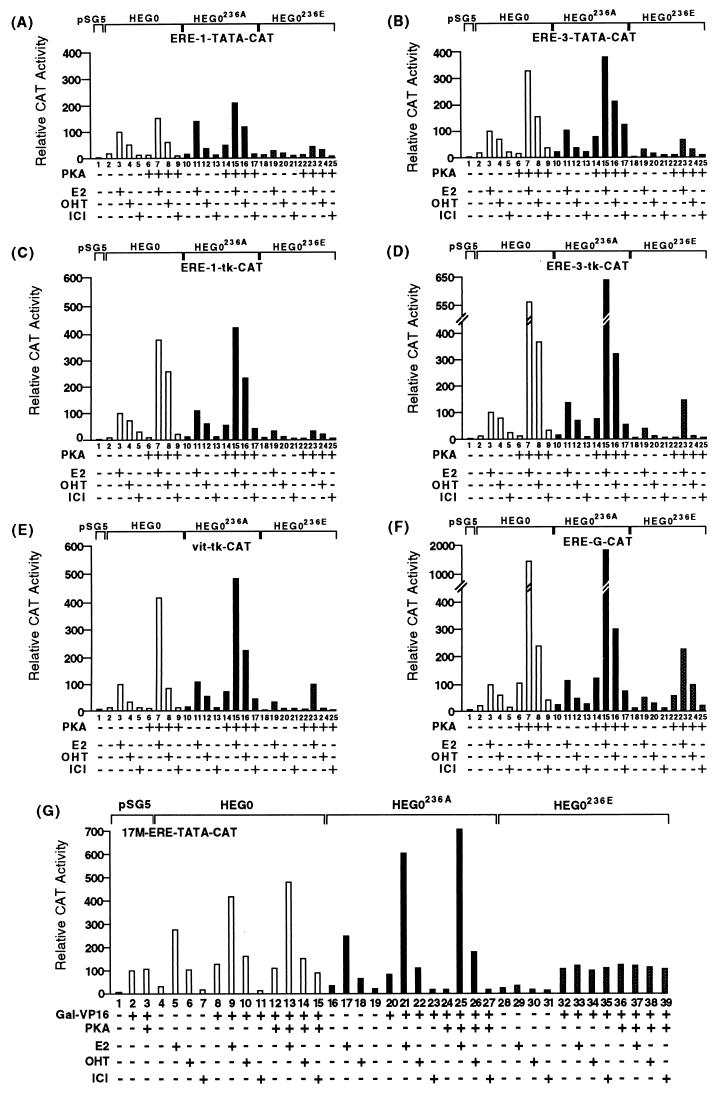
The results described above indicate that phosphorylation of serine 236 inhibits hERα dimerization in the absence of ligand or in the presence of ICI. In order to determine whether overexpression of PKA results in the prevention of transcriptional activation by hERα in a serine 236-dependent manner, we examined the ability of wild-type and mutant hERα to activate ERE-containing reporter genes. Previous studies have shown that stimulation of PKA results in a synergistic increase in transcriptional activation by hERα in the presence of E2 or OHT in a cell type-specific and promoter-independent manner, whereas PKA-induced transactivation by ERα in the absence of ligand is not observed in MCF7 cells (21) and in other cell types is promoter context dependent (42).
We examined transcriptional activation by HEG0, HEG0236A, and HEG0236E in COS-1 cells with a number of CAT-based reporters driven by minimal promoters containing one or three EREs (ERE-TATA-CAT and ERE-3-TATA-CAT) or more complex promoters comprising one or three EREs and the thymidine kinase (ERE-1-tk-CAT and ERE-3-tk-CAT) or the globin promoter (ERE-G-CAT) (Fig. 8). HEG0 activated each promoter in the presence of E2 and to a lesser extent in the presence of the partial agonist OHT (Fig. 8A to F, lanes 2 to 5). Little transactivation was observed in the presence of ICI or in the absence of ligand. In the case of each reporter levels of activation were similar for HEG0236A and HEG0 (Fig. 8A to F, compare lanes 2 to 5 with lanes 10 to 13). Transactivation by HEG0236E was lower than that observed for HEG0, although transcriptional activation in the presence of E2 and OHT was evident in the presence of E2 ranging between 23% (Fig. 8A) and 60% (Fig. 8F) relative to HEG0. Similar differences in the levels of trans-activation were observed for HEG0236E compared to HEG0 in the presence of OHT (Fig. 8A to F, compare lanes 2 to 5 with lanes 18 to 21).
FIG. 8.
Comparison of the transcriptional activities of hERα mutants on various estrogen-responsive reporter genes. (A to F) Transcriptional stimulation in COS-1 cells of six reporter genes by the wild-type human ERα (HEG0) and the serine 236 mutants HEG0236A and HEG0236E, together with pSG5 or pSG5-PKA, in the presence or absence of E2 (10 nM), OHT (100 nM), and ICI (100 nM) as indicated. The results are displayed in the form of a bar chart. The average values of at least three independent experiments (±10%) are given in each case. Transcriptional activation was determined with ERE-TATA-CAT (A), ERE-3-TATA-CAT (B), ERE-tk-CAT (C), ERE-3-tk-CAT (D), vit-tk-CAT (E), and ERE-G-CAT (F). For each reporter gene the level of transactivation by HEG0 in the presence of E2 was taken as 100%. In panel G COS-1 cells were cotransfected with the reporter gene 17M-ERE-TATA-CAT, HEG0, HEG0236A, or HEG0236E, together with either pSG5 or pSG5-PKA and GAL-VP16, as indicated. Ligands were added as described above. The results are displayed in the form of a bar chart. The average values of at least three independent experiments (±10%) are given in each case. The level of transactivation by GAL-VP16 was taken as 100%.
