Abstract
With the aging of the population, the incidence of osteoporosis (OP) is on the rise, but the ecology of immune cell subpopulations in OP is poorly understood. Therefore, identifying cell subpopulations involved in promoting the development of OP may facilitate the development of new treatments. Based on bioinformatics analysis, we constructed a single-cell landscape of the OP microenvironment and identified immune cell subpopulations in OP to further explore the role of different subpopulations in the abnormal bone microenvironment. Among macrophages (Mac), the Mac_OLR1 subpopulation has an M1-like phenotype and significantly activates cytokine and osteoclast differentiation pathways, interacting with osteoclasts via the HBEGF-CD9 axis. In neutrophils (Neut), the Neut_RSAD2 subpopulation significantly activated cytokine and osteoclast differentiation pathways and had a high neutrophil extracellular trap (NET) score, and H1FX was identified as its potential regulator. In effector memory T (Tem) cells, the Tem_CCL4 subpopulation significantly activated osteoclast differentiation and immune inflammation-related pathways and highly expressed proinflammatory molecules such as CCL4, CCL4L2, CCL5 and IFNG. In B cells, the abundance of the B_ACSM3 subpopulation was significantly increased in the OP group and the osteoclast differentiation pathway was significantly activated, and MYB was identified as its potential regulator. In summary, we identified several immune cell subpopulations that may be involved in promoting the formation of OP, further identified the transcription factors that regulate these subpopulations, and speculated that the development of OP may be accompanied by immune inflammatory responses mediated by these subpopulations. These findings provide candidate molecules and cells for future OP research and may help facilitate the development of new therapies.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80993-z.
Keywords: Single-cell sequencing, Abnormal bone microenvironment, Immune cells, Cell communication, Osteoporosis
Subject terms: Bioinformatics, Biomarkers, Diseases
Introduction
Osteoporosis (OP) is a systemic bone disease characterized by reduced bone mass, destruction of bone microarchitecture, and an increased risk of fractures1. OP-related fractures are particularly harmful and are among the leading causes of disability and mortality in elderly patients2,3. Additionally, the treatment and care required for OP and its associated fractures place a significant burden on families and society. Current treatments for OP primarily include drugs that inhibit bone resorption, such as bisphosphonates, and drugs that promote bone formation, such as parathyroid hormone analogs4–6. While these drugs are beneficial for bone health, long-term use can result in numerous adverse effects, including nausea, vomiting, joint pain, an increased risk of infection, and rashes. Moreover, the use of bisphosphonates may lead to osteonecrosis and atypical femoral fractures7,8 Therefore, it is necessary to develop molecular targeted drugs for OP.
The term “osteoimmunology” was introduced twenty years ago to describe the cross-regulation between the bone and immune systems9. Key research in this field has revealed the significance of the receptor activator of nuclear factor κB ligand (RANKL)-osteoprotegerin (OPG) axis in the pathophysiology of various bone-related diseases, including OP10. Recently, Srivastava et al. introduced the concept of “immunoporosis,” emphasizing the vital role of immune cells in the progression of OP11,12. Increasing evidence indicates that both innate and adaptive immune cells contribute to OP pathogenesis by producing proinflammatory mediators13,14. For instance, inflammatory molecules such as IL-6 and TNF-α can activate the NF-κB signaling pathway, leading to the upregulation of downstream inflammatory cytokines. This process promotes the survival and differentiation of osteoclasts (OCs), ultimately harming bone health15. The CXCL10-CXCR3 axis also plays a role, with T cells in the bone marrow (BM) promoting OCs differentiation through a RANKL-dependent mechanism16. Additionally, in postmenopausal OP, estrogen deficiency-induced changes in immune cells stimulate OCs activation to varying degrees17,18. In age-related OP, the aging process leads to chronic low-level activation of the immune system, disrupting immune balance and eventually resulting in OP19. In diabetic OP, hyperglycemia, caused by insulin deficiency or resistance, can lead to immune cell dysregulation and excessive production of proinflammatory factors, further aggravating OP20. These studies highlight the critical role of the immune response in the development and progression of OP.
The bone microenvironment is a complex system primarily composed of osteoblasts (OBs), OCs, and bone marrow mesenchymal stem cells (BMSCs). It also includes hematopoietic stem cells and various mature immune cells21,22, which play crucial roles in regulating bone formation and resorption23. For example, monocytes regulate bone remodeling by secreting various cytokines24; resting T cells protect bone health25; activated T cells promote OCs formation and subsequent bone loss26; and B cells can also regulate OCs generation27. However, most studies primarily focus on the functions of bone cells, with limited exploration into the specific roles of immune cell subpopulations in OP. This gap in research has restricted our understanding of how the immune microenvironment affects bone remodeling and the progression of OP. Additionally, existing studies often emphasize individual cell types, lacking a comprehensive analysis of the interactions between various immune cell subpopulations, especially their dynamic changes throughout disease progression. Moreover, many studies overlook the complex signaling pathways between cells, hindering a deeper understanding of the underlying disease mechanisms. Therefore, further investigation into the immune cell ecology within the OP microenvironment and identifying potential therapeutic targets is crucial.
