Abstract
Gongronella had accommodated only two species for more than half a century and as many as 17 new species have been described in this genus since 2015. However, no systematic studies were conducted for this genus so far. The distribution, substrate and morphology of all known species in Gongronella are analysed herein. Meanwhile, with the support of phylogenetic and morphological evidence, six new species (G.abortosporangiasp. nov., G.apophysatasp. nov., G.bawanglingensissp. nov., G.inconstanssp. nov., G.pingtangensissp. nov. and G.reniformissp. nov.) are proposed and G.pamphilae is recorded from China for the first time. The phylogenetic tree was constructed using ITS+LSU+TEF+ACT+RPB1 and the results were basically the same as ITS+LSU. All species of Gongronella, except G.namwonensis from fresh water, were isolated from soil. The genus is distributed worldwide, mainly in tropical and subtropical regions. A synoptic key is provided for a total of 24 species (18 species previously published and six species newly described herein), except for G.banzhaoae due to unavailable protologue, type and living culture. No morphologies were described when G.pamphilae was proposed. Thanks to the strains isolated in this study, G.pamphilae is included in the key and reported as a Chinese new record. This is the first comprehensive taxonomy and phylogeny of the genus Gongronella.
Key words: Mucoromycota, molecular phylogeny, new taxa, soil-borne fungi, taxonomy
Introduction
The genus Gongronella Ribaldi has a great potential in biological applications due to the ability of producing bioactive substance such as chitosan (Wang et al. 2008; Zhou et al. 2008), dissolving phosphate and degrading metalaxyl (Doilom et al. 2020; Martins et al. 2020). Gongronella sp. w5, a well-known strain in this genus, can induce fungi Panusrudis (Wei et al. 2010) and Coprinopsiscinerea (Pan et al. 2014; Hu et al. 2019; Liu et al. 2022) to produce laccase, secrete organic acids for improving the acquisition of phosphate in plants and thus promote their growth (Dong et al. 2018; Wang et al. 2021) and synthesise various bioactive enzymes, such as β-glucosidase and invertase (Zhou et al. 2020; Mai et al. 2021).
This genus was established in 1952 and typified with Gongronellaurceolifera Ribaldi (Ribaldi 1952). It belongs to Mucoromycota Doweld, Mucoromycetes Doweld, Mucorales Dumort, Cunninghamellaceae Naumov ex R.K. Benj. (Tedersoo et al. 2018). Before 2015, the taxonomy of Gongronella was stagnant, accommodating only two species G.urceolifera (= G.butleri) and G.lacrispora. Since 2015, as many as 17 species have been described successively (Hesseltine and Ellis 1961; Adamcik et al. 2015; Ariyawansa et al. 2015; Li et al. 2016; Tibpromma et al. 2017; Dong et al. 2019; Zhang et al. 2019; Crous et al. 2020; de Freitas et al. 2020; Doilom et al. 2020; Martins et al. 2020; Wang et al. 2023a; Zhao et al. 2023). At present, Gongronella contains 19 species, nearly half of which were initially found from China (Table 1). In the GlobalFungi database, there are a total of 3,039 sample records for the genus Gongronella covering Asia (1,566, 51.53%), North America (571, 18.79%), Europe (433, 14.58%), South America (261, 8.59%), Africa (123, 4.05%), Australia (64, 2.11%) and Atlantic Ocean (1, 0.03%) (https://globalfungi.com/, accessed on 18 October 2024). Considering geographical climate, most samples were collected from tropical and subtropical regions (https://globalfungi.com/, accessed on 17 October 2024). In conclusion, the species of Gongronella were distributed worldwide and mainly concentrated in tropical and subtropical regions in Asia.
Table 1.
The origin of taxonomic types in Gongronella.
| Countries | Type numbers | Percentage (%) |
|---|---|---|
| China | 9 | 47.4 |
| Korea | 3 | 15.8 |
| Brazil | 3 | 15.8 |
| Australia | 2 | 10.5 |
| Portugal | 1 | 5.3 |
| UK | 1 | 5.3 |
Note: These data are from the Index Fungorum (http://www.indexfungorum.org/, accessed on 9 December 2023) and Wang et al. (2023).
Regarding substrate of nomenclatural types within the genus Gongronella, G.namwonensis was isolated from fresh water and the other 18 species were all isolated from soil (Crous et al. 2020; Doilom et al. 2020). According to the GlobalFungi database, substrates include soil (1852, 60.94%), topsoil (475, 15.63%), root (403, 13.26%), rhizosphere soil (204, 6.71%), root + rhizosphere soil (52, 1.71%), litter (22, 0.72%), sediment (10, 0.33%), shoot (9, 0.3%) and deadwood (7, 0.23%), (https://globalfungi.com/, accessed on 19 October 2024). Although the GlobalFungi database showed more kinds of substrates of Gongronella, most strains were still isolated from a variety of soil samples.
In this study, 14 strains of the genus Gongronella were isolated from soil in Hainan, Yunnan, Sichuan and Guizhou Provinces from China. According to ITS+LSU+TEF+RPB1 molecular phylogenetic analyses and morphological comparisons, these strains were classified into six new species and one was identified as new record species to China. The morphological information of all described species of Gongronella was reviewed and compared.
Materials and methods
Isolation and morphology
Soil samples were collected in Hainan Province (April 2023 and October 2023), Sichuan Province (June 2023) and Guizhou Province (August 2023). Strains were isolated from the soil samples by a combination of soil dilution and single spore isolation.
About 1 g soil sample was mixed with 10 ml sterile water to prepare 10-1 soil suspension. One millilitre of 10-1 suspension was transferred to 9 ml of sterile water to obtain a 10-2 soil suspension. In the same way, 10-3 and 10-4 soil suspensions were made. The final 10-3 and 10-4 soil suspensions (200 ml) were pipetted on the surface of Rose-Bengal Chloramphenicol Agar (RBC: peptone 5.00 g/l, glucose 10.00 g/l, KH2PO4 1.00 g/l, MgSO4·7H2O 0.50 g/l, rose red 0.05 g/l, chloramphenicol 0.10 g/l, agar 15.00 g/l) (Corry et al. 1995), dispersed evenly with sterilised coating rods and cultured at 25 °C in the dark for 2–5 days. Upon colonies were visible, they were transferred onto Potato Dextrose Agar (PDA: glucose 20.00 g/l, potato 200.00 g/l, agar 20.00 g/l, pH 7). When sporangia were produced, sporangiospores were suspended with sterile water and streaked with a sterilised inoculation ring. The plates were cultured at 25 °C in darkness and single spore colonies were transferred on to a new PDA plate for subculturing. To ensure the formation of zygospores, pairing experiments were carried out by adding 0.1% lecithin to PDA and sealing Petri dishes to retain moisture. The microscopic morphological characteristics of fungi were observed with an optical microscope (Olympus BX53) and photographed with a high-definition colour digital camera (Olympus DP80). All strains were stored with 10% sterilised glycerine at 4 °C. Each morphological character was statistically calculated from 30 measurements (Zhang et al. 2022). Cultures were deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and the Shandong Agricultural University Culture Collection, Taian, China (SAUCC). Specimens were deposited in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (HMAS). Taxonomic information for the new taxa was registered in the Fungal Name repository (https://nmdc.cn/fungalnames/).
DNA extraction and amplification
Genomic DNA was extracted from mycelia using the CTAB method and BeaverBeads Plant DNA Kit (Cat. No.: 70409-20; BEAVER Biomedical Engineering Co., Ltd.) (Doyle et al. 1990; Guo et al. 2000; Wang et al. 2023b). ITS, LSU, TEF, ACT and RPB1 were amplified by polymerase chain reaction using ITS4/ITS5, LR0R/LR7, EF1-728F/EF1-986R, ACT-512F/ACT-783R and RPB1-Af/RPB1-Cr primer pairs, respectively (Table 2). Amplification was performed in a final volume of 20 μl, containing 10 μl 2× Hieff Canace® Plus PCR Master Mix (Yeasen Biotechnology, Cat No. 10154ES03), 0.5 μl of forward and reverse primers each (10 μM) (TsingKe, Beijing, China), 1 μl template genomic DNA (about 1 μM) and 8 μl distilled deionised water. Molecular loci, PCR primers and programmes used in this study are listed in Table 2. The PCR products were electrophoresed with 1% agarose gel. The DNA fragments were stained with GelRed and observed under ultraviolet light. Then a gel extraction kit (Cat# AE0101-C; Shandong Sparkiade Biotechnology Co., Ltd.) was used for gel recovery. Sanger sequencing was carried out by Biosune Company Limited (Shanghai, China). Consensus sequences were assembled using MEGA v.7.0 (Kumar et al. 2016). All sequences generated in this study were deposited at GenBank under the accession numbers in Table 3.
Table 2.
Molecular loci, PCR primers and programmes used in this study.
| Loci | PCR primers | Sequence (5’–3’) | PCR cycles | References |
|---|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | 95 °C 5 min; (95 °C 30 s, 55 °C 30 s, 72 °C 1 min) × 35 cycles; 72 °C 10 min | White et al. (1990) |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | |||
| LSU | LR0R | GTA CCC GCT GAA CTT AAG C | 95 °C 5 min; (95 °C 50 s, 47 °C 30 s, 72 °C 1.5 min) × 35 cycles; 72 °C 10 min | Vilgalys and Hester (1990) |
| LR7 | TAC TAC CAC CAA GAT CT | |||
| TEF | EF1-728F | CAT CGA GAA GTT CGA GAA GG | 95 °C 5 min; (95 °C 30 s, 55 °C 60 s, 72 °C 1 min) × 30 cycles; 72 °C 10 min | Carbone and Kohn (1999); O’Donnell et al. (1998) |
| EF2 | GGA RGT ACC AGT SAT CAT GTT | |||
| RPB1 | RPB1-Af | GAR TGY CCD GGD CAY TTY GG | 95 °C 3 min; (94 °C 40 s, 60 °C 40 s, 72 °C 2 min) × 9 (94 °C 45 s, 55 °C 1.5 min, 72 °C 2 min) × 37 cycles; 72 °C 10 min | Stiller and Hall (1997) |
| RPB1-Cr | CCN GCD ATN TCR TTR TCC ATR TA | |||
| ACT | ACT-512F | ATG TGC AAG GCC GGT TTC GC | 95 °C 3 min; (95 °C 1 min, 55 °C 1 min, 72 °C 1 min) × 30 cycles; 72 °C 10 min | Voigt and Wostemeyer (2000) |
| ACT-783R | TAC GAG TCC TTC TGG CCC AT |
Table 3.
Information of strains used in this study.