Cotransfection of HEG0 with pSG5-PKA resulted in a 1.6- to 14-fold increase (relative to the activity observed in the absence of PKA) in transactivation in the presence of E2 depending on the reporter gene used (Fig. 8A to F, compare lanes 3 and 7). PKA resulted in a 1.0- to 4.7-fold increase in OHT-induced transactivation by HEG0 (Fig. 8A to F, compare lanes 4 and 8). In the absence of ligand, cotransfection of pSG5-PKA did not yield any increase in transactivation in the case of the ERE-TATA-CAT, ERE-1-tk-CAT, ERE-3-tk-CAT, or the vit-tk-CAT reporter genes (Fig. 8A, C, D, and E), whereas 1.4- and 4.9-fold increases were observed for ERE-3-TATA-CAT and ERE-G-CAT (Fig. 8B and F), respectively. PKA also caused 1.5- and 2.2-fold increases in transactivation in the presence of the pure antiestrogen ICI in the case of ERE-3-TATA-CAT and ERE-G-CAT, respectively (Fig. 8B and F, compare lanes 5 and 9), but no stimulation was observed with the other reporter genes.
These results show that PKA synergizes with E2 and OHT to increase the transactivational ability of hERα. PKA-mediated transactivation in the absence of ligand is promoter dependent and, furthermore, PKA can also stimulate transactivation by hERα in the presence of the “pure” antiestrogen ICI, again in a promoter-specific manner. Similarly, in MCF7 cells transfection of PKA or the addition of 8-Br-cAMP resulted in increased transactivation in the presence of E2 and OHT from all of the reporter genes described above. However, no increase in transactivation in the presence of ICI or the absence of ligand was observed with any reporter gene, thus confirming previous reports indicating that ligand-independent transactivation mediated by PKA is cell type specific (data not shown) (9, 21, 42).
For HEG0236A, PKA-mediated increases in transactivation were similar to those observed for HEG0 in the presence of E2 and OHT (Fig. 8A to F, compare lanes 11 and 15 and lanes 12 and 16). Whereas PKA stimulated transactivation by HEG0 in the absence of ligand only when the ERE-G-CAT reporter was used, in the case of HEG0236A, increased transactivation by PKA was evident for all reporter genes (3.2- to 4.8-fold) (Fig. 8A to E, compare lanes 10 and 14). Similar increases were observed in the presence of ICI (Fig. 8A to F, lanes 13 and 17).
PKA also increased transactivation by HEG0236E in the presence of E2 and to a lesser extent in the presence of OHT (Fig. 8A to F, compare lanes 19 and 20 with lanes 23 and 24). As expected, no increase in transactivation was observed in the presence of ICI or in the absence of ligand, except that ERE-G-CAT (which gave a ligand-independent increase in transactivation of HEG0 by PKA) was stimulated to a similar extent (3.2-fold) in the absence of ligand (compare lanes 18 and 22) and in the presence of E2 or OHT (Fig. 8F, compare lanes 19 and 20 and lanes 23 and 24). Some increase (twofold) was also obtained in the presence of ICI (Fig. 8F, compare lanes 21 and 25).
Collectively, the above data indicate that phosphorylation of serine 236 does indeed inhibit the transcriptional activation by hERα in the absence of E2 or OHT. In order to directly show that PKA can inhibit DNA binding by hERα in vivo, we exploited a transcriptional interference assay which has previously been used to show that hERα can bind to an ERE in vivo in the presence of ICI (60). We used a reporter gene containing a binding site for the yeast transcriptional activator GAL4 (17M), an ERE, and the adenovirus major late promoter TATA box driving the CAT gene (79). Cotransfection of this reporter gene (17M-ERE-TATA-CAT) and GAL-VP16, in which the DBD (amino acids 1 to 147) of GAL4 is fused to the transcription activation function of the herpes simplex virus VP16 resulted in transcriptional activation (taken as 100%), which was unaffected by the coexpression of PKA (Fig. 8G lanes 2 and 3). Cotransfection of HEG0 and GAL-VP16 resulted in a 4-fold increase in CAT gene expression in the presence of E2 (lane 9) and a 1.8-fold increase in the presence of OHT (lane 10). In the presence of ICI, however, a fourfold decrease in expression was observed, a finding indicative of repression due to the specific DNA binding of IC-bound HEG0 (lane 11). It is not clear why the inhibition of expression is not seen in the absence of ligand (lane 8), but it may be a reflection of residual estrogens in the culture medium (see reference 60). Cotransfection of pSG5-PKA resulted in a further increase in transactivation in the presence of E2 and OHT but a repression of expression in the presence of ICI was lost (lane 15), thus showing that PKA inhibits DNA binding by HEG0 in the presence of ICI.