The advent of single-cell RNA sequencing (scRNA-seq) technology offers researchers a high-resolution perspective at the single-cell level, allowing for the identification of new cell subtypes and rare cell populations, the exploration of heterogeneity within cell populations, and the identification of cell states and dynamic transitions28–30. In this study, we constructed a single-cell landscape of the OP microenvironment using scRNA-seq data from human femoral heads. We identified several immune cell subpopulations potentially involved in OP formation and pinpointed the transcription factors (TFs) regulating these subpopulations. These findings provide potential molecular and cellular targets for future OP research and may contribute to the development of new therapeutic strategies.
Method
Data collection
OP-related scRNA-seq data were obtained from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). The GSE169396 dataset, based on the GPL11154 platform, includes scRNA-seq data from four live human femoral heads. Osteopenia is characterized by lower-than-normal bone density, while OP involves significant bone loss and structural deterioration. Osteopenia can be an early stage of OP, potentially progressing to the more advanced condition. To explore the cellular interactions and dynamic changes during the progression of OP, this study excluded one sample with an unspecified diagnosis and included one OP sample (61-year-old female) and two osteopenia samples (45-year-old female and 31-year-old male) for analysis. The corresponding clinical information is available in the GSE169396 dataset.
Unsupervised dimensionality reduction and clustering
To ensure data quality, this study implemented quality control measures on the nFeature (total gene count), nCount (total gene expression count), and mitochondrial gene expression ratio of Seurat subjects. Specifically, the top and bottom 1% of cells based on feature number were filtered out, as well as cells with a mitochondrial gene expression ratio exceeding 10%. After quality control, the cells were clustered using the Seurat package31,followed by dimensionality reduction and visualization using the Uniform Manifold Approximation and Projection (UMAP) algorithm32. The markers for each cell cluster were identified using the FindAllMarkers function (log fold change > 0.25). Based on previously reported classical cell markers, cell clusters with common marker expressions were merged and annotated into corresponding cell types. Each cell was referenced with more than three markers23,33, including: endothelial cells (VWF, PECAM1, ENG); pan-osteocytes (COL1A1, LEPR, RUNX2); erythrocytes (HBB, GYPA, GYPB); macrophages (HLA-DRA, CD14, CD68); neutrophils (CEACAM8, FCGR3B, CEACAM1); pDCs (IL3RA, CLEC4C, NRP1); T cells (CD3D, CD3E, CD3G); B cells (CD79A, CD79B, CD19); and plasma cells (XBP1, CD38, CD27). Additionally, pan-osteocytes were further annotated based on specific marker genes34–37, including pericytes (ACTA2, MCAM), OBs (RUNX2, BGLAP), chondrocytes (COL2A1, SOX9), MSCs (LEPR), and OCs (CTSK).
For the analysis of cell subpopulations, we first used the subset function to extract subsets of each cell type and performed clustering analysis on the scRNA-seq data using the Seurat package. Initially, we normalized the data and selected key features, followed by dimensionality reduction using PCA. Based on this, we applied the FindNeighbors and FindClusters functions to identify cell subpopulations, and subsequently used UMAP for visualization. The FindAllMarkers function was then employed to identify characteristic genes for each cell subpopulation, allowing for precise annotation of the subpopulations.
Functional enrichment analysis and gene set score calculation
To investigate the potential biological roles of immune cell subpopulations, the clusterProfiler package38was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Additionally, M1 and M2 macrophage feature gene sets were derived from previous studies39 and the M1 and M2 feature scores for macrophage subpopulations were calculated using the AddModuleScore function in the Seurat package. The neutrophil extracellular traps (NETs) scores for neutrophil subpopulations were calculated using the KEGG gene set (https://www.kegg.jp/entry/hsa04613).
Pseudotime trajectory analysis
Pseudotime analysis is a powerful approach that enables the reconstruction of cell lineage trajectories by analyzing gene expression changes across different cell subpopulations over time. In this study, we employed the Monocle 3 method40 to construct pseudotime trajectories for cell subpopulations within the human femoral head. Using default parameters, we generated these trajectories and visualized them through the UMAP method, providing a clear depiction of the developmental pathways. Furthermore, we explored the dynamic expression patterns of key genes of interest along the pseudotime trajectories, offering deeper insights into their roles during the progression of cellular development.
Gene regulatory network analysis
Gene regulatory networks (GRNs) were analyzed using the pySCENIC tool, a robust Python module designed for single-cell data analysis41. The process begins with a count matrix that quantifies the abundance of all genes across cells, followed by the inference of co-expression modules using the GRNBoost2 algorithm, which employs a per-target regression method to identify gene regulatory interactions. To refine these modules, indirect targets were filtered out through cis-regulatory motif discovery, utilizing the cisTarget approach. Finally, the activity of each regulator was quantified by calculating the enrichment scores (AUCell) of their target genes, providing a comprehensive view of the regulatory landscape within the cell populations.