| Species | Strains | Substrates | Countries | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | ACT | TEF | RPB1 | ||||
| Gongronellaabortosporangia | CGMCC 3.27028* | Soil | China | PP195847 | PP195948 | PP933938 | PP850088 | PP842883 |
| SAUCC 4064-2 | Soil | China | PP195848 | PP195949 | PP933939 | PP850089 | PP842882 | |
| G.apophysata | CGMCC 3.27031* | Soil | China | PP195853 | PP195954 | PP933947 | PP850099 | PP842878 |
| SAUCC 4846-3 | Soil | China | PP195854 | PP195955 | PP933948 | PP850100 | PP842877 | |
| G.banzhaoae | BRIP 75171a* | Soil | Australia | OR271908 | OR259049 | n.a. | n.a. | n.a. |
| G.bawanglingensis | CGMCC 3.27033* | Soil | China | PP195857 | PP195958 | PP933951 | PP850103 | PP883965 |
| SAUCC 6946-1 | Soil | China | PP195858 | PP195959 | PP933952 | PP850104 | PP883964 | |
| G.brasiliensis | URM 7487* | Soil | Brazil | NR_155148 | KY114932 | n.a. | n.a. | n.a. |
| URM 7488 | Soil | Brazil | KY114931 | KY114933 | n.a. | n.a. | n.a. | |
| G.butleri | CBS 216.58* | Soil | UK | JN206285 | MH869292 | n.a. | n.a. | n.a. |
| G.chlamydospora | CGMCC 3.16118* | Soil | China | OL678157 | n.a. | n.a. | n.a. | PP898292 |
| G.eborensis | MUM 10.262* | Soil | Portugal | KT809408 | MN947301 | n.a. | n.a. | n.a. |
| MUM 10.263 | Soil | Portugal | GU244500 | MN947302 | n.a. | n.a. | n.a. | |
| G.guangdongensis | CGMCC 2.15212* | Soil | China | NR_158464 | MN947303 | n.a. | n.a. | n.a. |
| CGMCC 2.15213 | Soil | China | KC462740 | MN947304 | n.a. | n.a. | n.a. | |
| G.hydei | KUMCC 18.0198* | Rhizosphere soil | China | NR_171964 | MT907273 | n.a. | n.a. | n.a. |
| G.inconstans | CGMCC 3.27029* | Soil | China | PP195849 | PP195950 | PP933941 | PP850091 | PP842874 |
| SAUCC 4113-3 | Soil | China | PP195850 | PP195951 | PP933942 | PP850092 | PP842873 | |
| G.koreana | EML-TS2Bp* | Soil | Korea | KP636529 | KP636530 | KP636527 | n.a. | n.a. |
| EML-TS2Bp-2 | Soil | Korea | KP835545 | KP835542 | KP835543 | n.a. | n.a. | |
| G.lacrispora | ATCC 24412* | Soil | Brazil | GU244498 | JN206609 | n.a. | n.a. | n.a. |
| G.multiramosa | CGMCC 3.26216* | Soil | China | OR733546 | OR733611 | PP933937 | PP850087 | PP842881 |
| SAUCC 4056-4 | Soil | China | OR733545 | OR733610 | n.a. | n.a. | n.a. | |
| G.multispora | CGMCC 3.16119* | Soil | China | OL678158 | n.a. | n.a. | n.a. | pm |
| G.namwonensis | CNUFC WW2-12* | Fresh water | Korea | NR_175640 | MN658482 | n.a. | n.a. | n.a. |
| G.oleae | CGMCC 3.26217* | Soil | China | OR742078 | OR733608 | PP933945 | PP850097 | PP850080 |
| SAUCC 4164-2 | Soil | China | OR742079 | OR733609 | PP933946 | PP850098 | PP850079 | |
| G.orasabula | EML-QF12-1* | Soil | Korea | NR_148087 | KT936263 | KT936265 | n.a. | n.a. |
| EML-QF12-2 | Soil | Korea | KT936270 | KT936264 | n.a. | n.a. | n.a. | |
| G.pamphilae | BRIP 74936a* | Soil | Australia | OR271909 | OR259050 | n.a. | n.a. | n.a. |
| CGMCC 3.27027 | Soil | China | PP195845 | PP195946 | PP933935 | PP850086 | PP850081 | |
| SAUCC 4031-2 | Soil | China | PP195846 | PP195947 | PP933936 | PP850085 | PP850082 | |
| G.pedratalhadensis | URM 8182* | Soil | Brazil | MN912512 | MN912508 | n.a. | n.a. | n.a. |
| G.pingtangensis | CGMCC 3.27032* | Soil | China | PP195855 | PP195956 | PP933949 | PP850101 | PP842880 |
| SAUCC 5676-2 | Soil | China | PP195856 | PP195957 | PP933950 | PP850102 | PP842879 | |
| G.qichaensis | CGMCC 3.26218* | Soil | China | OR733544 | OR733607 | n.a. | PP850093 | PP850084 |
| SAUCC 4137-3 | Soil | China | OR733543 | OR733606 | n.a. | PP850094 | PP850083 | |
| G.reniformis | CGMCC 3.27030* | Soil | China | PP195851 | PP195952 | PP933943 | PP850095 | PP842875 |
| SAUCC 4142-5 | Soil | China | PP195852 | PP195953 | PP933944 | PP850096 | PP842876 | |
| G.sichuanensis | CGMCC 3.19651* | Soil | China | MK813373 | MK813855 | MK820625 | n.a. | n.a. |
| CGMCC 3.19652 | Soil | China | MK813374 | MK813856 | MK820626 | n.a. | n.a. | |
| G.zunyiensis | CGMCC 3.19899* | Soil | China | MN453856 | MN453853 | n.a. | n.a. | n.a. |
| CGMCC 3.19900 | Soil | China | MN453857 | MN453854 | n.a. | n.a. | n.a. | |
| Cunninghamellaechinulata | CBS 156.28* | n.a. | n.a. | JN205895 | MH877699 | n.a. | n.a. | n.a. |
Notes: New species established in this study are in bold. Ex-type or ex-holotype strains are labelled with a star mark “*”. The abbreviation “n.a.” stands for “not available”
Relative sequences were obtained by BLAST search in the GenBank nucleotide database of NCBI website (Kumar et al. 2016). Sequences both generated herein and retrieved from GenBank (Table 3) were aligned using MAFFT 7 online service (http://mafft.cbrc.jp/alignment/server/, 20 October 2023) (Katoh et al. 2019). The ITS, LSU, TEF, ACT and RPB1 sequences were analysed individually and jointly. The optimal evolutionary model for each partition was determined and included in the analysis using MrModelTest v.2.3 (Nylander 2004). Phylogenetic history was reconstructed using Maximum Likelihood (ML) algorithm with RaxML-HPC2 on XSEDE 8.2.12 (Stamatakis 2014; Zhao et al. 2024) and Bayesian Inference (BI) algorithm with MrBayes 3.2.7a (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012). ML was performed for 1,000 bootstrap replicates with the GTRGAMMA model of nucleotide evolution. BI was performed using a quick start algorithm with an automatic stop option. The Bayesian analysis consisted of 5,000,000 generations with four parallel runs with the option of stopping rules and a sampling frequency of 100 generations. The burn-in score was set to 0.25 and the posterior probability (PP) was determined from the remaining trees. Initial adjustments of phylogenetic trees were made using FigTree v.1.4.4 (http://tree.bio.ed.ac) and the layout of the trees was finished using Adobe Illustrator CC 2019 (https://adobe.com/products/illustrator).
Results
Phylogenetic analyses
The sequence matrix included 43 strains in 25 species of Gongronella, with Cunninghamellaechinulata CBS 156.28 as outgroup. A total of 4,080 characters comprised ITS rDNA (1–989), LSU rDNA (990–1967), TEF (1968–2172), ACT (2173–2948) and RPB1 (2949–4080). Amongst these characters, 2,866 were constant, 562 variable, but parsimony non-informative and 652 parsimony informative characters (Suppl. material 1). MrModelTest suggested that the Dirichlet fundamental frequency and GTR+I+G evolution pattern for both partitions were adopted in Bayesian Inference. The topology of the Bayesian tree was consistent with that of the ML tree and, therefore, was used as a representative to summarise the evolutionary history within the genus Gongronella (Fig. 1). G.abortosporangia was closely related to G.multiramosa with a high support (BIPP = 0.95). G.pingtangensis was closely related to G.namwonensis with a high support (BIPP = 1). G.reniformis was closely related to G.pamphilae and G.brasiliensis with a high support (MLBV = 75, BIPP = 0.99). The G.bawanglingensis (MLBV = 100, BIPP = 1) is closely related to G.qichaensis and G.inconstans. G.inconstans (MLBV = 99, BIPP = 1) is closely related to G.qichaensis with a high support (BIPP = 0.96). G.apophysata is closely related to G.zunyiensis.
Figure 1.
A Maximum Likelihood (ML) phylogenetic consensus tree inferred from DNA sequences of ITS, LSU, TEF, ACT and RPB1, showing relationships amongst species of Gongronella with Cunninghamellaechinulata CBS 156.28 as outgroup. The Maximum Likelihood bootstrap value (MLBV) and Bayesian Inference posterior probability (BIPP) are successively shown at the nodes and separated by a slash “/”. Strains marked with a star “*” are ex-types or ex-holotypes. The strains isolated and sequenced in this study are shown in red. Branches shortened to fit the page are represented by double slashes “//” and folds “×”. The scale in the bottom centre indicates 0.2 substitutions per site.
Taxonomy
. Gongronella abortosporangia
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
21D8BDF9-EF04-5AF3-A57F-BA9041EF991A
Fungal Names: FN 571253
Figure 2.
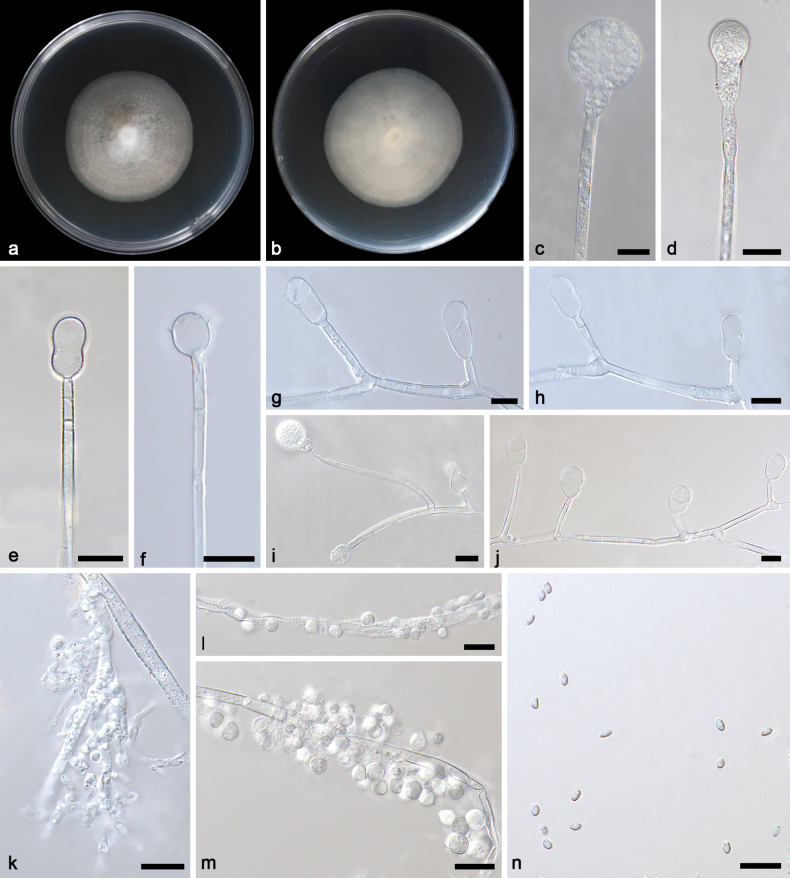
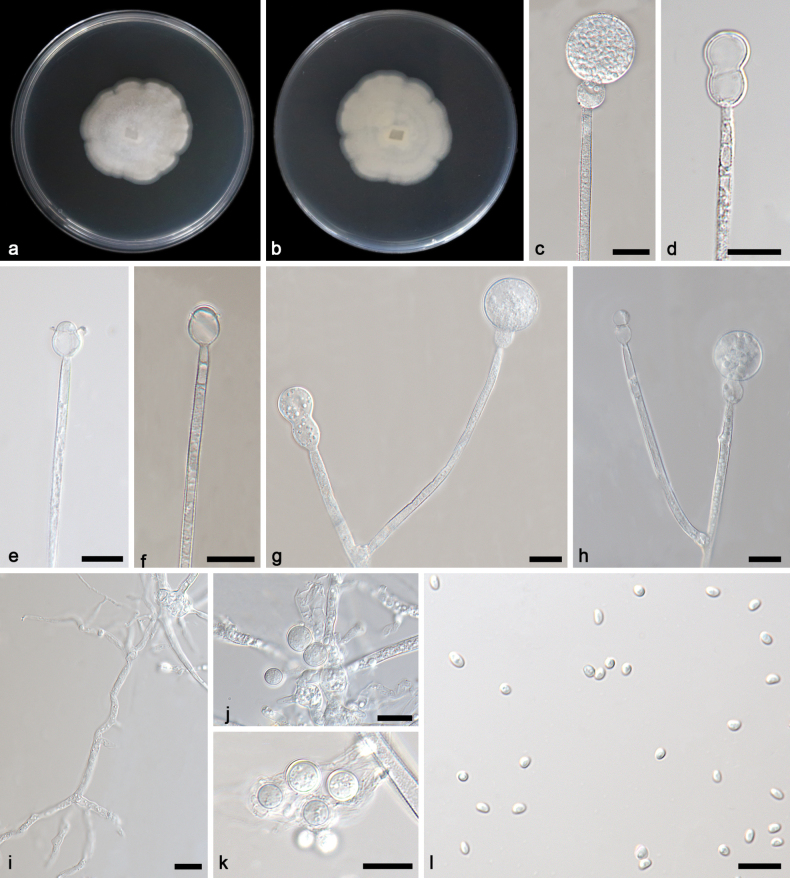
Gongronellaabortosporangia ex-holotype CGMCC 3.27028 a, b colonies on PDA (a obverse b reverse) c an unbranched sporangiophore with a mature sporangium d an unbranched sporangiophore with an immature sporangium e an aborted sporangium with two septa f columellae, collars and apophyses g, h, j branched sporangiophores with sterile (aborted) sporangia i a branched sporangiophore with a mature sporangium, columellae, collars and apophyses k rhizoids l, m giant cells n sporangiospores. Scale bars: 10 μm (c–n).
Etymology.
The epithet “abortosporangia” (Latin) refers to the abortive sporangia.
Type.
China • Hainan Province, Lingshui Li Autonomous County, Qixian Yaochi Yexi Hot Spring (18.70161°N, 109.69318°E), from soil sample, 10 April 2023, Yi-Xin Wang (holotype HMAS 352726, ex-holotype strain CGMCC 3.27028).
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 49.2–52.4 mm in diameter in seven days, white, regular at edge and cottony in the centre, reversely milky white. Rhizoids hyaline, branched, irregularly shaped, with oil droplets. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or branched (1–6 times), 4.0–96.8 × 1.0–4.2 μm, hyaline, smooth, mostly aseptate, sometimes one-septate and rarely two-septate, occasionally containing a line of oil droplets. Sterile (aborted) sporangia abundant, mainly on the top of short lateral branches of sporangiophores, mostly gourd-shaped, 11.6–16.7 × 5.5–17.7 μm, partially elliptical with a slight shrinkage, 12.5–18.0 × 6.7–10.6 μm, occasionally clavate, 20.1–22.7 × 9.5–10.4 μm. Fertile sporangia hyaline or light yellow, spherical, 7.0–23.2 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly hemispherical, 2.5–4.2 × 3.6–7.4 μm, sometimes sub-hemispherical, 1.3–3.9 × 3.6–5.5 μm, hyaline, smooth. Apophyses hyaline, smooth, variously shaped, mostly cup-shaped, 1.9–8.6 × 2.1–6.7 μm, partially hemispherical, 2.7–5.5 × 2.8–7.4 μm, occasionally pear-shaped, 8.2 × 7.2 μm. Sporangiospores not uniform, hyaline, smooth, ovoid, 2.6–3.5 × 1.7–2.1 μm, reniform, 2.9–3.5 × 1.7–2.3 μm. Chlamydospores gourd-shaped, 20.3–29.3 × 6.4–9.3 μm. Giant cells intercalary, globular, subglobular, 2.6–4.6 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Hainan Province, Lingshui Li Autonomous County, Benhao Town (18.70161°N, 109.69318°E), from soil sample, 10 April 2023, Yi-Xin Wang (living culture SAUCC 4064-2).
GenBank accession numbers.
CGMCC 327028 (ITS, PP195847; LSU, PP195948; TEF, PP850088; ACT, PP933938; RPB1, PP842883), SAUCC 4064-2 (ITS, PP195848; LSU, PP195949; TEF, PP850089; ACT, PP933939; RPB1, PP842882).