In the case of HEG0236A, ICI inhibited expression even in the presence of PKA (lane 27), indicating that DNA binding by HEG0236A is not inhibited by PKA. HEG0236E cotransfection with GAL-VP16 had little effect on expression in the absence or presence of PKA, suggesting that there is little DNA binding by HEG0236E (Fig. 8G, lanes 32 to 39), a finding in agreement with the results presented above.
DISCUSSION
hERα is phosphorylated on serine 236 by PKA.
Several studies have shown that the activation of PKA can increase transcriptional activation by ERα in an estrogen-independent manner (9, 19, 42) or in synergism with estrogen (21, 26, 42), as well as in the presence of antiestrogens (42). We have now shown that hERα (HEG0) is phosphorylated by PKA in vitro on at least two sites. Examination of the hERα amino acid sequence indicates that serine 236 is a potential substrate for PKA phosphorylation and that its mutation to alanine reduced, but did not abolish, phosphorylation of hERα by PKA, suggesting that serine 236 is a substrate for PKA phosphorylation.
Mutation of serine 236 to glutamic acid inhibits dimerization by hERα.
Serine 236 is located in the second zinc finger (CII) within the DBD. Although mutation of serine 236 to alanine had little effect on DNA binding, a mutation to glutamic acid dramatically reduced DNA binding by hERα, suggesting that phosphorylation of serine 236 inhibits DNA binding. Like other steroid receptors, ERα binds to specific EREs as a homodimer (12, 13, 34, 55, 56, 83). Recent studies have further shown that ERα can bind to EREs as a heterodimer with the newly discovered ERβ (22, 62, 63, 65). Extensive deletional and mutational analyses have shown that homodimerization by ERα is mediated by sequences in the DBD and the LBD, and determination of the crystal structures of the ERα DNA and LBDs have further delineated the amino acid residues involved in dimerization (17, 75). Since phosphorylation of serine 236 inhibits DNA binding, we investigated whether this is due to the inhibition of dimerization by hERα. Gelshifts performed with WCE of COS-1 cells transfected with full-length (HEG0, HEG0236A, or HEG0236E) and N-terminally deleted (HEG19, HEG19236A, or HEG19236E) receptors suggested that the wild-type and 236A receptors can heterodimerize, whereas the 236E mutants are unable to dimerize.
Inhibition of dimer formation was further demonstrated by using immunoprecipitations. HEG19 could be immunoprecipitated with HEG0 with an antibody recognizing an epitope absent in HEG19. HEG19 could not be coprecipitated with HEG0236E, nor was HEG19236E coprecipitated with HEG0, clearly showing that dimerization by the 236E mutant is impaired when compared to the wild-type and 236A mutant receptors.
As stated above, dimerization by ERα is mediated by sequences in the DNA and LBDs. Using the glutamic acid mutants, we have shown that sequences in the DBD are necessary for ERα dimerization and that serine 236 plays an important role in this dimerization. In the presence of E2, however, the 236E mutants bound DNA and could dimerize, albeit to a lower extent when compared to wild-type or 236A mutants. These results indicate that dimerization by ERα in the absence of ligand is mediated largely by the DBD and that the addition of estrogen enables dimerization by the LBD. Our results further suggest that the DBD or the LBD (in the presence of E2) is sufficient for dimerization, thereby promoting binding to an ERE.
Dimerization by HEG0236E could also be obtained by the addition of the partial agonist OHT. Addition of the pure antiestrogen ICI 182,780, however, did not enable dimerization by HEG0236E, indicating that ICI prevents the activation of dimerization by the LBD. This is in agreement with a previous report showing that dimerization by mouse ERα synthesized in insect cells was inhibited by ICI (27).
Dimerization of hERα is inhibited by PKA in a ligand-dependent manner.
Inhibition of ERα dimerization by the replacement of serine 236 with negatively charged glutamic acid suggested that PKA phosphorylation inhibits dimer formation. This was confirmed by the finding that DNA binding by wild type but not by the 236A mutant was inhibited after treatment with activators of PKA and, indeed, overexpression of the catalytic subunit of PKA or activation of endogenous PKA resulted in reduced dimerization as determined by immunoprecipitations for HEG0 but not HEG0236A, indicating that the reduction in DNA binding results from the inhibition of dimerization by PKA-mediated phosphorylation of serine 236. The inhibition of dimerization could be prevented by the addition of the specific PKA inhibitor H89. As observed for HEG0236E, E2 and OHT prevented inhibition of dimerization by PKA, whereas ICI treatment did not relieve the inhibition. Replacement of serine 236 with alanine prevented inhibition of dimerization by PKA and obviated the need for ligand, further demonstrating the inhibitory effect of phosphorylation on dimerization.
Phosphorylation of serine 236 of hERα regulates heterodimer formation with hERβ.