Cell communication analysis
Ligand-receptor binding is a crucial mechanism for signal transduction, facilitating communication between neighboring and distant cells. To delve deeper into the synergistic roles of immune cells in the development of an abnormal bone microenvironment, this study utilized the iTALK package42 to identify high-confidence ligand-receptor interactions between different cell populations. These interactions were categorized based on their primary functions into four distinct groups: cytokines/chemokines, immune checkpoint genes, growth factors, and other signaling molecules. This classification allows for a more detailed understanding of how immune cells contribute to the complex signaling network within the bone microenvironment.
Data analysis and statistics
All bioinformatics analyses in this study were performed based on the Bioinforcloud platform (http://www.bioinforcloud.org.cn). P value < 0.05 was considered to indicate statistical significance.
Result
Abnormal single-cell landscape in osteoporosis patients
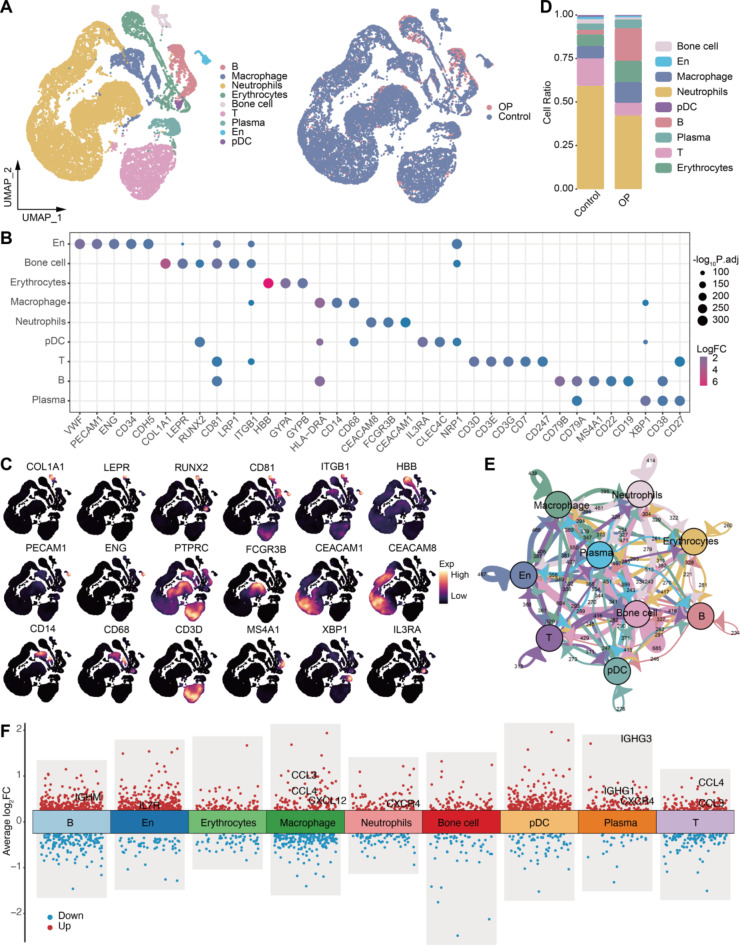
In this study, we constructed a single-cell RNA landscape of the abnormal bone microenvironment in OP patients, identifying immune cell subpopulations and investigating their synergistic roles in promoting this pathological state. After rigorous data quality control and standardization, we obtained a total of 20,734 high-quality single-cell expression profiles. These cells were classified into nine major types (Fig. 1A), including osteocytes, endothelial cells (En), plasmacytoid dendritic cells (pDCs), macrophages, B cells, and T cells. Each cell type was identified using more than three specific markers (Fig. 1B), such as En: VWF, PECAM1, and ENG; osteocytes: COL1A1, LEPR, and RUNX2; erythrocytes: HBB, GYPA, and GYPB; macrophages: HLA − DRA, CD14, and CD68; neutrophils: CEACAM8, FCGR3B, and CEACAM1; pDCs: IL3RA, CLEC4C, and NRP1; T cells: CD3D, CD3E, and CD3G; B cells: CD79A, CD79B, and CD19; and plasma cells: XBP1, CD38, and CD27. These markers were exclusively expressed in the corresponding cell types, aligning with previous single-cell and laboratory studies (Fig. 1C).
Fig. 1.
Single-cell overview of the abnormal bone microenvironment. (A) Single-cell landscape of the abnormal bone microenvironment. En: endothelial cells; pDCs: plasmacytoid dendritic cells; OP: osteoporosis. (B) Bubble plot showing cell markers guiding cell annotation. (C) Density plot mapping of cell marker expression in different cell types. (D) Bar plot showing differences in cell abundance between the control and osteoporosis groups. (E) Overview of cell communication in the bone microenvironment. (F) Differentially expressed genes in cells of the bone microenvironment.
Compared to the control group, OP patients exhibited a decrease in the proportions of neutrophils and T cells, while the proportions of macrophages, B cells, and plasma cells increased (Fig. 1D). Cell communication analysis revealed frequent ligand-receptor interactions among these cell types (Fig. 1E). Additionally, immune cells such as macrophages, T cells, and plasma cells in the OP group showed significant upregulation of immune-inflammatory molecules compared to the control group (Fig. 1F, Table S1). These findings suggest that the interaction between immune cells and immune inflammation plays a crucial role in fostering the formation of an abnormal bone microenvironment.