Notes.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species Gongronellaabortosporangia formed an independent clade with high supports (MLBV = 100; Fig. 1), which is closely related to G.multiramosa (BIPP = 0.95; Fig. 1). This new species differs morphologically from G.multiramosa in sporangium, septum, columella, collar and apophysis (Wang et al. 2023a). The G.abortosporangia is different from G.multiramosa in shape and size of sterile sporangia, the former being variously shaped, mostly gourd-shaped, 11.6–16.7 × 5.5–17.7 μm, partially elliptical with a slight shrinkage, 12.5–18.0 × 6.7–10.6 μm, occasionally clavate, 20.1–22.7 × 9.5–10.4 μm, while the latter being only ovoid, 9.6 × 6.2 µm in diameter. In fertile sporangia, G.abortosporangia has a smaller minimum diameter than G.multiramosa (7.0 μm vs. 15.5 μm). G.abortosporangia has more septa on sporangiophores compared to G.multiramosa (0–2 vs. 0–1). Although G.abortosporangia is similar in shape of columellae to G.multiramosa, it is shorter in length (hemispherical, 3.6–7.4 µm vs. 8.0–9.8 µm, sub-hemispherical, 3.6–5.5 μm vs. 7.6–10.0 µm). The G.abortosporangia has shorter collars than G.multiramosa, 0.6–3.9 μm vs. 1.3–7.2 µm. The G.abortosporangia is similar in shape of apophyses to G.multiramosa. However, they are different from each other in main pattern and size: The former mostly cup-shaped (1.9–8.6 × 2.1–6.7 μm vs. 4.6–7.0 × 8.5–10.0 µm) and partially hemispherical (2.7–5.5 × 2.8–7.4 μm vs. 4.4–5.6 × 8.5–9.0 µm) and the latter opposite. Combining morphological and molecular phylogenetic analyses, we classified the two isolates as a new species G.abortosporangia allied to G.multiramosa.
. Gongronella apophysata
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
7FE20022-07AB-54AB-9323-FBC903DC5A81
Fungal Names: FN 571631
Figure 3.
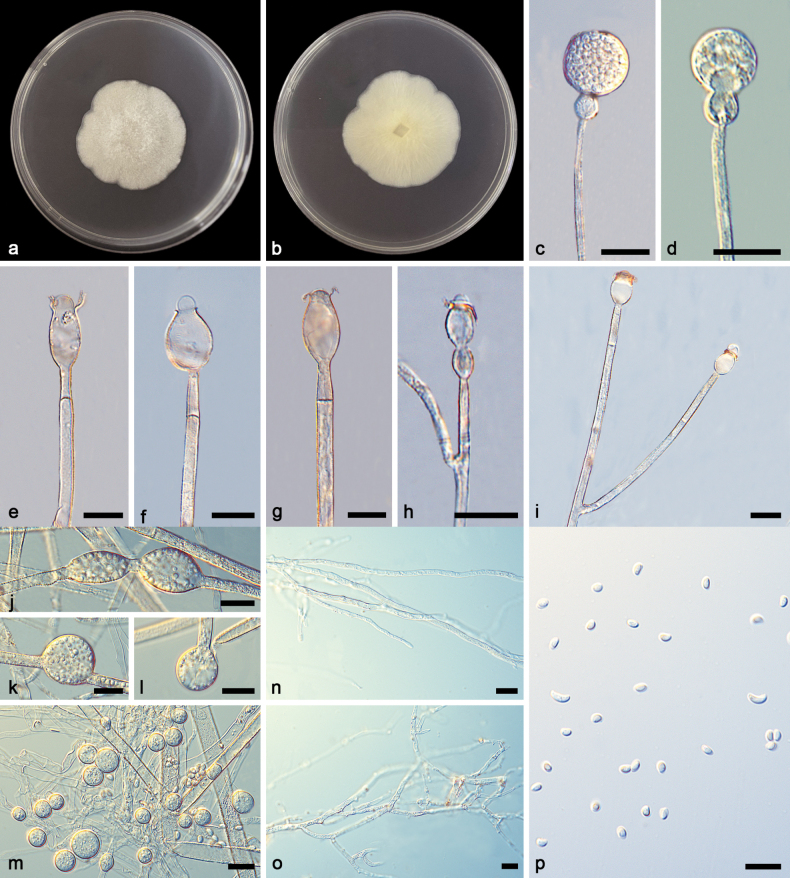
Gongronellaapophysata ex-holotype CGMCC 3.27031 a, b colonies on PDA (a obverse b reverse) c an unbranched sporangiophore with a fertile sporangium d an unbranched sporangiophore with an immature sporangium e–g columellae, collars, apophyses and septa h branched sporangiophores with columellae, collars and apophyses i branched sporangiophores with columellae, collars and apophyses j–l chlamydospores m giant cells n, o rhizoids p sporangiospores. Scale bars: 10 μm (c–p).
Etymology.
The epithet “apophysata” (Latin) refers to various shapes of apophyses.
Type.
China • Sichuan Province, Emeishan City, Leshan City, Ehong Road, near the Xu family residence (29.59211°N, 103.37776°E), from soil sample, 25 June 2023, Yi-Xin Wang (holotype HMAS 352728, ex-holotype strain CGMCC 3.27031).
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 35.8–42.4 mm in diameter in seven days, white, irregular at edge and cottony in the centrr, reversely milky white. Rhizoids hyaline, branched, irregularly shaped. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or slightly branched (1–2 times), 11.2–190.9 × 1.6–3.9 μm, hyaline, smooth, mostly aseptate or one-septate, occasionally two-septate. Sterile (aborted) sporangia predominantly on the top of short lateral branches of sporangiophores, gourd-shaped, 14.0 × 8.3 μm. Fertile sporangia hyaline or light yellow, spherical, 12.5–40.5 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae elliptic, 2.6–4.0 × 2.1–5.5 μm, sub-hemispherical, 1.4–2.7 × 2.2–4.3 μm, hyaline, smooth. Apophyses hyaline, smooth, variously shaped, mostly ellipsoidal to olive-shaped, 2.3–17.3 × 2.4–10.0 μm, partially subglobose, 4.6–10.2 × 4.3–10.0 μm, occasionally gourd-shaped, 11.4 × 4.9 μm. Sporangiospores not uniform, hyaline, smooth, mostly reniform, 3.2–5.5 × 1.7–3.1 μm, ovoid, 2.5–3.7 × 1.7–2.6 μm, occasionally subglobose, 1.7–2.5 μm. Chlamydospores present, not uniform, gourd-shaped, ellipsoidal and suborbicular, mostly gourd-shaped, 23.5–35.4 × 10.8–14.0 μm, partially ellipsoidal, 18.6–21.4 × 10.3–18.5 μm. Giant cells in the rhizoids, intercalary, globose, 4.4–10.5 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Sichuan Province, Emeishan City, Leshan City, Ehong Road, near the Xu family residence (29.59211°N, 103.37776°E), from soil sample, 25 June 2023, Yi-Xin Wang (living culture SAUCC 4846-3).
GenBank accession numbers.
CGMCC 3.27031 (ITS, PP195853; LSU, PP195954; TEF, PP850099; ACT, PP933947; RPB1, PP842878), SAUCC 4846-3 (ITS, PP195854; LSU, PP195956; TEF, PP850100; ACT, PP933948; RPB1, PP842877).
Notes.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species Gongronellaapophysata form an independent clade with high support (MLBV = 98; Fig. 1), which is closely related to G.zunyiensis. In ITS, G.apophysata differs from the type species of G.zunyiensis by 13 base pairs. This new species differs morphologically from G.zunyiensis in sporangium, columellae, apophyses and chlamydospore (Dong et al. 2019). G.apophysata has larger sporangia than G.zunyiensis (12.5–40.5 μm vs. 11.0–19.5 μm). G.apophysata differs from G.zunyiensis in the shape of columellae, the former being elliptic and the latter being hemispherical and globose. As for apophyses, G.apophysata and G.zunyiensis are remarkably different in shape and size, the former variously shaped, mostly ellipsoidal to olivary, 2.3–17.3 × 2.4–10.0 μm, partially subglobose, 4.6–10.2 × 4.3–10.0 μm, occasionally gourd-shaped, 11.4 × 4.9 μm and the latter hemispherical, 1.5–3.5 × 1.0–3.0 μm. G.apophysata is remarkably different from G.zunyiensis in shape and size of chlamydospores, the former being not uniform, mostly gourd-shaped, 23.5–35.4 × 10.8–14.0 μm, partially ellipsoidal, 18.6–21.4 × 10.3–18.5 μm and the latter being terminal or lateral, globose or subglobose, 7.0–10.5 μm in diameter. Combining morphological and molecular phylogenetic analyses, we classified the two isolates together as a new species G.apophysata allied to G.zunyiensis.
. Gongronella bawanglingensis
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
D23F69CC-3F5F-52A8-8049-F7DCD0A0CC78
Fungal Names: FN 571903
Figure 4.
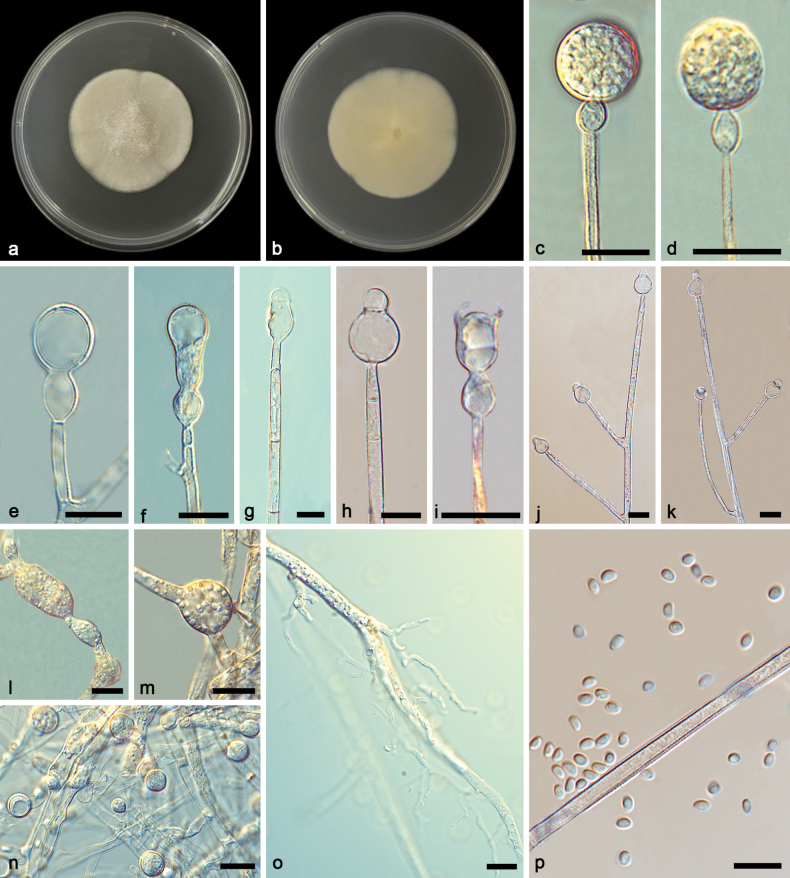
Gongronellabawanglingensis ex-holotype CGMCC 3.27033 a, b colonies on PDA (a obverse b reverse) c, d an unbranched sporangiophore with a fertile sporangium e branched sporangiophores with sterile (aborted) sporangia f branched sporangiophores with immature sporangia g–i columellae, collars, apophyses and septa j, k branched sporangiophores with columellae, collars and apophyses l, m chlamydospores n giant cells o rhizoids p sporangiospores. Scale bars: 10 μm (c–p).
Etymology.
The epithet “bawanglingensis” (Latin) refers to the location where the type was collected, Bawangling National Forest Park.
Type.
China • Hainan Province, Changjiang Li Autonomous County, Bawangling National Forest Park (19.08593°N, 109.12275°E), from soil sample, 14 October 2023, Yi-Xin Wang (holotype HMAS 352730, ex-holotype strain CGMCC 3.27033).
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 45.6–48.8 mm in diameter in seven days, white, cottony in the centre, on the reverse milky white. Rhizoids hyaline, branched, irregularly shaped. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or slightly branched (up to 3 times), sympodially branched, 1.3–4.5 μm in width, hyaline, smooth, mostly aseptate or one-septate, no more than four-septate. Sterile (aborted) sporangia mainly on the top of short lateral branches of sporangiophores, mostly gourd-shaped. Fertile sporangia hyaline or light yellow, spherical, 4.2–18.5 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly hemispherical, 1.6–5.1 × 2.1–7.2 μm, sometimes arch-shaped, 1.4–3.7 × 2.6–8.8 μm, occasionally spherical, 2.3–6.1 × 2.5–8.1 μm, hyaline, smooth. Collars mostly distinct, 0.7–5.9 μm. Apophyses hyaline, smooth, variously shaped, mostly oval, 3.9–20.6 × 3.3–12.9 μm, sometimes subglobose, 4.8–12.2 × 4.7–12.3 μm, occasionally gourd-shaped. Sporangiospores not uniform, hyaline, smooth, mostly ovoid, 2.5–3.6 × 1.7–2.6 μm, partially reniform, 2.6–3.3 × 1.9–2.2 μm. Chlamydospores not uniform, gourd-shaped, 15.1–24.6 × 7.4–12.9 μm, ellipsoidal, 15.1–18.6 × 8.3–14.0 μm, suborbicular, 12.6–13.5 μm in diameter. Giant cells intercalary, globular, 3.2–6.9 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Hainan Province, Changjiang Li Autonomous County, Bawangling National Forest Park (19.08593°N, 109.12275°E), from soil sample, 14 October 2023, Yi-Xin Wang (living culture SAUCC 6946-1).
GenBank accession numbers.
CGMCC 3.27033 (ITS, PP195857; LSU, PP195958; TEF, PP50103; ACT, PP933951; RPB1, PP883965), and SAUCC 6946-1 (ITS, PP1195858; LSU, PP195959; TEF, PP850104; ACT, PP933952; RPB1, PP883964).