As with homodimerization by hERα, heterodimerization between hERα and hERβ was prevented by a mutation of serine 236 of hERα to glutamic acid or by overexpression of PKA in an estrogen-dependent manner, indicating that similar mechanisms operate for the heterodimerization by hERα and hERβ as for the homodimerization by hERα. However, while heterodimer formation between HEG0 and hERβ1 was not observed in the absence of ligand or in the presence of ICI, heterodimers also did not form in the presence of OHT when PKA was overexpressed. These results suggest that while OHT enables the dimerization of the hERα LBD, it also inhibits or prevents the conformational changes required for the activation of the LBD dimerization function. In the case of hERα, tamoxifen and its metabolite OHT have been shown to have partial agonistic activity which correlates with activation of AF-1 but with inhibition of AF-2. Our findings are indicative of mechanistic differences between hERα and hERβ with regard to the action of OHT. Indeed, Tremblay et al. (80, 81) found that OHT had no agonistic activity with mouse ERβ in a cell line in which it had agonistic activity with mouse ERα, although it is clearly possible that OHT acts as a partial agonist with hERβ in other cell types or in activating promoters other than the ones used in these studies.
PKA modulates transcriptional activation by hERα.
Previous studies have shown that PKA synergizes with estrogen to increase transcriptional activation by ERα. However, the ligand-independent increase in transcription activation by PKA appears to be promoter and cell type specific (9, 21, 42). The results presented here confirm those findings. We show that in COS-1 cells PKA increases transactivation by hERα in the presence of E2 and OHT. A PKA-mediated increase in transactivation in the absence of ligand or in the presence of ICI was, however, restricted to two reporter genes, ERE-G-CAT and, to a lesser extent, ERE-3-TATA-CAT. Perhaps interactions with other transcription factors can stabilize DNA binding by hERα and overcome the inhibition of dimerization caused by phosphorylation of serine 236. The results with ERE-3-TATA-CAT suggest that multiple EREs may also be sufficient to provide some stabilization of DNA binding, although this was not observed in the case of ERE-3-tk-CAT.
Mutation of serine 236 to alanine, which stabilizes dimerization in the absence of ligand or in the presence of ICI, led to PKA-mediated ligand-independent transactivation in the case of all the reporter genes studied, whereas HEG0236E was only activated in the absence of ligand or in the presence of ICI in the case of ERE-G-CAT. These results provide evidence to support our findings that phosphorylation of serine 236 of hERα results in inhibition of dimerization.
Transcriptional interference of GAL-VP16-induced expression of the 17M-ERE-TATA-CAT reporter gene by HEG0 (but not HEG0236A) in the presence of ICI and its prevention by PKA are further indicative of a role for serine 236 phosphorylation in regulating dimerization and thereby DNA binding by hERα. The inability to determine the loss of DNA binding by the unliganded receptor in this assay system may be the result of residual E2 in the E2-stripped cell culture medium and does not allow an unequivocal determination of the inhibitory effect of serine 236 phosphorylation on hERα function in the absence of ligand.
Conclusion.
Our results demonstrate that hERα is phosphorylated by PKA on serine 236. Phosphorylation at this site can inhibit dimerization in the absence of estrogen, and the addition of estrogen can overcome this inhibition. Crystallization of the hERα DBD showed the DBD dimers to be present in two forms (75). In one form of DBD dimers, ordered water molecules provide bridging contacts between serine 236 in one DBD molecule and methionine 220 in the other molecule (see Fig. 1B). Phosphorylation of serine 236 would inhibit the formation of the dimerization interface by disrupting the network of water molecules at the dimer interface due to the steric hindrance between monomers at the interface by the presence of phosphates and/or by electrostatic repulsion between monomers, since the phosphates would be brought close together in the dimer. The second, “loose” dimer type seen would probably be able to accommodate the phosphorylation but might nevertheless not be as stable. The fact that the 236A mutant does not disrupt dimerization but would not support the same water network as seen in the crystal and the finding that a larger charged glutamic acid residue blocks dimerization suggest that either the steric and/or the electrostatic factors are important, rather than simply the disruption of favorable networks of water molecules. In any case, ligand binding would be likely to allow dimer formation independently of the DBD, perhaps by enabling the “loose” DBD conformation (74a).
Whilst ER binding to ERE in vitro does not require ligand, a number of studies indicate that DNA binding by the estrogen receptor is ligand-dependent in vivo. In yeast ligand binding is required for DNA binding and induction of changes in chromatin structure by hERα (30, 48, 66) and in chickens in vivo footprinting of the vitellogenin II and apoVLDL II genes demonstrated protection in the presence of E2 (67, 87). In vivo, phosphorylation of serine 236 could be important in preventing estrogen receptor binding to EREs in gene promoters and in preventing interference with gene transcription in the absence of ligand, since the binding of the transcriptionally inactive estrogen receptor could repress the estrogen receptor-independent transcription of responsive genes (e.g., see reference 28) in a manner similar to the inhibition by hERα of transcriptional activation by GAL4-VP16 described here. Alternatively, inhibition of DNA binding may be important for preventing alteration of chromatin structure in the absence of ligand (48).