There is an imbalance in the proportion of bone cells in osteoporosis
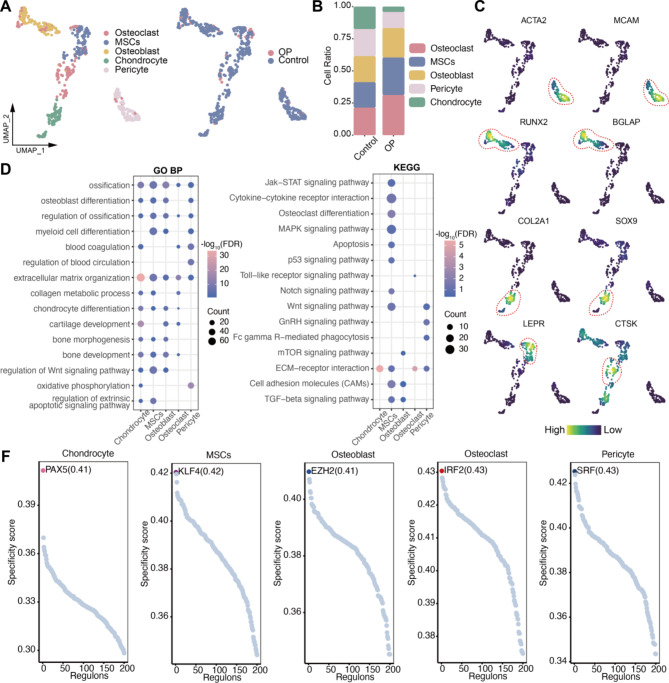
We further categorized bone cells into five groups: OBs, OCs, chondrocytes, mesenchymal stem cells (MSCs), and pericytes (Fig. 2A). In OP patients, the abundance of MSCs and OCs increased, while the numbers of chondrocytes and pericytes decreased compared to the control group (Fig. 2B). During bone metabolism, OBs form new bone, while OCs resorb old bone. Thus, an increase in OCs abundance may disrupt the balance between OCs and OBs, contributing to the development of OP. The markers expressed by these bone cells align with previous studies: pericytes: ACTA2 and MCAM; OBs: RUNX2 and BGLAP; chondrocytes: COL2A1 and SOX9; MSCs: LEPR; and OCs: CTSK (Fig. 2C).
Fig. 2.
Bone cell subpopulations in the abnormal bone microenvironment. (A) Bone cell subpopulations in the bone microenvironment. (B) Bar plot showing differences in cell abundance between the control and osteoporosis groups. (C) Density plot mapping of cell marker expression in the bone cell subpopulations. (D-E) Bubble plots showing biological processes and pathways that are significantly activated in bone cell subpopulations. GO BP: Gene Ontology Biological Process; KEGG: Kyoto Encyclopedia of Genes and Genomes. (F) Regulator activity scores of bone cell subpopulations. RAS: regulator activity score.
Additionally, functional enrichment analysis revealed distinct roles for these cells: chondrocytes primarily activated pathways related to chondrocyte differentiation and cartilage development; MSCs and OBs were involved in myeloid cell differentiation, ossification, and bone development; and pericytes were linked to the regulation of blood circulation, consistent with their known biological functions. To further investigate the key regulators governing the differentiation and development of these cells, we conducted GRN analysis. This identified the most specific regulators for each cell type, including PAX5, KLF4, EZH2, IRF2, and SRF, based on the regulator specificity score (RSS). These regulators are likely to play crucial roles in the biological activities of bone cells.