Notes.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species Gongronellabawanglingensis form an independent clade with full support (MLBV = 100, BIPP = 1; Fig. 1), which is closely related to G.inconstans and G.qichaensis. In ITS, G.bawanglingensis differs from G.inconstans by 21 base pairs. This new species differs morphologically from G.inconstans in columella, apophysis, colour and sporangiospore. G.bawanglingensis and G.inconstans are similar in the dominant shape of columellae, but the former is longer than that of the latter (2.1–7.2 μm vs. 2.0–3.9 μm). As for apophyses, G.bawanglingensis and G.inconstans are remarkably different from each other in shape and size, the former mostly oval, 3.9–20.6 × 3.3–12.9 μm, sometimes subglobose, 4.8–12.2 × 4.7–12.3 μm, occasionally gourd-shaped, the latter mostly long fusiform, 7.6–17.4 × 5.4–4.7 μm, sometimes oval, 5.5–8.8 × 4.4–6.3 μm, rarely egg-shaped, 5.0–6.4 × 4.2–5.7 μm. As for collars, the G.inconstans are more distinct than G.bawanglingensis (2.0–17.0 μm vs. 0.7–5.9 μm). As for sporangiospores, G.bawanglingensis and G.inconstans are similar in dominant shape, but the former is smaller in size than the latter (ovoid, 2.5–3.6 × 1.7–2.6 μm vs. 2.7–4.9 × 1.8–3.5 μm, reniform, 2.6–3.3 × 1.9–2.2 μm vs. 3.1–4.1 × 2.0–4.5 μm). Additionally, the G.inconstans has more shapes, except ovoid and reniform. In ITS, G.bawanglingensis differs from G.qichaensis by 28 base pairs. This new species differs morphologically from G.qichaensis in sporangium, columellae and apophysis (Wang et al. 2023a). The G.bawanglingensis has evidently smaller sporangia than G.qichaensis, 4.2–18.5 μm vs. 7.9–36.7 μm. In columella and apophysis, the two species have evident differences in shape. Combining morphological and molecular phylogenetic analyses, we classified the two isolates together as a new species G.bawanglingensis allied to G.inconstans and G.qichaensis.
. Gongronella inconstans
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
92DF5D15-CA6D-5A3F-B2E7-49765B7D306D
Fungal Names: FN 571905
Figure 5.
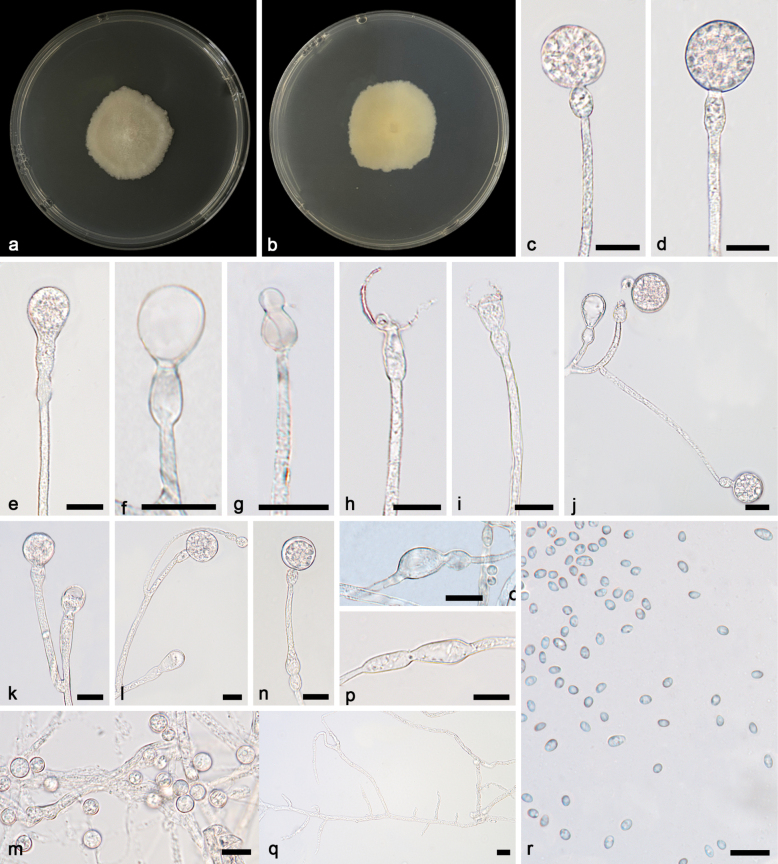
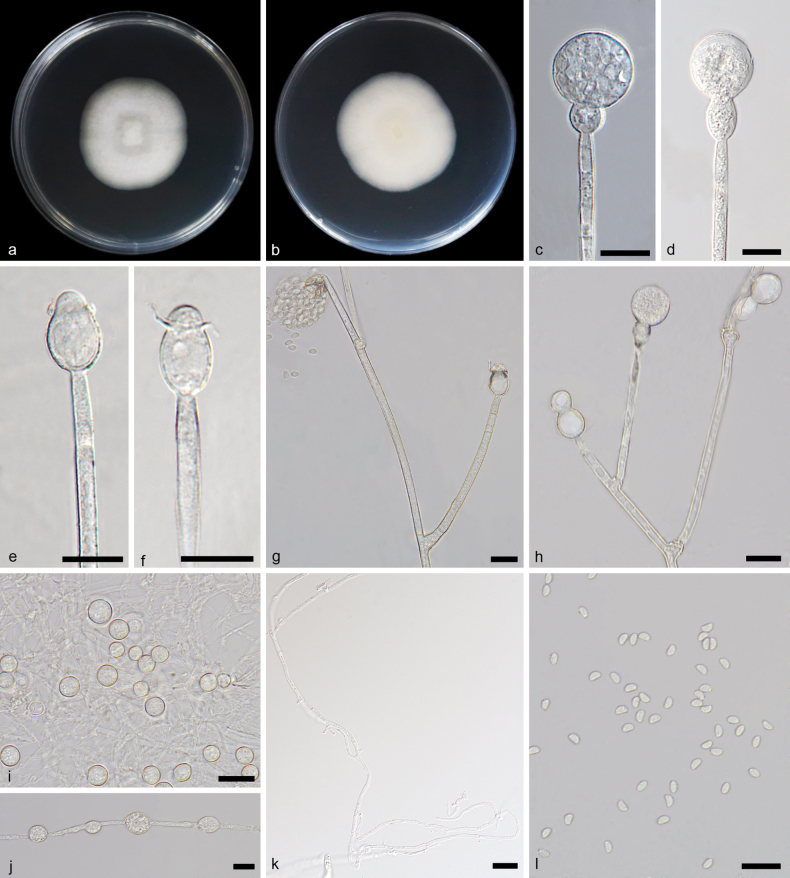
Gongronellainconstans ex-holotype CGMCC 3.27029 a, b colonies on PDA (a obverse b reverse) c, d an unbranched sporangiophore with a fertile sporangium e an unbranched sporangiophore with a premature sporangium f branched sporangiophores with aborted sporangia g–i columellae, collars, apophyses j–l branched sporangiophores with fertile sporangia, columellae, collars and apophyses n fertile sporangium with protuberance o, p chlamydospores m giant cells q rhizoids r sporangiospores. Scale bars: 10 μm (c–r).
Etymology.
The epithet “inconstans” (Latin) refers to the inconstant shape of apophyses.
Type.
China • Hainan Province, Lingshui Li Autonomous County (18.69850°N, 109.88098°E), from soil sample, 7 Apr 72023, Yi-Xin Wang (holotype HMAS 352731, ex-holotype strain CGMCC 3.27029).
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 31.2–36.8 mm in diameter in seven days, white, regular at edge and cottony, reversely milky white. Rhizoids hyaline, branched, irregular, ubiquitous. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or slightly branched (2–3 times), 1.7–3.9 μm width, hyaline, smooth, mostly aseptate. Fertile sporangia hyaline or light yellow, spherical, 8.8–21.4 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly hemispherical, 1.2–2.4 × 2.0–3.9 μm, sometimes spherical, 3.2–7.2 × 3.4–7.2 μm, hyaline, smooth. Collars distinct, 2.0–17.0 μm wide. Apophyses hyaline, smooth, variously shaped, mostly long fusiform, 7.6–17.4 × 4.7–5.4 μm, sometimes oval, 5.5–8.8 × 4.4–6.3 μm, rarely egg-shaped, 5.0–6.4 × 4.2–5.7 μm. Sporangiospores not uniform, hyaline, smooth, mostly ovoid, 2.7–4.9 × 1.8–3.5 μm, sometimes reniform, 3.1–4.1 × 2.0–4.5 μm or subglobose, 2.4–4.1 μm in diameter, occasionally irregular, 5.0–8.0 × 2.5–3.2 μm. Chlamydospores present, gourd-shaped and irregular. Giant cells intercalary, globular, 4.2–8.0 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Hainan Province, Lingshui Li Autonomous County (18.69850°N, 109.88098°E), from soil sample, 7 April 2023, Yi-Xin Wang (living culture SAUCC 4113-1).
GenBank accession numbers.
CGMCC 3.27029 (ITS, PP1955849; LSU, PP195950; TEF, PP850091; ACT, PP933941; RPB1, PP842874), and SAUCC 4113-1 (ITS, PP105850; LSU, PP195951; TEF, PP850092, ACT, PP933942; RPB1, PP842873).
Note.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species Gongronellainconstans form an independent clade with full support (MLBV = 100, BIPP = 1; Fig. 1), which is closely related to G.qichaensis with high support (BIPP = 0.96; Fig. 1). In ITS, G.inconstans differs from G.qichaensis by 28 base pairs. This new species differs morphologically from G.inconstans in sporangium, columellae and apophysis. As for sporangium, the G.inconstans is smaller than the G.qichaensis, 8.8–21.4 μm vs. 7.9–36.7 μm. The G.inconstans and G.qichaensis are different in size and shape of columellae (Wang et al. 2023a). The G.inconstans mostly hemispherical, 1.2–2.4 × 2.0–3.9 μm, sometimes spherical, 3.2–7.2 × 3.4–7.2 μm. Additionally, the columellae of G.qichaensis is mostly ellipsoidal, 0.8–6.5 × 1.2–8.1 µm, sometimes sub-hemispherical to curved, 1.0–2.0 × 2.5–4.5 µm. G.inconstans and G.qichaensis are evidently different in apophysis shape. The former mostly long fusiform, sometimes oval-shaped and rarely egg shaped. The latter mostly pear-shaped to oval, partially elliptical or sub-globose. Combining morphological and molecular phylogenetic analyses, we classified the two isolates together as a new species: G.inconstans allied to G.qichaensis.
. Gongronella pamphilae
Y.P. Tan, Bishop-Hurley & R.G. Shivas
B30FB7F4-63FA-5B58-88DD-1A927935385D
Fungal Names: FN 900776
Figure 6.
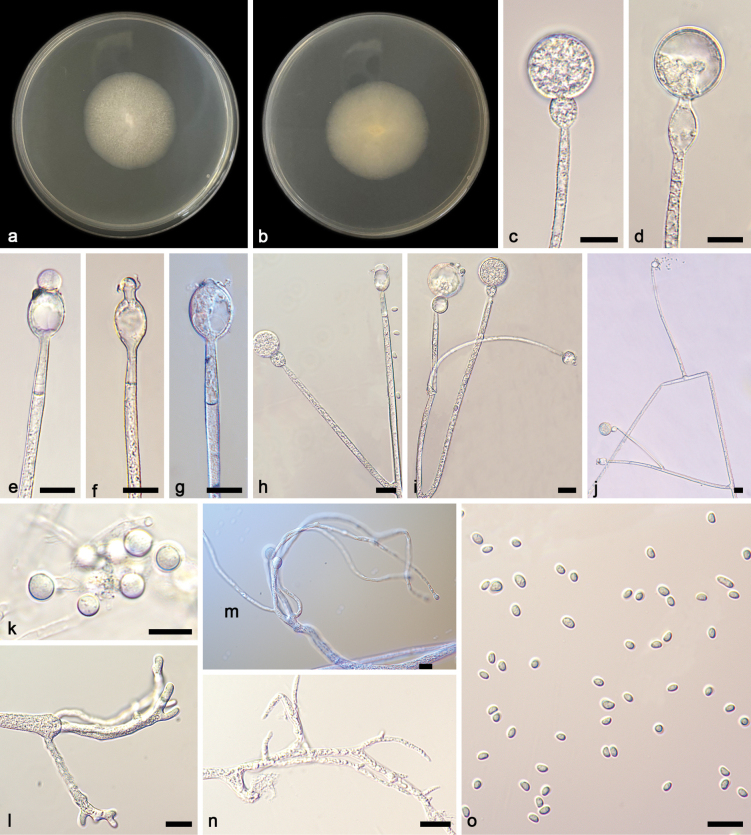
Gongronellapamphilae ex-living culture CGMCC 3.27027 a, b colonies on PDA (a obverse b reverse) c an unbranched sporangiophore with a fertile sporangium d an unbranched sporangiophore with an aborted sporangium e, f an unbranched sporangiophore with columellae, apophyses and collars g, h branched sporangiophores with columellae, collars, apophyses i Rhizoids j, k giant cells l sporangiospores. Scale bars: 10 μm (c–l).
Etymology.
Named after Pamphilae of Epidaurus (ca. 1st century AD), a historian of Egyptian descent who lived in Greece.