Interaction of the estrogen receptor with several transcriptional regulators, including AP1, NF-kB, and Brn-3a and -3b, leading to modulation of their activity or that of the estrogen receptor, has been described. In some cases the DBD is dispensable for the interaction (e.g., AP1 [74]). The repression of the interleukin-6 promoter by the estrogen receptor is mediated by NF-kB and C-EBP beta and requires the presence of the ERα DBD, although high-affinity DNA binding by ERα is not required (70, 76). Interaction between the DBD of ERα and the POU domain of Brn-3a and Brn-3b is ligand independent and modulates transcription activation by ERα (18). It is possible that phosphorylation of serine 236 is important for the regulation of these interactions.
In summary, we have shown that hERα is phosphorylatable on serine 236 within the CII zinc finger of the DBD. Phosphorylation by PKA or mutation of serine 236 to glutamic acid impairs dimerization by the DBD, indicating that the DBD is important for ERα dimerization. In the presence of E2 or OHT, however, dimer formation occurs despite PKA overexpression, showing that the estrogen receptor contains two dimerization domains, one in the DBD and one in the LBD. Dimerization by the LBD is ligand dependent, whereas dimerization mediated by the DBD is ligand independent. Our results further indicate that sequences within the DBD or the LBD are sufficient for dimerization, although there are likely to be mechanistic differences between dimerization by the DBD and/or the LBD. Our results are in agreement with previous studies which have indicated that the antagonistic activity of tamoxifen (and OHT) is not mediated in any part by inhibition of dimerization and/or DNA binding but is mediated solely through inhibition of AF-2. However, we show that ICI, which inhibits both AF-1 and AF-2 and reduces the half-life of ERα, also prevents dimerization by the LBD, the dimer formation being mediated, in the presence of ICI solely by the DBD. Finally, phosphorylation of serine 236 provides a mechanism for E2 regulation of DNA binding by the estrogen receptor and may modulate its association with other transcription factors.
ACKNOWLEDGMENTS
We thank P. Chambon, D. Metzger, S. Mader, and J. White for the ER plasmids and M.-P. Gaub and C. Rochette-Egly for the pSG5-PKA plasmid. We are particularly grateful to J. W. R. Schwabe for his evaluation of our findings with reference to the crystal structure of the ERα DBD.
This work was supported by the Cancer Research Campaign.
REFERENCES
- 1.Ali S, Metzger D, Bornert J-M, Chambon P. Phosphorylation of the human oestrogen receptor: identification of a phosphorylation site required for trans-activation. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S, Lutz Y, Bellocq J-P, Chenard-Neu M-P, Rouyer N, Metzger D. Production and characterisation of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 3.Arnold S F, Obourn J D, Jaffe H, Notides A C. Serine 167 is the major estradiol-induced phosphorylation site on the human estrogen receptor. Mol Endocrinol. 1994;8:1208–1214. doi: 10.1210/mend.8.9.7838153. [DOI] [PubMed] [Google Scholar]
- 4.Arnold S F, Obourn J D, Jaffe H, Notides A C. Phosphorylation of the human estrogen receptor by mitogen-activated protein kinase and casein kinase II: consequence on DNA binding. J Steroid Biochem Mol Biol. 1995;55:163–172. doi: 10.1016/0960-0760(95)00177-2. [DOI] [PubMed] [Google Scholar]
- 5.Arnold S F, Obourn J D, Yudt M R, Carter T H, Notides A C. In vivo and in vitro phosphorylation of the human estrogen receptor. J Steroid Biochem Mol Biol. 1995;52:159–171. doi: 10.1016/0960-0760(94)00166-j. [DOI] [PubMed] [Google Scholar]
- 6.Arnold S F, Obourn J D, Jaffe H, Notides A C. Phosphorylation of the human estrogen receptor on tyrosine 537 in vivo and by src family tyrosine kinases in vitro. Mol Endocrinol. 1995;9:24–33. doi: 10.1210/mend.9.1.7539106. [DOI] [PubMed] [Google Scholar]
- 7.Arnold S F, Vorojeikina D P, Notides A C. Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. J Biol Chem. 1995;270:30205–30212. doi: 10.1074/jbc.270.50.30205. [DOI] [PubMed] [Google Scholar]
- 8.Arnold S F, Melamed M, Vorojeikina D P, Notides A C, Sasson S. Estradiol-binding mechanism and binding capacity of the human estrogen receptor is regulated by tyrosine phosphorylation. Mol Endocrinol. 1997;11:48–53. doi: 10.1210/mend.11.1.9876. [DOI] [PubMed] [Google Scholar]
- 9.Aronica S M, Katzenellenbogen B S. Stimulation of estrogen receptor-mediated transcription and alteration in the phosphorylation state of the rat uterine estrogen receptor by estrogen, cyclin adenosine monophosphate and insulin-like growth factor-1. Mol Endocrinol. 1993;7:743–752. doi: 10.1210/mend.7.6.7689695. [DOI] [PubMed] [Google Scholar]
- 10.Auricchio F. Phosphorylation of steroid receptors. J Steroid Biochem. 1989;32:613–622. doi: 10.1016/0022-4731(89)90397-x. [DOI] [PubMed] [Google Scholar]
- 11.Bai W, Weigel N L. Phosphorylation of Ser-211 in the chicken progesterone receptor modulates its transcriptional activity. J Biol Chem. 1996;271:12801–12806. doi: 10.1074/jbc.271.22.12801. [DOI] [PubMed] [Google Scholar]