Identification of myeloid cell subpopulations in the osteoporotic microenvironment
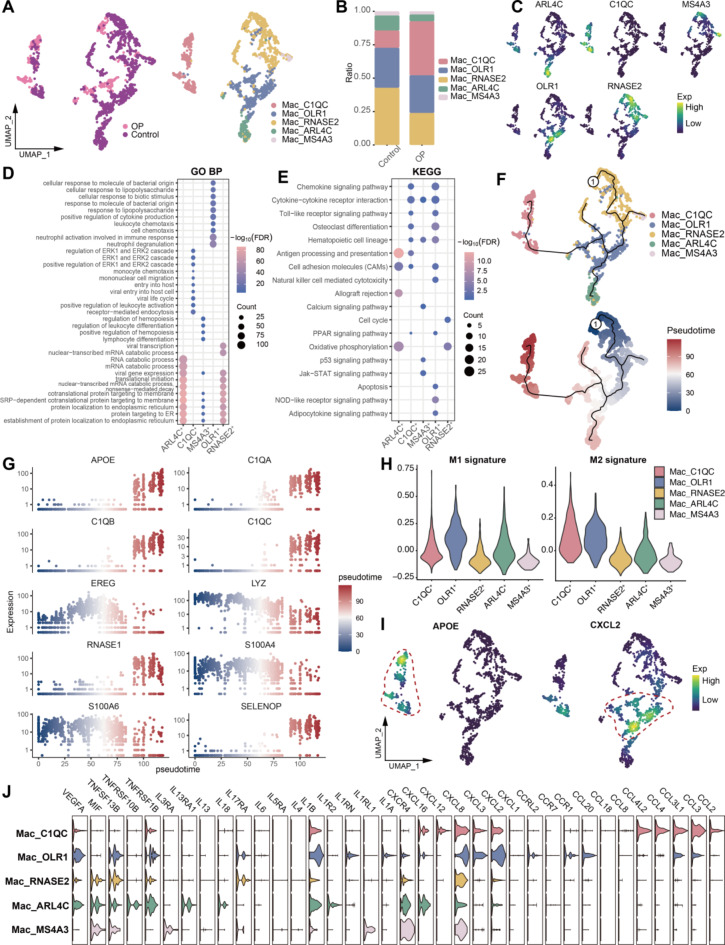
To further investigate the role of immune cells in the abnormal bone microenvironment, we first identified subpopulations of myeloid macrophages and neutrophils. We identified five macrophage subpopulations (Fig. 3A, Table S2), with the Mac_C1QC subpopulation showing a significantly increased abundance in OP patients (Fig. 3B). These subpopulations expressed specific markers, such as ARL4C, C1QC, and MS4A3 (Fig. 3C). Functional enrichment analysis revealed that the Mac_OLR1 subpopulation significantly activated cytokine- and osteoclast-related pathways (Fig. 3D-E). Pseudotime analysis further explored the developmental trajectory of these macrophage subpopulations within the abnormal bone microenvironment, revealing that the Mac_C1QC and Mac_ARL4C subpopulations were located at the terminal stages of this trajectory (Fig. 3F). Additionally, the expression of genes like APOE, C1QA, C1QB, and C1QC increased with the progression of pseudotime (Fig. 3G).
Fig. 3.
Identification and analysis of macrophage subpopulations. (A) Macrophage subpopulations in the OP microenvironment. (B) Bar graph showing the difference in macrophage abundance between control and osteoporosis patients. (C) Density map mapping of cell subpopulation-specific marker expression in macrophage subpopulations. (D-E) Bubble plots showing biological processes and pathways that are significantly activated in macrophage subpopulations. GO BP: Gene Ontology Biological Process; KEGG: Kyoto Encyclopedia of Genes and Genomes. (F) Pseudotime developmental trajectories of macrophage subpopulations. (G) Dynamic expression of genes of interest along the differentiation trajectory of macrophage subpopulations. (H) Expression of M1 and M2 signatures of macrophage subpopulations. (I) Density map of the expression of APOE and CXCL2 in macrophage subpopulations. (J) Violin plots show cytokine expression of macrophage subpopulations.
When evaluating macrophage phenotype scores, we found that the Mac_ARL4C subpopulation had a higher M1 score, the Mac_C1QC subpopulation had a higher M2 score, and the Mac_OLR1 subpopulation exhibited both elevated M1 and M2 scores (Fig. 3H). This suggests that the Mac_OLR1 subpopulation may represent an intermediate state of M1-to-M2 transdifferentiation. Moreover, APOE, a gene with a crucial regulatory role in osteocytes, was specifically expressed in the Mac_C1QC subpopulation, while the inflammatory chemokine CXCL2 was specifically expressed in the Mac_OLR1 subpopulation (Fig. 3I). This distinction reflects the differential roles of these subpopulations in modulating inflammation. Through further analysis, we found that the Mac_OLR1 subpopulation may play a pro-inflammatory role through cytokine and chemokine ligands such as IL1B, CXCL2, CXCL3 and CXCL8 (Fig. 3J).
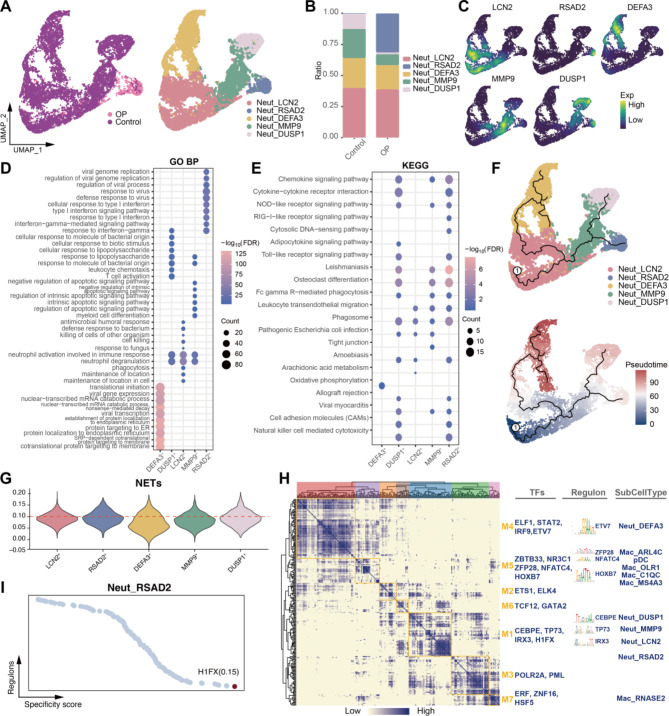
Additionally, we identified five neutrophil subpopulations (Fig. 4A, Table S3), with the Neut_RSAD2 subpopulation showing a significantly increased abundance in OP patients (Fig. 4B). These subpopulations expressed specific markers, including LCN2, RSAD2, and DEFA3 (Fig. 4C). Functional enrichment analysis revealed that the Neut_RSAD2 subpopulation significantly activated cytokine- and osteoclast-related pathways (Fig. 4D-E), suggesting a potential role in promoting inflammation and the progression of OP. Pseudotime analysis indicated that the Neut_RSAD2 subpopulation was located at the terminal end of one developmental trajectory branch (Fig. 4F) and exhibited high NETs scores (Fig. 4G), implicating NETs formation in the progression of OP. Through GRN analysis (Fig. 4H), we identified several TFs regulating each myeloid subpopulation, with HIFX emerging as the most specific TF for the Neut_RSAD2 subpopulation (Fig. 4I), highlighting it as a potential therapeutic target for OP.