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 36.6–44.6 mm in diameter in seven days, white, regular at edge and cottony in the centre, reversely milky white. Rhizoids hyaline, branched, irregular. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or slightly branched (1–2 times), 3.7–154.9 × 1.4–4.1 μm, hyaline, smooth, mostly aseptate, no more than two-septate. Fertile sporangia hyaline or light yellow, spherical, 13.8–30.8 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly hemispherical, 1.8–4.7 × 2.0–7.7 μm, sometimes arc-shaped, 0.5–1.6 × 3.3–4.6 μm, occasionally subglobose, 4.8–6.4 × 5.9–6.9 μm, hyaline, smooth. Collars distinct, 1.0–5.1 μm wide. Apophyses hyaline, smooth, variously shaped, mostly subglobose, 5.7–8.1 × 5.6–9.0 μm, sometimes ellipsoidal, 4.8–6.9 × 4.8–6.1 μm. Sporangiospores not uniform, hyaline, smooth, reniform, 3.0–5.5 × 1.8–3.4 μm, ovoid, 2.5–5.6 × 1.8–3.7 μm. Chlamydospores present, ellipsoidal. Giant cells intercalary, globose, 4.0–8.1 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Hainan Province, Lingshui Li Autonomous County, Shizhi Village Road (18.70178°N, 109.83679°E), from soil sample, 10 April 2023, Yi-Xin Wang (specimen HMAS 352732, living culture CGMCC 3.27027, SAUCC 4031-2).
GenBank accession numbers.
CGMCC 3.27027 (ITS, PP195845; LSU, PP195946; TEF, PP850086; ACT, PP933935; RPB1, PP850081), and SAUCC 4031-2 (ITS, PP195846; LSU, PP195947; TEF, PP850085; ACT, PP933936; RPB1, PP850082).
Note.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 DNA sequences, the two isolates of the new record species Gongronellapamphilae form an independent clade with full support (MLBV = 100; Fig. 1), which is closely related to G.pamphilae (MLBV = 100; BI = 1, Fig. 1). In ITS, the two isolates differ from G.pamphilae by only 2 base pairs. As no morphological descriptions were provided for the G.pamphilae in its protologue, we classified the two isolates together as members of G.pamphilae just based on molecular phylogenetic analyses. Consequently, we provide herein a supplemental description for the species.
. Gongronella pingtangensis
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
2BEBB187-42DE-5630-A2D9-2BEA6C838CA6
Fungal Names: FN 571904
Figure 7.
Gongronellapingtangensis ex-holotype CGMCC 3.27032 a, b colonies on PDA (a obverse b reverse) c, d an unbranched sporangiophore with a fertile sporangium e–g columellae, collars, apophyses and septa h–j branched sporangiophores with fertile sporangia, columellae, collars, apophyses and septa k giant cells i–n rhizoids o sporangiospores. Scale bars: 10 μm (c–o).
Etymology.
The epithet “pingtangensis” (Latin) refers to the location where the type was collected, Pingtang County.
Type.
China • Qiannan Buyi and Miao Autonomous Prefecture, Pingtang County, Kapu Maonan Town (25.79510°N, 107.38631°E), from soil sample, 7 August 7 2023, Yi-Xin Wang (holotype HMAS 352732, ex-holotype strain CGMCC 3.27032).
Description.
Colonies growing slowly on PDA in darkness at 25 °C, reaching 38.8–45.6 mm in diameter in seven days, white, cottony, in reverse milky white. Rhizoids hyaline, branched, irregular. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched, or slightly branched (1–4 times), sympodially branched, 1.4–5.9 μm in width, hyaline, smooth, mostly aseptate or one-septate. Sterile (aborted) sporangia predominantly on the top of short lateral branches of sporangiophores. Fertile sporangia hyaline or light yellow, spherical, 14.2–27.1 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly hemispherical, 2.3–4.0 × 2.8–6.9 μm, partially arch-shaped, 0.9–1.5 × 4.1–4.9 μm, rarely spherical, 4.4–6.0 × 5.1–6.9 μm, hyaline, smooth. Collars mostly distinct, 0.6–8.7 μm wide. Apophyses hyaline, smooth, variously shaped, mostly oval, 7.1–19.8 × 6.9–15.9 μm, partially bowling pin-shaped, 15.6–17.5 × 8.5–9.4 μm, rarely egg-shaped, 4.6–9.8 × 3.6–8.7 μm. Sporangiospores not uniform, hyaline, smooth, mostly ovoid, 2.8–3.9 × 2.0–2.5 μm, sometimes reniform, 2.9–3.6 × 1.9–2.4 μm and globose, 2.1–2.7 μm in diameter, occasionally irregular, 4.8–6.2 × 2.1–2.8 μm. Chlamydospores absent. Giant cells in rhizoids, intercalary, globose, 5.2–6.8 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Qiannan Buyi and Miao Autonomous Prefecture, Pingtang County, Kapu Maonan Town (25.79510°N, 107.38631°E), from soil sample, 7 August 2023, Yi-Xin Wang (living culture SAUCC 5676-2).
GenBank accession numbers.
CGMCC 3.27032 (ITS, PP195855; LSU, PP195956; TEF, PP850101; ACT, PP933949; RPB1, PP842880), and SAUCC 5676-4 (ITS, PP195856; LSU, PP195957; TEF, PP850102; ACT, PP933950; RPB1, PP842879).
Note.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species G.pingtangensis form an independent clade with high support (MLBV = 100, BIPP = 0.84; Fig. 1), which is closely related to G.namwonensis with high support (BIPP = 1; Fig. 1). In ITS, G.pingtangensis differs from G.namwonensis by 14 base pairs. This new species differs morphologically from G.namwonensis in columellae, apophysis and giant cell (Crous et al. 2020). G.pingtangensis and G.namwonensis greatly differ from each other in shape of columellae, the former being mostly hemispherical, partially arch-shaped, rarely spherical and the latter being globose, subglobose, hemispherical, nipple-like and ellipsoidal. As for apophyses, G.pingtangensis and G.namwonensis obviously differ from each other in shape, the former being mostly oval, partially bowling pin-shaped, rarely egg-shape and the latter being subglobose and ellipsoid, sometimes with a truncated base. As for giant cells, the G.namwonensis varies in shape more than G.pingtangensis. Combining morphological and molecular phylogenetic analyses, we classified the two isolates together as a new species G.pingtangensis allied to G.namwonensis.
. Gongronella reniformis
Yi Xin Wang, H. Zhao & X.Y. Liu sp. nov.
02DD5FD9-57CE-5086-9663-46C81352A6F0
Fungal Names: FN 571630
Figure 8.
Gongronellareniformis ex-holotype CGMCC 3.27030 a, b colonies on PDA (a obverse b reverse) c an unbranched sporangiophore with a fertile sporangium d an unbranched sporangiophore with an immature sporangium e, f columellae, collars and apophyses g branched sporangiophores with shedding sporangia, columellae, collars, apophyses and septa h branched sporangiophores with fertile sporangia and sterile (aborted) sporangia i giant cells j chlamydospore k rhizoids l sporangiospores. Scale bars: 10 μm (c–l).
Etymology.
The epithet “reniformis “ (Latin) refers to the reniform sporangiospores.
Type.
China • Hainan Province, Changjiang Li Autonomous County, Qicha Town (19.11750°N, 109.15000°E), from soil sample, 11 April 2023, Yi-Xin Wang (holotype HMAS 352727, ex-holotype strain CGMCC 3.27030).
Description.
Colonies on PDA in darkness at 25 °C growing slowly, reaching 39.4–41.8 mm in diameter in seven days, white, regular at edge and cottony in the centre, on reverse milky white. Rhizoids hyaline, branched, irregular, sometimes with giant cells in the terminal. Stolons absent. Sporangiophores on aerial mycelia, erect or slightly curved, unbranched or slightly branched (1–3 times), 3.4–157.9 × 0.8–3.4 μm, hyaline, smooth, mostly aseptate, partially no more than two-septate. Sterile (aborted) sporangia predominantly on the top of short lateral branches of sporangiophores, gourd-shaped, 15.0–19.9 × 3.1–10.9 μm. Fertile sporangia hyaline or light yellow, spherical, 7.9–26.0 μm in diameter, smooth and deliquescent-walled, leaving a collar after releasing sporangiospores. Columellae mostly elliptic, 1.7–4.6 × 1.4–5.2 μm, sometimes sub-hemispherical, 1.4–2.6 × 3.3–4.9 μm, hyaline, smooth. Collars distinct, 2.1–4.3 μm. Apophyses hyaline, smooth, variously shaped, pear-shaped, 3.3–8.5 × 3.0–7.3 μm, ellipsoidal, 4.6–10.1 × 2.9–7.8 μm. Sporangiospores not uniform, hyaline, smooth, mostly reniform, 2.8–3.5 × 1.8–2.3 μm, occasionally ovoid, 3.1–3.4 × 1.7–2.0 μm. Chlamydospores, mostly ellipsoidal, 7.3–12.5 × 6.1–11.2 μm, sometimes irregular. Giant cells intercalary, globose, 3.5–10.0 μm in diameter. Zygospores not found.
Additional specimen examined.
China • Hainan Province, Changjiang Li Autonomous County, Qicha Town (19.11750°N, 109.15000°E), from soil sample, 11 April 2023, Yi-Xin Wang (living culture SAUCC 4142-5).
GenBank accession numbers.
CGMCC 3.27030 (ITS, PP195851; LSU, PP195952; TEF, PP850095; ACT, PP933943; RPB1, PP84875), SAUCC 4142-5 (ITS, PP195852; LSU, PP195953; TEF, PP850096; ACT, PP933944; RPB1, PP842876).
Notes.
Based on phylogenetic analyses of ITS+LSU+TEF+ACT+RPB1 sequences, the two isolates of the new species Gongronellareniformis form an independent clade with full support (MLBV = 100, BIPP = 1; Fig. 1), which is close to G.pamphilae and G.brasiliensis with a high support (MLBV = 89, BIPP = 1; Fig. 1). Comparing ITS sequences showed that G.reniformis is relatively closely related to G.pamphilae (44 bp of dissimilarity) and G.brasiliensis (40 bp of dissimilarity). There were no morphological descriptions of G.pamphilae in its protologue, so the morphological comparison was made between G.reniformis and the G.pamphilae strains identified in this study. This new species differs morphologically from G.pamphilae in sporangium, columellae, apophysis, sporangiospore. The sporangium of G.reniformis is smaller than that of G.pamphilae (7.9–26.0 μm vs. 13.8–30.8 μm). G.reniformis and G.pamphilae are different from each other mainly in shape and size of columellae, the former being mostly elliptic, 1.7–4.6 × 1.4–5.2 μm, sometimes sub-hemispherical, 1.4–2.6 × 3.3–4.9 μm and the latter being mostly hemispherical, 1.8–4.7 × 2.0–7.7 μm, sometimes arc-shaped, 0.5–1.6 × 3.3–4.6 μm. The G.reniformis and G.pamphilae are different from each other in dominant shape and size of apophyses, the former being pear-shaped, 3.3–8.5 × 3.0–7.3 μm and ellipsoidal, 4.6–10.1 × 2.9–7.8 μm, the latter being spherical, 5.7–8.1 × 5.6–9.0 μm and ellipsoidal, 4.8–6.9 × 4.8–6.1 μm. The sporangiospores of G.reniformis are smaller than those of G.pamphilae (reniform, 2.8–3.5 × 1.8–2.3 μm vs. 3.0–5.5 × 1.8–3.4 μm, ovoid, 3.1–3.4 × 1.7–2.0 μm vs. 2.5–5.6 × 1.8–3.7 μm). This new species differs morphologically from G.brasiliensis in sporangiophore, columellae and giant cells (Tibpromma et al. 2017). In sporangiophores, the G.renformis differs from the G.brasiliensis in size, 3.4–157.9 × 0.8–3.4 μm vs. 26.5–320.0 × 2.5–5.0 μm. As for columellae, the G.renformis and G.brasiliensis are different in shape. The former mostly elliptic, sometimes sub-hemispherical. The latter globose, subglobose and conical-cylindrical. The G.renformis is evidently smaller than G.brasiliensis in giant cells, 3.5–10.0 μm vs. up to 48 μm. Combining morphological and molecular phylogenetic analyses, we classified the two isolates as a new species G.reniformis.
Morphological comparisons and key to the species of Gongronella
Together with the six new species proposed in this study, a total of 25 species of Gongronella have been described worldwide. Except G.banzhaoae, morphological comparisons were made amongst 18 species published before and six species newly proposed in this study (Table 4). We provide herein a synoptic key for these species. Characteristics adopted in the key include colonies, sporangiophores, sporangia, columellae, apophyses, sporangiospores and giant cells.
Table 4.