- 12.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 13.Beato M, Herlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 14.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berthois Y, Katzenellenbogen J A, Katzenellenbogen B S. Phenol red in tissue culture media is a weak estrogen: implications concerning the study of estrogen-responsive cells in culture. Proc Natl Acad Sci USA. 1986;89:1218–1222. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–148. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 17.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Green G L, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism of the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 18.Budhram-Mahadeo V, Parker M, Latchman D S. POU transcription factors Brn-3a and Brn-3b interact with the estrogen receptor and differentially regulate transcriptional activity via an estrogen response element. Mol Cell Biol. 1998;18:1029–1041. doi: 10.1128/mcb.18.2.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunone G, Briand P-A, Miksicek R J, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 20.Castoria G, Migliaccio A, Green S, Di-Domenico M, Chambon P, Aurrichio F. Properties of a purified estradiol-dependent calf uterus tyrosine kinase. Biochemistry. 1993;32:1740–1750. doi: 10.1021/bi00058a007. [DOI] [PubMed] [Google Scholar]
- 21.Cho H, Katzenellenbogen B S. Synergistic activation of estrogen receptor-mediated transcription by estradiol and protein kinase activators. Mol Endocrinol. 1993;7:441–452. doi: 10.1210/mend.7.3.7683375. [DOI] [PubMed] [Google Scholar]
- 22.Cowley S M, Hoare S, Mosselman S, Parker M G. Estrogen receptor α and β form heterodimers on DNA. J Biol Chem. 1997;272:19858–19862. doi: 10.1074/jbc.272.32.19858. [DOI] [PubMed] [Google Scholar]
- 23.Dauvois S, Danielian P S, White R, Parker M G. The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J Cell Sci. 1993;103:1377–1388. doi: 10.1242/jcs.106.4.1377. [DOI] [PubMed] [Google Scholar]
- 24.Dauvois S, Danielian P S, White R, Parker M G. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc Natl Acad Sci USA. 1992;89:4037–4041. doi: 10.1073/pnas.89.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeFranco D B, Qi M, Boffor K C, Garabedian M J, Brautigan D L. Protein phosphatase types 1 and/or 2A regulate nucleocytoplasmic shuttling of glucocorticoid receptors. Mol Endocrinol. 1991;5:1215–1528. doi: 10.1210/mend-5-9-1215. [DOI] [PubMed] [Google Scholar]
- 26.El-Tanani M K, Green C D. Two separate mechanisms for ligand-independent activation of the estrogen receptor. Mol Endocrinol. 1997;11:928–937. doi: 10.1210/mend.11.7.9939. [DOI] [PubMed] [Google Scholar]
- 27.Fawell S E, White R, Hoare S, Sydenham M, Page M, Parker M G. Inhibition of estrogen receptor-DNA binding by the “pure” antiestrogen ICI 164,384 appears to be mediated by impaired receptor dimerization. Proc Natl Acad Sci USA. 1990;8:6883–6887. doi: 10.1073/pnas.87.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaub M-P, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;63:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- 29.Gibson M K, Nemmers L A, Beckman W C, Davis V L, Curtis S W, Korach K S. The mechanism of ICI 164,384 antiestrogenicity involves rapid loss of estrogen receptor in uterine tissue. Endocrinology. 1991;129:2000–2010. doi: 10.1210/endo-129-4-2000. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert D M, Losson R, Chambon P. Ligand dependence of estrogen receptor induced changes in chromatin structure. Nucleic Acids Res. 1992;20:4525–4531. doi: 10.1093/nar/20.17.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glineur C, Zenke M, Beug H, Ghysdael J. Phosphorylation of the v-erbA protein is required for its function as an oncogene. Genes Dev. 1990;4:1663–1676. doi: 10.1101/gad.4.10.1663. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg Y, Glineur C, Gesquiere J C, Ricouart A, Sap J, Vennstrom B, Ghysdael J. Activation of protein kinase C or cAMP-dependent protein kinase increases phosphorylation of the c-erbA-encoded thyroid hormone receptor and of the v-erbA-encoded protein. EMBO J. 1988;7:2425–2433. doi: 10.1002/j.1460-2075.1988.tb03088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green S, Isseman I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;10:369–370. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gronemeyer H. Transcription activation by estrogen and progesterone receptors. Annu Rev Biochem. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 35.Hidaka H, Watanabe M, Kobayashi R. Properties and use of H-series compounds as protein kinase inhibitors. Methods Enzymol. 1991;201:328–339. doi: 10.1016/0076-6879(91)01029-2. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh J-C, Jurutka P W, Galligan M A, Terpenning C M, Haussler C A, Samuels D S, Shimizu Y, Shimizu N, Haussler M R. Phosphorylation of the human vitamin D receptor by protein kinase C. Biochemical and functional evaluation of the serine 51 recognition site. Proc Natl Acad Sci USA. 1991;88:9315–9319. doi: 10.1073/pnas.88.20.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh J-C, Jurutka P W, Shigeo N, Galligan M A, Haussler C A, Shimizu Y, Shimizu N, Whitfield G K, Haussler M R. Human vitamin D receptor is selectively phosphorylated by protein kinase C on serine 51, a residue crucial to its transactivation function. J Biol Chem. 1993;268:15118–15126. [Google Scholar]