Fig. 4.
Identification and analysis of neutrophil subpopulations. (A) Neutrophil subpopulations in the OP microenvironment. (B) Bar graph showing the difference in neutrophil abundance between controls and osteoporosis patients. (C) Density plot mapping of cell subpopulation-specific marker expression in neutrophil subpopulations. (D-E) Bubble plots showing biological processes and pathways that are significantly activated in neutrophil subpopulations. GO BP: Gene Ontology Biological Process; KEGG: Kyoto Encyclopedia of Genes and Genomes. (F) Pseudotime developmental trajectories of neutrophil subpopulations. (G) Violin plots showing neutrophil extracellular traps (NET) scores of neutrophil subpopulations. (H) Coexpression modules of transcription factors in myeloid cell subpopulations. (I) Most specific regulators of the Neut_RSAD2 subpopulation.
Identification of lymphocyte subpopulations in the osteoporotic microenvironment
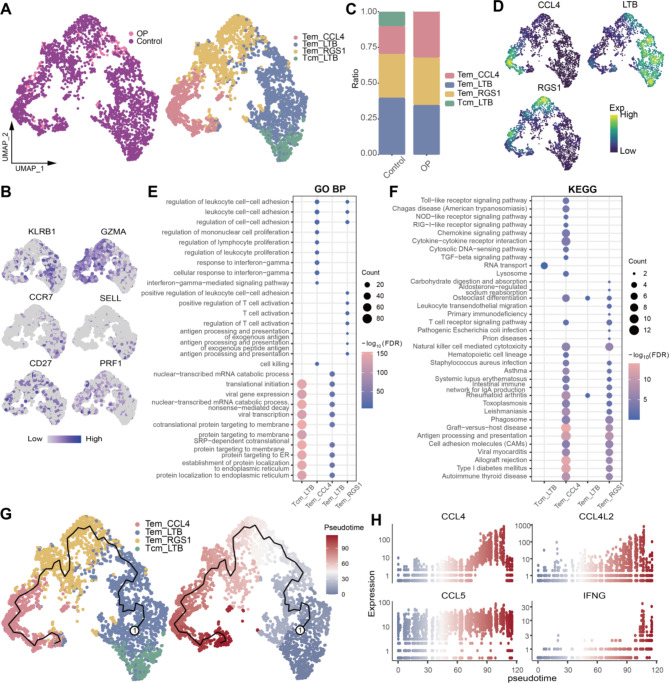
We also investigated the roles of lymphocyte subpopulations, including B cells and T cells. Cluster analysis initially identified four T-cell subpopulations (Fig. 5A, Table S4), which were classified into effector memory T cells (Tem) and central memory T cells (Tcm) based on cell markers (Fig. 5B). Specifically, the Tcm_LTB subpopulation was only observed in the control group, while the percentage of the Tem_CCL4 subpopulation significantly increased in the OP group compared to the control group (Fig. 5C). These T-cell subpopulations expressed specific markers, such as CCL4, LTB, and RGS1 (Fig. 5D). Notably, the Tem_CCL4 subpopulation significantly activated pathways related to immune inflammation and osteoclast (OC) differentiation (Fig. 5E-F) and was positioned at the terminal stage in the T-cell development trajectory (Fig. 5G). Immune-related genes like CCL4, CCL4L2, CCL5, and IFNG were significantly upregulated at this stage (Fig. 5H), suggesting that this subpopulation may play a crucial role in enhancing the inflammatory microenvironment in OP.
Fig. 5.
Identification and analysis of T-cell subpopulations. (A) T-cell subpopulations in the OP microenvironment. (B) Single-cell plot mapping of T-cell subpopulation marker expression. (C) Bar graph showing the difference in T-cell abundance between controls and osteoporosis patients. (D) Density map of the expression of cell subpopulation-specific markers in T-cell subpopulations. (E-F) Bubble plots showing biological processes and pathways that are significantly activated in T-cell subpopulations. GO BP: Gene Ontology Biological Process; KEGG: Kyoto Encyclopedia of Genes and Genomes. (G) Pseudotime developmental trajectories of T-cell subpopulations. (H) Dynamic expression of genes of interest along the differentiation trajectory of T-cell subpopulations.