Morphological comparisons of Gongronella species.
| Species | Colonies | Sporangiophores | Sporangia | Columellae | Apophyses | Sporangiospores | Giant cell | Reference |
|---|---|---|---|---|---|---|---|---|
| G.abortosporangia | PDA: dark 25 °C 7 d 24.6–26.2 mm diam., white, regular at edge and cottony in the centre, in reverse milky white | unbranched or branched 1–6 times, 4.0–96.8 × 1.0–4.2 μm, mostly aseptate, partially 1-septate, rarely 2-septate, occasionally containing a line of oil droplets | Aborted: mostly gourd-shape, 11.6–16.7 × 5.5–17.7 μm, partially elliptical with slight shrinkage, 12.5–18.0 × 6.7–10.6, occasionally clavate, 20.1–22.7 × 9.5–10.4 μm; Fertile: 7.0–23.2 μm diam | mostly hemispherical, 2.5–4.2 × 3.6–7.4 μm, sometimes sub-hemispherical, 1.3–3.9 × 3.6–5.5 μm | mostly cup-shaped, 1.9–8.6 × 2.1–6.7 μm, partially hemispherical, 2.7–5.5 × 2.8–7.4 μm, occasionally pear-shaped, 8.2 × 7.2 μm | ovoid, 2.6–3.5 × 1.7–2.1 μm, reniform, 2.9–3.5 × 1.7–2.3 μm | intercalary, globular, subglobular, 2.6–4.6 μm diam. | This study |
| G.apophysata | PDA: dark 25 °C 7 d 17.9–21.2 mm in diam., white, irregular at edge and cottony in centre, in reverse milky white | unbranched or branched 1–2 times, 11.2–190.9 × 1.6–3.9 μm, mostly aseptate or 1-septate, occasionally 2-septate | Aborted: gourd-shape, 14.0 × 8.3 μm; Fertile: spherical, 12.5–40.5 μm diam. | elliptic, 2.6–4.0 × 2.1–5.5 μm, sub-hemispherical, 1.4–2.7 × 2.2–4.3 μm | mostly ellipsoidal to olivary, 2.3–17.3 × 2.4–10.0 μm, partially subglobose, 4.6–10.2 × 4.3–10.0 μm, occasionally gourd-shaped, 11.4 × 4.9 μm | mostly reniform, 3.2–5.5 × 1.7–3.1 μm, ovoid, 2.5–3.7 × 1.7–2.6 μm, occasionally sub-orbicular, 1.7–2.5 μm | intercalary, globular, 4.4–10.5 μm diam. | This study |
| G.bawanglingensis | PDA: dark 25 °C 7 d 22.8–24.4 mm diam., white, cottony in centre, in reverse milky white | unbranched or sympodially branched 1–3 times, 1.3–4.5 μm wide, mostly aseptate or 1-septate, occasionally up to 4-septate | Aborted: mostly gourd-shaped; Fertile: spherical, 4.2–18.5 μm diam. | mostly hemispherical, 1.6–5.1 × 2.1–7.2 μm, some arch-shaped, 1.4–3.7 × 2.6–8.8 μm, spherical, 2.3–6.1 × 2.5–8.1 μm | oval-shaped, 3.9–20.6 × 3.3–12.9 μm, subglobose-shaped, 4.8–12.2 × 4.7–12.3 μm, occasionally gourd-shaped | mostly ovoid, 2.5–3.6 × 1.7–2.6 μm, reniform, 2.6–3.3 × 1.9–2.2 μm | intercalary, globular, 3.2–6.9 μm diam. | This study |
| G.brasiliensis | MEA: 25 °C 7 d 1.0–2.0 mm high 60.0 mm diam., white, cottony, irregular at edge, reverse cream to buff | 26.5–320.0 × 2.5–5.0 µm, solitary, arising from stolons or in whorls of two, often with a single branch, 1- or 2-septate below apophyses | Aborted: globose, 5.0–17.0 µm diam.; Fertile: globose, subglobose, 9.5–30.0 μm diam. | globose, subglobose, (3.0–)4.0–8.0(–9.0) μm, conical-cylindrical, 1.5–2.5 × 2.0–3.0 μm, some very small, up to 1 μm diam. | globose, (3.0–)4.0–5.0(–6.0) μm diam., vase-shaped, (3.0–)4.0 × 12.0(–14.5) μm, ellipsoidal, 5.0–10.0(–12.0) × 3.0–7.0(–8.5) μm | reniform, 1.5–4.0 × 1.5–2.5 μm, ellipsoid to fusiform, 2.0–6.5 × 1.5–3.0 μm, ellipsoid with a flattened end, 2.5–7.5 × 1.5–4.0 μm | globose, subglobose, ovoid, some hypha-like, irregularly swollen, up to 48.0 μm diam. | Tibpromma et al. (2017) |
| G.butleri | White turf | simply or irregularly branched, 2.1–3.1 μm wide, always 1-septate | Fertile: globose, 16.5–22.7 μm | swollen, oval-shaped, 7.0–10 × 8.0–8.7 μm | oval to flattened on one side to reniform, 2.5–7.2 × 1.7–4.7 μm | Ribaldi (1952), Babu et al. (2015) | ||
| G.chlamydospora | PDA: 27 °C 11 d 90.0 mm diam., floccose, at first white, then drab grey | unbranched or simply branched, hyaline, slightly constricted at top | Fertile: globose, 8.5–17.0 μm diam. | ovoid to depressed subglobose, 3.0–5.5 × 3.5–6.5 μm | urn-shaped to subglobose, 6.0–12.0 × 6.0–10.0 μm | ellipsoid, reniform or irregular, 2.0–3.0 × 1.0–2.0 μm | Zhao et al. (2023) | |
| G.eborensis | PDA: 25 °C 5 d 28.0–32.0 mm diam. | 46.0–94.0 × 1.5–3.0 µm, irregularly or simply branched, always 1-septate under apophyses | Fertile: globose to subglobose, 7.5–16.0 × 7.0–13.0 µm | hemispherical to subglobose, 11.5–5.5 × 8.2–3.2 µm | globose to subglobose, 3.5–6.5 × 3.0–7.0 µm | reniform to fusiform-elliptical, 2.6–3.8 × 1.2–1.6 µm | Martins et al. (2020) | |
| G.guangdongensis | PDA: 25 °C 13 d 1–2 mm high, 50.0 mm diam., white or pale, irregular at edge; in reverse buff to honey | irregularly or simply branched, 28.0–100.0 × 2.0–2.5 μm, always 1-septate | Aborted: sometimes present; Fertile: always globose, 14.0–21.5 μm diam. | hemispherical, spherical or ovoid, 2.5–12.0 × 2.0–12.0 μm | hemispherical, 5.5–9.0 μm in diam. | globose, 2.0–3.0 μm in diam. | Adamcik et al. (2015) | |
| G.hydei | PDA: 25 °C 7 d 60.0–65.0 mm diam., circular, entire at edge, flat or effuse, dense, white; | up to 120.0 µm long, 1.6–3.2 µm wide, mostly unbranched, occasionally branched, mostly 1-septate | Fertile: globose to subglobose, 10.5–18.8 × 10.0–17.5 µm | hemispherical, sometimes tiny, 1.7–4.7 × 2.2–6.3 µm | cuboid-shaped with truncate at the base, 2.5–3.9 × 3.5–5.1 µm; cup-shaped rounded at the base, 2.7–6.2 × 3.8–7.8 µm; cup-shaped truncate at the base, 3.7–7.3 × 3.8–7.3 µm | reniform, 2.4–3.8 × 1.5–2.3 µm, ellipsoidal to fusiform, 2.6–3.4 × 1.8–3.4 µm | globose, guttulate, up to 25.0 µm diam. | Doilom et al. (2020) |
| G.inconstans | PDA: dark 25 °C 7 d 15.6–18.4 mm diam., white, regular at edge, cottony, in reverse milky white | unbranched or branched 2–3 times, 1.7–3.9 μm wide, mostly aseptate | Aborted: existence; Fertile: spherical, 8.8–21.4 μm diam. | mostly hemispherical, 1.2–2.4 × 2.0–3.9 μm, sometimes spherical, 3.2–7.2 × 3.4–7.2 μm | variously shaped, mostly long fusiform, 7.6–17.4 × 4.7–5.4 μm, sometimes oval-shaped, 5.5–8.8 × 4.4–6.3 μm, rarely egg-shaped, 5.0–6.4 × 4.2–5.7 μm | ovoid, 2.7–4.9 × 1.8–3.5 μm, reniform, 3.1–4.1 × 2.0–4.5 μm, some subcircular, 2.4–4.1 μm, occasionally irregular, 5.0–8.0 × 2.5–3.2 μm | intercalary, globular, 4.2–8.0 μm diam. | This study |
| G.koreana | PDA: 25 °C 7 d 31.5–33.0 mm diam., light white at first, cotton white with age, reverse from light-coloured to white | 2.5–2.8 μm wide, mostly branched, 1-septate | Fertile: globose, 12.3–15.5 × 12.4–15.6 μm diam. | hemispherical, 1.2–2.3 × 2.6–3.3 μm | typically pyriform, 5.4–6.5 × 5.9–7.1 μm | mostly bean-shaped, 1.7–2.1 × 2.1–2.8 μm | Ariyawansa et al. (2015) | |
| G.lacrispora | 25 °C 13 d 50.0 mm in diam., 1–3 mm high, thickly floccose to felty, irregular at edge, at first white, then grey or pale grey, later pale wine colour | up to 6.5 μm wide, rarely septate | Aborted: sometimes present; Fertile: typically perfectly globose, 13.0–41.0 μm diam | dorsiventrally flattened to spherical, 2.5–13.0 × 4.5–20.0 μm | hemispherical, 4.0–8.6 μm. in diam. | lacrymoid to narrowly napiform, 2.8-4.5 × 5.5-9.0 μm | intercalary, globose to irregular, often with vacuoles or oil droplets, 20.0-37.0 × 60.0 μm | Hesseltine and Ellis (1961) |
| G.multiramosa | PDA: dark 25 °C 7 d 21.6–25.6 mm diam., white, regular at edge, cottony in centre, reverse milky white | unbranched or sympodially branched up to 7 times, 4.7–128.4 × 2.6–3.9 µm, usually 1-septate, occasionally containing a line of oil droplets | Aborted: ovoid, 9.6 × 6.2 µm diam.; Fertile: spherical, 15.5–23.2 µm diam.; | mostly hemispherical, 3.6–5.7 × 8.0–9.8 µm, sometimes sub-hemispherical, 3.0; –3.9 × 7.6–10.0 µm | mostly hemispherical, 4.4–5.6 × 8.5–9.0 µm, partially cup-shaped,4.6–7.0 × 8.5–10.0 µm | subspherical, 1.7–2.6 µm, ovoid, 2.6–3.3 × 1.7–2.3 µm, few reniform, 2.7–3.4 × 1.3–1.9 µm | globular, sub-spherical, 3.0–6.7 µm diam. | Wang et al. (2023a) |
| G.multispora | PDA: 27 °C 10 d 80.0 mm diam., 10.0 mm high, from white to yellowish, in reverse crusty, yellow | unbranched or sympodially branched, 2–3 in whorls and swollen on the base, 1 to several septate | Fertile: globose, 12.0–17.0 μm diam. | hemispherical, 2.0–4.5 × 2.0–4.0 μm | pyriform to subglobose, 8.0–12.0 × 7.0–9.5 μm | ellipsoid, fusiform, cylindrical, reniform subglobose to globose or irregular, 2.5–3.5 × 1.5–2.5 μm | Zhao et al. (2023) | |
| G.namwonensis | MEA:25 °C 7 d 55.0 mm diam. (28 °C 5 d 90.0 mm diam), white, in reverse cream | simply or sympodially or monopodially branched, up to 1 mm long and 5.0 μm wide, in whorls of 2 or 3 times, mostly 1-septate | Aborted: sometimes formed; Fertile: globose, up to 30.0 µm diam. | globose, subglobose, 3.5–7.0 µm diam., hemispherical,; 1.8–5.5 × 2.5–8.5 µm, nipple-like, ellipsoidal, 2.0–3.8 × 2.0–5.0 µm | globose (2.5–)5.0–9.5(–12.0); µm, subglobose and ellipsoid, some with a truncated base,; 7.5–14.5 × 5.5–12.0 µm | reniform, ellipsoidal, some ovoid, 2.5–3.5 × 1.7–2.5 µm, rarely irregular, up to 6 × 2.5 µm | globose, subglobose and branched | Crous et al. (2020) |
| G.oleae | PDA: dark 25 °C 7 d, 16.3–17.0 mm diam., white, regular at edge, cottony in centre, inreverse milky white | unbranched or branched 3–4 times, 7.0–96.8 × 0.9–3.5 µm, mostly aseptate, sometimes 1-septate | Aborted: 7.0–7.8 µm diam.; Fertile: spherical, 8.8–24.5 µm diam.; | mostly sub-spherical or ovoid, 2.6–5.2 × 3.2–6.5 µm, sometimes hemi-spherical, 0.4–3.3 × 2.8–5.3 µm | pear-shaped, 4.4–5.6 × 8.5–9.0 µm, cup-shaped, 4.6–7.0 × 8.5–10.0 µm, elliptical or subspherical, 2.7–8.0 × 2.8–9.1 µm | ovoid, 2.40–3.34 × 1.51–2.35 µm, reniform, 2.58–4.99 × 1.48–2.24 µm | terminal, globular, sub-spherical, 3.2–6.5 µm diam. | Wang et al. (2023a) |
| G.orasabula | SMA: 25 °C 5 d, 33.0–35.0 mm, initial white, later off-white, irregular at edge, in reverse white | 35.0–200.0 × 2.5–4.0 μm, simply branched 1–3 times | Fertile: globose to subglobose or calabash vase-shaped, 12.0–20.0 × 12.5–22.0 μm | hemispherical, 2.0–3.0 × 3.0–4.0 μm | globose, subglobose to pyriform, 5.0–10.0 × 4.5–8.5 μm | mostly bean-shaped, 2.0–3.5 × 2.0–2.5 μm | Li et al. (2016) | |
| G.pamphilae | PDA: dark 25 °C 7 d 18.3–22.3 mm in diam., white, regular at edge and cottony in centre, in reverse milky white | unbranched or branched 1–2 times, 3.7–154.9 × 1.4–4.1 μm, mostly aseptate, occasionally 1- or 2-septate | Aborted: existence; Fertile: spherical, 13.8–30.8 μm diam. | mostly hemispherical, 1.8–4.7 × 2.0–7.7 μm, sometimes arc-shaped, 0.5–1.6 × 3.3–4.6 μm, spherical, 4.8–6.4 × 5.9–6.9 μm | spherical, 5.7–8.1 × 5.6–9.0 μm, ellipsoidal, 4.8–6.9 × 4.8–6.1 μm | reniform, 3.0–5.5 × 1.8–3.4 μm, ovoid, 2.5–5.6 × 1.8–3.7 μm | intercalary, globular, 4.0–8.1 μm diam | This study |
| G.pedratalhadensis | PDA: 25 °C 7 d 5.5 mm high, 45.0 mm diam., white, irregular at edge, in reverse pale | sympodially branched 1–2 times, 9.5–30.0 × 2.5–7.0 μm, mostly 1-septate below sporangia, rarely two or more septate | Aborted: existence; Fertile: globose 17.0–35.0(40.0) μm diam. | mostly hemispherical, some short hemispherical or subglobose, 5.0–15.0 × 4.0–21.5 μm | vasiform, short or long, 5.0–15.0 × 4.5–15.0 μm | bean-shaped, 2.5–3.5 × 1.5–2.5 μm, rarely irregular, 2.5–3.5 × 2.0–3.0 μm | de Freitas et al. (2020) | |