- 38.Hsu S C, de Franco D B. Selectivity of cell cycle regulation of glucocorticoid receptor function. J Biol Chem. 1995;270:3359–3364. doi: 10.1074/jbc.270.7.3359. [DOI] [PubMed] [Google Scholar]
- 39.Hu J M, Bodwell J E, Munck A. Cell cycle-dependent glucocorticoid receptor phosphorylation and activity. Mol Endocrinol. 1994;8:1709–1713. doi: 10.1210/mend.8.12.7708059. [DOI] [PubMed] [Google Scholar]
- 40.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 41.Ignar-Trowbridge D, Teng C T, Ross K A, Parker M G, Korach K S, McLachlan J A. Peptide growth factors elicit estrogen receptor-dependent transcriptional activation of an estrogen-responsive element. Mol Endocrinol. 1993;7:992–998. doi: 10.1210/mend.7.8.8232319. [DOI] [PubMed] [Google Scholar]
- 42.Ince B A, Montano M M, Katzenellenbogen B S. Activation of transcriptionally inactive human estrogen receptors by cyclin adenosine 3′,5′-monophosphate and ligands including antiestrogens. Mol Endocrinol. 1994;8:1397–1406. doi: 10.1210/mend.8.10.7531820. [DOI] [PubMed] [Google Scholar]
- 43.Jackson S P. Regulating transcription factor activity by phosphorylation. Trends Cell Biol. 1992;2:104–108. doi: 10.1016/0962-8924(92)90014-e. [DOI] [PubMed] [Google Scholar]
- 44.Joel P B, Traish A M, Lannigan D A. Estradiol and phorbol ester cause phosphorylation of serine 118 in the human estrogen receptor. Mol Endocrinol. 1995;9:1041–1052. doi: 10.1210/mend.9.8.7476978. [DOI] [PubMed] [Google Scholar]
- 45.Joel P B, Smith J, Sturgill T W, Fisher T L, Blenis J, Lannigan D A. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of Ser-167. Mol Cell Biol. 1998;18:1978–1984. doi: 10.1128/mcb.18.4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karin M. Signal transduction from the cell surface to the nucleus through phosphorylation of transcription factors. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 47.Kato S, Endoh E, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 48.Kladde M P, Xu M, Simpson R T. Direct study of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 1996;15:6290–6300. [PMC free article] [PubMed] [Google Scholar]
- 49.Krust A, Green S, Argos P, Kumar V, Walter P, Bornert J-M, Chambon P. The chicken oestrogen receptor sequence: homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986;5:891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar V, Green S, Staub A, Chambon P. Localisation of the oestradiol-binding and putative DNA-binding domains of the human oestrogen receptor. EMBO J. 1986;5:2231–2236. doi: 10.1002/j.1460-2075.1986.tb04489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar V, Green S, Stack G, Berry M, Jin J R, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 52.Lees J A, Fawell S E, Parker M G. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989;17:5477–5488. doi: 10.1093/nar/17.14.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Goff P, Montano M M, Schodin D J, Katzenellenbogen B S. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269:4458–4466. [PubMed] [Google Scholar]
- 54.Ma Z Q, Santagati S, Patrone C, Pollio G, Vegeto E, Maggi A. Insulin-like growth factors activate estrogen receptor to control the growth and differentiation of the human neuroblastorna cell line SK-ER3. Mol Endocrinol. 1994;8:910–918. doi: 10.1210/mend.8.7.7984152. [DOI] [PubMed] [Google Scholar]
- 55.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 56.Mangelsdorf D J, Thummel C, Beato M, Hefflich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;8:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonnell D P, Clemm D L, Hermann T, Goldman M E, Pike J W. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- 58.McInerney E M, Katzenellenbogen B S. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 59.Metzger D, Ali S, Bornert J-M, Chambon P. Characterization of the N-terminal transcriptional activation function (AF-1) of the human oestrogen receptor in animal and yeast cells. J Biol Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 60.Metzger D, Berry M, Ali S, Chambon P. Effect of antagonists on DNA binding properties of the human oestrogen receptor in vitro and in vivo. Mol Endocrinol. 1995;9:579–591. doi: 10.1210/mend.9.5.7565805. [DOI] [PubMed] [Google Scholar]
- 61.Newton C J, Buric R, Trapp T, Brockmeier S, Pagotto U, Stalla G K. The unliganded estrogen receptor (ER) transduces growth factor signals. J Steroid Biochem Mol Biol. 1994;48:481–486. doi: 10.1016/0960-0760(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 62.Ogawa S, Inoue S, Watanabe T, Hiroi H, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. The complete primary structure of human estrogen receptor β (hERβ) and its heterodimerization with ERα in vivo and in vitro. Biochem Biophys Res Commun. 1998;243:122–126. doi: 10.1006/bbrc.1997.7893. [DOI] [PubMed] [Google Scholar]