Additionally, we identified four major B-cell subpopulations (Fig. 6A, Table S5). Among these, the abundance of the B_ACSM3 subpopulation significantly increased in the OP group (Fig. 6B). These B-cell subpopulations expressed specific markers, such as ACSM3, DNTT, PLPP5, and GPR183 (Fig. 6C). Functional enrichment analysis indicated that the B_ACSM3 subpopulation activated pathways related to B-cell activation, cytokine signaling, OCs differentiation, and B-cell receptor signaling (Fig. 6D-E). Pseudotime analysis revealed that this subpopulation was in the middle of the B-cell development trajectory, suggesting that this subpopulation may be in an active proliferation stage (Fig. 6F). GRN analysis revealed several TFs regulating each lymphoid subpopulation (Fig. 6G). Among them, MYB was the TF with the highest regulatory score in the B_ACSM3 subpopulation (Fig. 6H) and was specifically expressed in this subpopulation (Fig. 6I).
Fig. 6.
Identification and analysis of B-cell subpopulations. (A) B-cell subpopulations in the OP microenvironment. (B) Bar graph showing the difference in neutrophil abundance between controls and osteoporosis patients. (C) Density map of the expression of cell subpopulation-specific markers in B-cell subpopulations. (D-E) Bubble plots showing biological processes and pathways that are significantly activated in B-cell subpopulations. GO BP: Gene Ontology Biological Process; KEGG: Kyoto Encyclopedia of Genes and Genomes. (F) Pseudotime developmental trajectories of B-cell subpopulations. (G) Coexpression modules of transcription factors in lymphoid cell subpopulations. (H) Specific regulators of the B_ACSM3 subpopulation. (I) Densified expression of MYB in B-cell subpopulations.
Overview of cell communication in the abnormal bone microenvironment
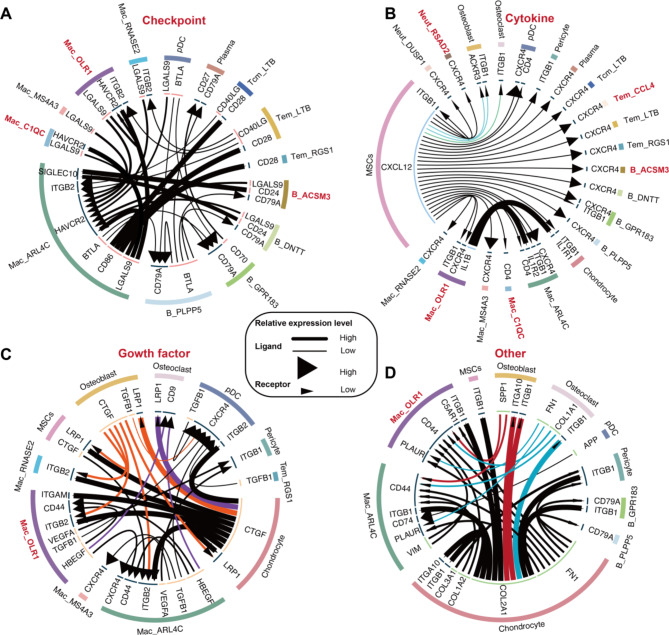
Although we identified multiple OP-related immune cell subpopulations and outlined their potential roles in promoting OP and modulating immunity, the crosstalk between these cells remains unclear. To address this, we conducted cell communication analysis to further explore the interactions between these myeloid and lymphoid immune cell subpopulations and bone cells. Among the ligand-receptor pairs associated with immune checkpoints (Fig. 7A), the LGALS9-HAVCR2 axis was commonly observed across immune cell subpopulations. Regarding cytokine/chemokine interactions (Fig. 7B), mesenchymal stem cells (MSCs) may interact with osteoclasts (OCs) and osteoblasts (OBs) through the CXCL12-ITGB1 axis. Additionally, the Mac_OLR1 subpopulation interacts with chondrocytes via the IL1B-IL1R1 axis, indicating that the proinflammatory effects of the Mac_OLR1 subpopulation may disrupt chondrocyte homeostasis. In the context of growth factor-related interactions (Fig. 7C), OBs communicate with various macrophage subpopulations through the CTGF-ITGB2 axis, and the Mac_OLR1 subpopulation may interact with OCs via the HBEGF-CD9 axis. Since excessive OCs activity is a key factor in bone loss in OP, macrophages may promote osteoclast activation and maintenance through HBEGF signaling. Chondrocytes, which specialize in producing extracellular matrix (ECM) to form cartilage tissue, primarily interact with OBs, OCs, MSCs, and other cells through the COL2A1-ITGB1 and FN1-ITGB1 axes (Fig. 7C). In summary, our findings suggest that the roles of myeloid and lymphoid immune cell subpopulations in contributing to bone microenvironment abnormalities are interrelated.
Fig. 7.