| G.pingtangensis | PDA: dark 25 °C 7 d 19.4–22.8 mm diam., white, cottony, in reverse milky white | unbranched or sympodially branched 1–4 times, 1.4–5.9 μm wide, aseptate or 1-septate | Aborted: existence; Fertile: spherical, 14.2–27.1 μm diam.; | mostly hemispherical, 2.3–4.0 × 2.8–6.9 μm, some arch-shaped, 0.9–1.5 × 4.1–4.9 μm, spherical, 4.4–6.0 × 5.1–6.9 μm | mostly oval-shaped, 7.1–19.8 × 6.9–15.9 μm, some bowling pin-shaped, 15.6–17.5 × 8.5–9.4 μm, egg-shaped, 4.6–9.8 × 3.6–8.7 μm | mostly ovoid, 2.8–3.9 × 2.0–2.5 μm, some reniform, 2.9–3.6 × 1.9–2.4 μm, spherical, 2.1–2.7 μm, occasionally large irregularly shaped, 4.8–6.2 × 2.1–2.8 μm | intercalary, globular, 5.2–6.8 μm diam. | This study |
| G.qichaensis | PDA: dark, 25 °C 7 d 20.3–22.7 mm diam., white, cottony, regular at edge, in reverse milky white | unbranched or branched 1–2 times, 17.3–141.2 × 0.7–4.3 µm, usually aseptate, occasionally 2-septate | Aborted: ovoid,12.2–13.7 µm in diam.; Fertile: spherical, 7.9–36.7 µm diam. | ellipsoidal, 0.8–6.5 × 1.2–8.1 µm, sometimes sub-hemispherical to curved, 1.0–2.0 × 2.5–4.5 µm | mostly pear-shaped to oval, 4.6–13.4 × 3.4–10.7 µm, partially elliptical or sub-spherical, 6.0–11.3 × 4.8–9.0 µm | mostly ellipsoidal, 3.0–4.2 × 2.1–2.8 µm, sometimes reniform, 2.8–3.7 × 2.3–2.8 µm, few spherical, 2.4–3.3 µm | intercalary or terminal, globular, sub-spherical, 3.5–6.7 µm diam. | Wang et al. (2023a) |
| G.reniformis | PDA: dark 25 °C 7 d 19.7–20.9 mm diam., white, regular at edge and cottony in centre in reverse milky white | unbranched or branched 1–3 times, 3.4–157.9 × 0.8–3.4 μm, mostly aseptate, occasionally 1- or 2-septate | Aborted: gourd-shape, 15.0–19.9 × 3.1–10.9 μm; Fertile: spherical, 7.9–26.0 μm diam. | mostly elliptic, 1.7–4.6 × 1.4–5.2 μm, sometimes sub-hemispherical, 1.4–2.6 × 3.3–4.9 μm | pear-shaped, 3.3–8.5 × 3.0–7.3 μm, ellipsoidal, 4.6–10.1 × 2.9–7.8 μm | mostly reniform, 2.8–3.5 × 1.8–2.3 μm, ovoid, 3.1–3.37 × 1.7–2.0 μm | intercalary, globular, 3.5–10.0 μm diam. | This study |
| G.sichuanensis | PDA: 25 °C 14 d 4.0–5.0 mm high, 67.0–68.0 mm diam., white, regular at edge, in reverse grey | solitary or simply branched, 28.0–46.5 × 1.0–3.0 μm, 1- or 2-septate | Fertile: globose, subglobose, 10.5–26.5 μm diam. | hemispherical, 1.5–3.5 × 1.0–3.0 μm | ellipsoidal to subglobose, 4.5–8.5 × 4.5–6.0 μm in diam. | reniform, ovoid or ellipsoidal, 1.5–2.0 × 1.0–1.5 μm | Zhang et al. (2019) | |
| G.zunyiensis | PDA: 25 °C 14 d 3.0–6.0 mm high, 70.0–75.0 mm diam., white, villiform, irregular at edge, in reverse grey-white | 1.5–4.0 μm wide, branched several times, usually aseptate | Fertile: subglobose to globose, 11.0–19.5 μm diam. | hemispherical and globose, 2.0–3.0 × 3.5–7.0 μm | subglobose, 3.5–9.5 μm, conical-cylindrical, 4.0–7.0 × 5.0–9.0 μm | subglobose, reniform, 1.5–2.0 × 2.0–3.5 μm | Dong et al. (2019) |
| 1 | Giant cells known | 2 |
| – | Giant cells unknown | 15 |
| 2 | Aborted sporangia known | 3 |
| – | Aborted sporangia unknown | G.hydei |
| 3 | Fertile sporangia > 25 μm diameter | 4 |
| – | Fertile sporangia < 25 μm diameter | 10 |
| 4 | Sporangiospores mainly not reniform | 5 |
| – | Sporangiospores mainly reniform | 7 |
| 5 | Columellae mainly ellipsoidal | G.qichaensis |
| – | Columellae mainly not ellipsoidal | 6 |
| 6 | Fertile sporangia 14.2–27.1 μm | G.pingtangensis |
| – | Fertile sporangia, 13.0–41.0 μm | G.lacrispora |
| 7 | Sporangiospores > 4 μm wide | 8 |
| – | Sporangiospores < 4 μm wide | 10 |
| 8 | Sporangiophores branched ≥ 3 times | G.namwonensis |
| – | Sporangiophores branched < 3 times | 9 |
| 9 | Columellae mainly globose and subglobose, 4.0–8.0 μm | G.brasiliensis |
| – | Columellae mainly hemispherical, 1.8–4.7 × 2.0–7.7 μm | G.pamphilae |
| 10 | Apophyses mainly reniform, 2.8–3.5 × 1.8–2.3 μm | G.reniformis |
| – | Apophyses mainly reniform, 3.2–5.5 × 1.7–3.1 μm | G.apophysata |
| 11 | Sporangiophores branched > 3 times | 12 |
| – | Sporangiophores branched ≤ 3 times | 14 |
| 12 | Giant cells > 6 μm diameter | 13 |
| – | Giant cells < 6 μm diameter | G.abortosporangia |
| 13 | Columellae mainly subspherical and ovoid, 2.6–5.2 × 3.2–6.5 μm | G.oleae |
| – | Columellae mainly hemispherical, 4.4–5.6 × 8.5–9.0 μm | G.multiramosa |
| 14 | Apophyses oval, subglobose and gourd-shaped | G.bawanglingensis |
| – | Apophyses long fusiform, oval and egg-shaped | G.inconstans |
| 15 | Fertile sporangia > 25 μm diameter | 16 |
| – | Fertile sporangia < 25 μm diameter | 17 |
| 16 | Apophyses vasiform, 5.0–15.0 × 4.5–15.0 μm | G.pedratalhadensis |
| – | Apophyses ellipsoidal to subglobose, 4.5–8.5 × 4.5–6.0 μm | G.sichuanensis |
| 17 | Columellae hemispherical | 18 |
| – | Columellae not hemispherical | 19 |
| 18 | Apophyses urn-shaped to subglobose, 6.0–12.0 × 6.0–10.0 μm | G.chlamydospora |
| – | Apophyses oval, 7.0–10 × 8.0–8.7 μm | G.butleri |
| 19 | Sporangiospores ≥ 3.5 μm long | 20 |
| – | Sporangiospores < 3.5 μm long | 21 |
| 20 | Apophyses globose to subglobose, 3.5–6.5 × 3.0–7.0 μm | G.eborensis |
| – | Apophyses pyriform to subglobose, 8.0–12.0 × 7.0–9.5 μm | G.multispora |
| 21 | Sporangiospores ≥ 4 μm width, mostly not bean-shaped | 22 |
| – | Sporangiospores < 4 μm width, mostly bean-shaped | 23 |
| 22 | Columellae hemispherical, spherical or ovoid, 2.5–12.0 × 2.0–12.0 μm | G.guangdongensis |
| – | Columellae hemispherical and globose, 2.0–3.0 × 3.5–7.0 μm | G.zunyiensis |
| 23 | Sporangiospores 2.0–3.5 × 2.0–2.5 μm | G.orasabula |
| – | Sporangiospores 1.7–2.1 × 2.1–3.8 μm | G.koreana |
Discussion
Southern China is located in tropical and subtropical areas, which belong to tropical monsoon climate and subtropical monsoon climate. All the samples used in this study were collected from these areas, including Hainan, Sichuan, Yunnan and Guizhou Provinces. This is consistent with the geographical distribution of the species of Gongronella, mainly inhabiting tropical and subtropical regions.
The genus Gongronella was established in 1952 and its type Gongronellaurceolifera was synonymised with Gongronellabutleri whose basionym is Absidiabutleri (Ribaldi 1952). Numbers of this genus have increased rapidly recently, with as many as 17 species being described between 2015 and 2024 and the genus currently has a total of 25 members including the six new species proposed herein, all of which are listed in Table 3. However, there are no systematic analyses of the morphological characteristics of the species of Gongronella. In this study, the morphological characteristics of the 24 species of Gongronella were comparatively analysed (Table 4), except G.banzhaoae. Since G.banzhaoae only has molecular data and no morphological description, it is not compared in this study.
Since 2019, phylogenetic analyses of Gongronella have mainly been conducted on the basis of morphological characteristics and ITS+LSU sequence (Zhang et al. 2019). In this study, new TEF, ACT and RPB1 protein-coding sequences were added for the construction of phylogenetic trees and the results were basically consistent with previous studies based on ITS+LSU. Twelve strains were grouped into six individual clades and two strains were grouped along with G.pamphilae. Compared with G.multiramosa, G.abortosporangia has more abundant and various aborted sporangia, smaller fertile sporangia and smaller columellae (Wang et al. 2023a). Compared with G.pamphilae, G.reniformis has smaller sporangia and sporangiospores, as well as different shapes of columellae and apophyses. Compared with G.brasiliensis, G.reniformis has smaller sporangiophores, different columella shapes and smaller giant cells (Tibpromma et al. 2017). Compared with G.zunyiensis, G.apophysata has larger sporangia, as well as different shapes of columellae, apophyses and chlamydospores (Dong et al. 2019). Compared with G.inconstans, G.bawanglingensis has smaller sporangiospores, larger columellae, different shapes and sizes of apophyses. Compared with G.qichaensis, the G.bawanglingensis has smaller sporangia, different columellae and apophysis shapes (Wang et al. 2023a). G.pingtangensis and G.namwonensis are different in size and shape of columellae (hemispherical vs. globose) and apophyses (oval vs. subglobose). G.namwonensis has more shapes of giant cells (Crous et al. 2020). These significant morphological differences, coupled with those phylogenetically independent clades, ensure their novelty (Wang et al. 2023a). As for G.pamphilae, two strains were grouped into an independent separate clade and there are only two base pairs of difference in ITS rDNA sequences. As no morphological descriptions were provided for G.pamphilae in its protologue, we classified the two isolates together as the new record species of G.pamphilae only based on molecular phylogenetic analyses.
In summary, the molecular phylogenetic and morphological results support the identification of the six new species for the 12 strains cultured in this study, namely G.abortosporangia, G.reniformis, G.apophysata, G.bawanglingensis, G.pingtangensis, G.inconstans and two strains as new record species of G.pamphilae, complementing the morphological description of G.pamphilae. TFE, ACT and RPB1 protein-coding sequences were newly added to construct the phylogenetic evolutionary tree and the results were basically consistent with ITS+LSU results. The morphology of members of the genus Gongronella was systematically described herein, with a morphological description table being established for the described strains of Gongronella and the new strains described in this study (Table 4).
Supplementary Material
Acknowledgements
We thank Zhao-Xue Zhang (Shandong Agricultural University), Xin-Yi Wang (Shandong Normal University) and Shu-Bin Liu (Beijing Forestry University) for soil collection.
Citation
Wang Y-X, Zhao H, Jiang Y, Liu X-Y, Tao M-F, Liu X-Y (2024) Unveiling species diversity within early-diverging fungi from China III: Six new species and a new record of Gongronella (Cunninghamellaceae, Mucoromycota). MycoKeys 110: 287–317. https://doi.org/10.3897/mycokeys.110.130260
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 32170012, 32300011, 32470004), Ji’nan City’s ‘New University 20 Policies’ Initiative for Innovative Research Teams Project (202228028), Innovative Agricultural Application Technology Project of Jinan City (CX202210) and Key Technological Innovation Program of Shandong Province, China (2022CXGC020710)
Author contributions
Y.X. Wang took charge of the microscopy, DNA sequencing, data analyses and drafted the paper; H. Zhao made specimens, proposed new species and revised the paper; Y. Jiang, Xin-Y. Liu and M.F. Tao collected samples and isolated cultures; Xiao-Y. Liu proposed new species, revised the paper and provided funding.
Author ORCIDs
Yi-Xin Wang https://orcid.org/0009-0001-5231-914X
Heng Zhao https://orcid.org/0000-0003-2938-5613
Yang Jiang https://orcid.org/0009-0003-1292-610X
Xin-Ye Liu https://orcid.org/0009-0004-9396-1763
Xiao-Yong Liu https://orcid.org/0000-0002-8808-010X
Data availability
The sequences were deposited in the GenBank database.
Supplementary materials
The combined ITS+LSU+TEF+ACT+RPB1 sequence matrix used in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Yi-Xin Wang
Data type
fas
Explanation note
The sequence matrix included 43 strains in 25 species of Gongronella, with Cunninghamellaechinulata CBS 156.28 as outgroup. A total of 4,080 characters comprised ITS rDNA (1–989), LSU rDNA (990–1967), TEF rDNA (1968–2172), ACT rDNA (2173–2948) and RPB1 rDNA (2949–4080). Among them, there were 2866 constant, 562 variable but parsimony non-informative and 652 parsimony informative characters.