- 63.Pace P E, Taylor J, Suntharalingam S, Coombes R C, Ali S. Human estrogen receptor beta binds DNA in a manner similar to and dimerises with estrogen receptor alpha. J Biol Chem. 1997;272:25832–25838. doi: 10.1074/jbc.272.41.25832. [DOI] [PubMed] [Google Scholar]
- 64.Patrone C, Ma Z Q, Pollio G, Agrati P, Parker M G, Maggi A. Cross-coupling between insulin and estrogen receptor in human neuroblastoma cells. Mol Endocrinol. 1996;10:499–507. doi: 10.1210/mend.10.5.8732681. [DOI] [PubMed] [Google Scholar]
- 65.Pettersson K, Grandien K, Kuiper G G, Gustafsson J A. Mouse estrogen receptor β forms estrogen response element-binding heterodimers with estrogen receptor α. Mol Endocrinol. 1997;11:1486–1496. doi: 10.1210/mend.11.10.9989. [DOI] [PubMed] [Google Scholar]
- 66.Pham T A, Elliston J F, Nawaz Z, McDonnell D P, Tsai M-J, O’Malley B W. Antiestrogens can establish nonproductive receptor complexes and alter chromatin structure at target enhancers. Proc Natl Acad Sci USA. 1991;88:3125–3129. doi: 10.1073/pnas.88.8.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Philipsen J N J, Hennis B C, Ab G. In vivo footprinting of the estrogen-inducible vitellogenin II gene from chicken. Nucleic Acids Res. 1988;16:9663–9676. doi: 10.1093/nar/16.20.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pietras R J, Arboleda J, Reese D M, Wongvipat N, Pelgram M D, Ramos L, Gorman C M, Parker M G, Sliwkowski M X, Slamon D J. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10:2435–2446. [PubMed] [Google Scholar]
- 69.Power R F, Mani S K, Codina J, Conneely O M, O’Malley B W. Dopaminergic and ligand-independent activation of steroid hormone receptors. Science. 1991;254:1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- 70.Ray A, Prefontaine K E, Ray P. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–12946. [PubMed] [Google Scholar]
- 71.Reese J C, Katzenellenbogen B S. Examination of the DNA-binding ability of estrogen receptor in whole cells: implications for hormone-independent transactivation and the actions of antiestrogens. Mol Cell Biol. 1992;12:4531–4538. doi: 10.1128/mcb.12.10.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rochette-Egly C, Adam S, Rossignol M, Egly J-M, Chambon P. Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 73.Rochette-Egly C, Oulad-Abdelghani M, Staub A, Pfister V, Scheuer I, Chambon P, Gaub M-P. Phosphorylation of the retinoic acid receptor a by protein kinase A. Mol Endocrinol. 1995;9:860–871. doi: 10.1210/mend.9.7.7476969. [DOI] [PubMed] [Google Scholar]
- 74.Saha S, Brickman J M, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 74a.Schwabe, J. W. R. Personal communication.
- 75.Schwabe J W R, Chapman L, Finch J T, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;70:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 76.Stein B, Yang M X. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C-EBP beta. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takimoto G S, Hovland A R, Tasset D M, Melville M Y, Tung L, Horwitz K B. Role of phosphorylation on DNA binding and transcriptional functions of human progesterone receptors. J Biol Chem. 1996;271:13308–13316. doi: 10.1074/jbc.271.23.13308. [DOI] [PubMed] [Google Scholar]
- 78.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 80.Tremblay G B, Tremblay A, Copeland N G, Gilbert D J, Jenkins N A, Labrie F, Giguere V. Cloning, chromosomal localization and functional analysis of the murine estrogen receptor β. Mol Endocrinol. 1997;11:353–365. doi: 10.1210/mend.11.3.9902. [DOI] [PubMed] [Google Scholar]
- 81.Tremblay A, Tremblay G B, Labrie C, Labrie F, Giguere V. EM800, a novel antiestrogen, acts as a pure antagonist of the transcriptional functions of estrogen receptors α and β. Endocrinology. 1998;139:111–118. doi: 10.1210/endo.139.1.5702. [DOI] [PubMed] [Google Scholar]
- 82.Truss M, Beato M. Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocrine Rev. 1993;14:459–479. doi: 10.1210/edrv-14-4-459. [DOI] [PubMed] [Google Scholar]
- 83.Tsai M J, O’Malley B W. Molecular mechanisms of action of steroid/thyroid/receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 84.Webster N J G, Green S, Jin J R, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 85.Weis K E, Ekena K, Thomas J A, Lazenneac G, Katzenellenbogen B S. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 86.White R, Sjoberg M, Kalkhoven E, Parker M G. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wijnholds J, Philipsen J N J, Ab G. Tissue-specific and steroid-dependent interaction of transcription factors with the oestrogen-inducible apoVLDL II promoter in vivo. EMBO J. 1988;7:2757–2763. doi: 10.1002/j.1460-2075.1988.tb03130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Beck C A, Poletti A, Clement IV J P, Prendergast P, Yip T-Y, Hutchens T W, Edwards D P, Weigel N L. Phosphorylation of human progesterone receptor by cyclin-dependent kinase 2 on three sites that are authentic basal phosphorylation sites in vivo. Mol Endocrinol. 1997;11:823–832. doi: 10.1210/mend.11.6.0006. [DOI] [PubMed] [Google Scholar]