References
- Adamcik S, Cai L, Chakraborty D, Chens X, Cotter HV, Dai D, Daii Y, Das K, Deng C, Ghobad-Nejhad M, Hyde K, Langer E, Deepna LKP, Liu F, Liu S, Liu T, Lv W, Lv S, Machado A, Buyck B. (2015) Fungal biodiversity profiles 1–10. Cryptogamie. Mycologie 36(2): 121–166. 10.7872/crym/v36.iss2.2015.121 [DOI] [Google Scholar]
- Ariyawansa H, Hyde K, Jayasiri S, Buyck B, Kandawatte T, Dai D, Dai Y, Daranagama D, Jayawardena R, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala K, Voigt K, Zhao R, Li G, Doilom M, Boonmee S, Yang Z, Chen X. (2015) Fungal diversity notes 111–252: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75(1): 27–274. 10.1007/s13225-015-0346-5 [DOI] [Google Scholar]
- Babu AG, Kim SW, Adhikari M, Yadav DR, Um YH, Kim C, Lee HB, Lee YS. (2015) A new record of Gongronellabutleri isolated in Korea. Mycobiology 43(2): 166–169. 10.5941/MYCO.2015.43.2.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Corry J E L, Curtis G D W, Baird R M, 1995. Rose bengal chloramphenicol (RBC) agar, in Progress in Industrial Microbiology. Elsevier, 431–433. 10.1016/S0079-6352(05)80079-7 [DOI]
- Crous P, Wingfield M, Chooi Y, Gilchrist C, Lacey E, Pitt J, Roets F, Swart W, Cano J, Valenzuela-Lopez N, Hubka V, Shivas R, Stchigel AM, Holdom D, Jurjević Ž, Kachalkin A, Lebel T, Lock C, Martín M, Groenewald JZ. (2020) Fungal Planet description sheets: 1042-1111. Persoonia 44(1): 301–459. 10.3767/persoonia.2020.44.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Freitas LMS, de Oliveira RJV, Leite TRC, Nguyen TTT, Lim HJ, Lee HB, de Santiago ALCMA. (2020) Gongronellapedratalhadensis, a new species of Mucorales (Mucoromycota) isolated from the Brazilian Atlantic Forest, with an identification key for the genus. Sydowia 73: 61–68. [Google Scholar]
- Doilom M, Guo J, Phookamsak R, Mortimer PE, Karunarathna SC, Dong W, Liao C, Yan K, Pem D, Suwannarach N, Promputtha I, Lumyong S, Xu J. (2020) Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Frontiers in Microbiology 11: 585215. 10.3389/fmicb.2020.585215 [DOI] [PMC free article] [PubMed]
- Dong Y, Sun Q, Zhang Y, Wang X, Liu P, Xiao Y, Fang Z. (2018) Complete genome of Gongronella sp. w5 provides insight into its relationship with plant. Journal of Biotechnology 286: 1–4. 10.1016/j.jbiotec.2018.08.022 [DOI] [PubMed] [Google Scholar]
- Dong CB, Zhang ZY, Chen WH, Han YF, Huang JZ, Liang ZQ. (2019) Gongronellazunyiensis sp. nov. (Cunninghamellaceae, Mucorales) isolated from rhizosphere soil in China. Phytotaxa 425: 290–296. 10.11646/phytotaxa.425.5.4 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL, Brown AHD. (1990) Chloroplast DNA phylogenetic affinities of newly described species in Glycine (Leguminosae: Phaseoleae). Systematic Botany 15(3): 466–471. 10.2307/2419362 [DOI] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistonachinensis based on morphology and rDNA sequences. The New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Hesseltine CW, Ellis JJ. (1961) Notes on Mucorales, especially Absidia. Mycologia 53(4): 406–426. 10.1080/00275514.1961.12017970 [DOI]
- Hu J, Zhang Y, Xu Y, Sun Q, Liu J, Fang W, Xiao Y, Kües U, Fang Z. (2019) Gongronella sp. w5 elevates Coprinopsiscinerea laccase production by carbon source syntrophism and secondary metabolite induction. Applied Microbiology and Biotechnology 103(1): 411–425. 10.1007/s00253-018-9469-4 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England) 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, Baghela A, Bahkali AH, Beug M, Bhat DJ, Bojantchev D, Boonpratuang T, Bulgakov TS, Camporesi E, Boro MC, Ceska O, Chakraborty D, Chen JJ, Chethana KWT, Chomnunti P, Consiglio G, Cui BK, Dai DQ, Dai YC, Daranagama DA, Das K, Dayarathne MC, De Crop E, De Oliveira RJV, de Souza CAF, de Souza JI, Dentinger BTM, Dissanayake AJ, Doilom M, Drechsler-Santos ER, Ghobad-Nejhad M, Gilmore SP, Góes-Neto A, Gorczak M, Haitjema CH, Hapuarachchi KK, Hashimoto A, He MQ, Henske JK, Hirayama K, Iribarren MJ, Jayasiri SC, Jayawardena RS, Jeon SJ, Jerônimo GH, Jesus AL, Jones EBG, Kang JC, Karunarathna SC, Kirk PM, Konta S, Kuhnert E, Langer E, Lee HS, Lee HB, Li WJ, Li XH, Liimatainen K, Lima DX, Lin CG, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Lücking R, Lumbsch HT, Lumyong S, Leaño EM, Marano AV, Matsumura M, McKenzie EHC, Mongkolsamrit S, Mortimer PE, Nguyen TTT, Niskanen T, Norphanphoun C, O’Malley MA, Parnmen S, Pawłowska J, Perera RH, Phookamsak R, Phukhamsakda C, Pires-Zottarelli CLA, Raspé O, Reck MA, Rocha SCO, de Santiago ALCMA, Senanayake IC, Setti L, Shang QJ, Singh SK, Sir EB, Solomon KV, Song J, Srikitikulchai P, Stadler M, Suetrong S, Takahashi H, Takahashi T, Tanaka K, Tang LP, Thambugala KM, Thanakitpipattana D, Theodorou MK, Thongbai B, Thummarukcharoen T, Tian Q, Tibpromma S, Verbeken A, Vizzini A, Vlasák J, Voigt K, Wanasinghe DN, Wang Y, Weerakoon G, Wen HA, Wen TC, Wijayawardene NN, Wongkanoun S, Wrzosek M, Xiao YP, Xu JC, Yan JY, Yang J, Da Yang S, Hu Y, Zhang JF, Zhao J, Zhou LW, Peršoh D, Phillips AJL, Maharachchikumbura SSN. (2016) Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78(1): 1–237. 10.1007/s13225-016-0366-9 [DOI] [Google Scholar]
- Liu J, Peng C, Han Q, Wang M, Zhou G, Ye B, Xiao Y, Fang Z, Kües U. (2022) Coprinopsiscinerea uses laccase Lcc9 as a defense strategy to eliminate oxidative stress during fungal‐fungal interactions. Applied and Environmental Microbiology 88(1): e01760–e21. 10.1128/AEM.01760-21 [DOI] [PMC free article] [PubMed]
- Mai Z, Wang L, Zeng Q. (2021) Characterization of a novel isoflavone glycoside-hydrolyzing β-glucosidase from mangrove soil metagenomic library. Biochemical and Biophysical Research Communications 569: 61–65. 10.1016/j.bbrc.2021.06.086 [DOI] [PubMed] [Google Scholar]
- Martins MR, Santos C, Soares C, Santos C, Lima N. (2020) Gongronellaeborensis sp. nov., from vineyard soil of Alentejo (Portugal). International Journal of Systematic and Evolutionary Microbiology 70(5): 3475–3482. 10.1099/ijsem.0.004201 [DOI] [PubMed] [Google Scholar]
- Nylander J. (2004) MrModeltest V2. program distributed by the author. Bioinformatics (Oxford, England) 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K, Zhao N, Yin Q, Zhang T, Xu X, Fang W, Hong Y, Fang Z, Xiao Y. (2014) Induction of a laccase Lcc9 from Coprinopsiscinerea by fungal coculture and its application on indigo dye decolorization. Bioresource Technology 162: 45–52. 10.1016/j.biortech.2014.03.116 [DOI] [PubMed] [Google Scholar]
- Ribaldi M. (1952) Sopra un interessante zigomicete terricolo: Gongronellaurceolifera n. gen. et n. sp. Rivista di Biologia 44(1): 157–166. [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, Hall BD. (1997) The origin of red algae: Implications for plastid evolution. Proceedings of the National Academy of Sciences of the United States of America 94(9): 4520–4525. 10.1073/pnas.94.9.4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Sánchez-Ramírez S, Kõljalg U, Bahram M, Döring M, Schigel D, May T, Ryberg M, Abarenkov K. (2018) High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity 90(1): 135–159. 10.1007/s13225-018-0401-0 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu J, Bhat DJ, Jones EBG, McKenzie EHC, Camporesi E, Bulgakov TS, Doilom M, de Azevedo Santiago ALCM, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang J-B, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KNA, Latha KPD, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon DS, Li J, Dayarathne MC, Perera RH, Xiao Y, Wanasinghe DN, Senanayake IC, Goonasekara ID, de Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KWT, Luo Z-L, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Mešić A, Dutta AK, de Souza CAF, Richter C, Lin C-G, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, da Silva GA, Plautz Jr HL, Ariyawansa HA, Lee H, Kušan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, Mikšík M, Samarakoon MC, Wijayawardene NN, Kim NK, Matočec N, Singh PN, Tian Q, Bhatt RP, de Oliveira RJV, Tulloss RE, Aamir S, Kaewchai S, Marathe SD, Khan S, Hongsanan S, Adhikari S, Mehmood T, Bandyopadhyay TK, Svetasheva TY, Nguyen TTT, Antonín V, Li W-J, Wang Y, Indoliya Y, Tkalčec Z, Elgorban AM, Bahkali AH, Tang AMC, Su H-Y, Zhang H, Promputtha I, Luangsa-ard J, Xu J, Yan J, Ji-Chuan K, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJL, Nontachaiyapoom S, Wen T-C, Karunarathna SC. (2017) Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83(1): 1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Wostemeyer J. (2000) Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological Research 155(3): 179–195. 10.1016/S0944-5013(00)80031-2 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou W, Yuan H, Wang Y. (2008) Characterization of a novel fungal chitosanase Csn2 from Gongronella sp. JG. Carbohydrate Research 343(15): 2583–2588. 10.1016/j.carres.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Wang X, Fang J, Liu P, Liu J, Fang W, Fang Z, Xiao Y. (2021) Mucoromycotina fungi possess the ability to utilize plant sucrose as a carbon source: Evidence from Gongronella sp. w5. Frontiers in Microbiology 11: 591697. 10.3389/fmicb.2020.591697 [DOI] [PMC free article] [PubMed]
- Wang YX, Zhao H, Ding ZY, Ji XY, Zhang ZX, Wang S, Zhang XG, Liu XY. (2023) Three new species of Gongronella (Cunninghamellaceae, Mucorales) from soil in Hainan, China based on morphology and molecular phylogeny. Journal of Fungi (Basel, Switzerland) 9(12): 1182. 10.3390/jof9121182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu X, Xiong C, Gao S, Xu W, Zhao L, Song C, Liu X, James TY, Li Z, Zhang X. (2023) ASF1 regulates asexual and sexual reproduction in Stemphyliumeturmiunum by DJ-1 stimulation of the PI3K/AKT signaling pathway. Fungal Diversity 123(1): 159–176. 10.1007/s13225-023-00528-1 [DOI] [Google Scholar]
- Wei F, Hong Y, Liu J, Yuan J, Fang W, Peng H, Xiao Y. (2010) Gongronella sp. induces overproduction of laccase in Panusrudis. Journal of Basic Microbiology 50(1): 98–103. 10.1002/jobm.200900155 [DOI] [PubMed]
- White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, Sninsky J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, in PCR protocols: a guide to methods and applications, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zhang ZY, Han YF, Chen WH, Liang ZQ. (2019) Gongronellasichuanensis (Cunninghamellaceae, Mucorales), a new species isolated from soil in China. Phytotaxa 416: 167–174. 10.11646/phytotaxa.416.2.4 [DOI] [Google Scholar]
- Zhang ZX, Liu RY, Liu SB, Mu TC, Zhang XG, Xia JW. (2022) Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 88: 171–192. 10.3897/mycokeys.88.82229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Nie Y, Zong T, Wang K, Lv M, Cui Y, Tohtirjap A, Chen J, Zhao C, Wu F, Cui B, Yuan Y, Dai Y, Liu X. (2023) Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Diversity 123(1): 49–157. 10.1007/s13225-023-00525-4 [DOI]
- Zhao H, Nie Y, Huang B, Liu XY (2024, October 14) Unveiling species diversity within early-diverging fungi from China I: Three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 109: 285–304. 10.3897/mycokeys.109.126029 [DOI] [PMC free article] [PubMed]
- Zhou W, Yuan H, Wang J, Yao J. (2008) Production, purification and characterization of chitosanase produced by Gongronella sp. JG. Letters in Applied Microbiology 46(1): 49–54. 10.1111/j.1472-765X.2007.02262.x [DOI] [PubMed] [Google Scholar]
- Zhou G, Peng C, Liu X, Chang F, Xiao Y, Liu J, Fang Z. (2020) Identification and immobilization of an invertase with high specific activity and sucrose tolerance ability of Gongronella sp. w5 for high fructose syrup preparation. Frontiers in Microbiology 11: 633. 10.3389/fmicb.2020.00633 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The combined ITS+LSU+TEF+ACT+RPB1 sequence matrix used in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Yi-Xin Wang
Data type
fas
Explanation note
The sequence matrix included 43 strains in 25 species of Gongronella, with Cunninghamellaechinulata CBS 156.28 as outgroup. A total of 4,080 characters comprised ITS rDNA (1–989), LSU rDNA (990–1967), TEF rDNA (1968–2172), ACT rDNA (2173–2948) and RPB1 rDNA (2949–4080). Among them, there were 2866 constant, 562 variable but parsimony non-informative and 652 parsimony informative characters.
Data Availability Statement
The sequences were deposited in the GenBank database.