Cell subpopulation communication in the abnormal bone microenvironment. (A) Cell subpopulations based on immune checkpoint ligand‒receptor interactions. (B) Cell subpopulations based on chemokine ligand‒receptor interactions. (C) Cell subpopulations based on growth factor ligand‒receptor interactions. (D) Cell subpopulations based on extracellular matrix ligand‒receptor interactions.
Discussion
In this study, we constructed a single-cell landscape based on OP-related scRNA-seq data and identified immune cell subpopulations potentially involved in promoting OP development, including Mac_OLR1, Neut_RSAD2, B_ACSM3, and Tem_CCL4, which predominantly exhibit proinflammatory phenotypes. Previous studies have shown that OP can increase the risk of osteoarthritis (OA), and conversely, OA can accelerate the onset of OP43, highlighting the role of inflammation in OP formation. Since chondrocyte aging promotes OA development, the reduced abundance of chondrocytes observed in OP in this study may be related to OA progression. Additionally, the abundance of MSCs in the OP group increased compared to the control group. Under normal conditions, MSCs differentiate into OBs, playing a crucial role in bone formation and homeostasis. However, under abnormal microenvironmental conditions, the differentiation function of MSCs may be impaired, reducing their osteogenic capacity and leading to an imbalance between OBs and OCs.
The interaction between macrophages and OBs is vital for bone formation and repair44, and they play a key role in inflammation-driven bone diseases45. Tissue damage from various factors can activate macrophages, disrupting bone homeostasis and leading to bone destruction46. Moreover, macrophages can promote osteoporosis by supporting OC-mediated bone resorption47. In this study, the Mac_OLR1 subpopulation significantly activated cytokine- and OC-related pathways and may be in a state of M1 polarization. Oxidized low-density lipoprotein receptor 1 (OLR1) serves as a molecular marker for OC activity48, and OLR1 knockout impairs cell-cell fusion in OC-like cells49, indicating that the Mac_OLR1 subpopulation may contribute to a proinflammatory environment by mediating abnormal bone microenvironment formation and stimulating OC formation and activation. Additionally, this study revealed a significant increase in the Mac_C1QC subpopulation in the OP group, which specifically expressed APOE. As a key apolipoprotein, ApoE can indirectly influence bone metabolism by regulating hyperlipidemia50. Its anti-inflammatory and antioxidant effects may inhibit OP development. The C1QC-positive macrophage subpopulation has been previously identified in various diseases and exhibits an M2 phenotype51–53. Therefore, the increased abundance of this subpopulation may be related to the body’s immune response to the microenvironment, and promoting its differentiation could help regulate the abnormal inflammatory microenvironment.
Among neutrophils, we identified the Neut_RSAD2 subpopulation, which may be associated with the formation of an abnormal bone microenvironment. The protein encoded by RSAD2 is an interferon-induced antiviral protein involved in cellular antiviral responses and innate immune signaling54. Downregulation of RSAD2 has been shown to inhibit osteoclastogenesis55, suggesting that the Neut_RSAD2 subpopulation may affect bone homeostasis by promoting OC formation, leading to an abnormal bone microenvironment. We also identified a specific regulator of this subpopulation, H1FX, a recently discovered and less-characterized member of the H1 histone family. Histone H1 plays an important role in the structure of chromatin and in regulating gene expression. It regulates the compaction and decompression of chromatin by binding to nucleosomes, thereby affecting the accessibility of DNA. Previous studies have shown that inhibition of H1FX triggers DNA damage, thereby inducing cellular senescence56. Our study found that H1FX is involved in regulating the development and differentiation of the Neut_RSAD2 subpopulation and may be a potential target for OP treatment.
Among lymphocyte subpopulations, the Tem_CCL4 subpopulation showed a significant increase in abundance in the OP group. CCL4 is a CC chemokine known for promoting tumor development and progression by recruiting regulatory T cells and tumor-promoting macrophages57. It is also closely related to CD8 T cell infiltration58. Our study revealed that this subpopulation significantly activated immune-inflammatory pathways, indicating its involvement in fostering an inflammatory environment. We also observed a significant increase in the B_ACSM3 subpopulation in the OP group, which activated the OC differentiation pathway. ACSM3 expression is associated with various tumors59. This study preliminarily confirmed that this subpopulation may be related to bone diseases and potentially regulated by MYB. MYB plays a crucial role in hematopoietic regulation and has been found to be abnormally expressed in various malignant tumors60. Moreover, c-Myb plays a critical role in lymphocyte specification and B-cell development61,62. c-Myb controls the upregulation of a subset of lymphocyte genes in multipotent progenitors and is required for the normal development of committed B-cell progenitors63. Additionally, c-Myb precisely regulates the spatiotemporal expression of the Rag genes during B lymphocyte development by inhibiting the expression of the Rag activator Foxo1 and directly occupying the Erag enhancer, preventing untimely DNA damage64. Therefore, as a regulator of the B_ACSM3 subpopulation, MYB may also be a potential therapeutic target for OP.
In summary, this study identified several ligand-receptor axes as potential therapeutic targets for OP. However, it has several limitations. First, the small sample size due to limited data may affect the generalizability and reliability of the findings. Second, the participants in the control group were not matched in terms of age, gender, and other potential confounders that could influence the bone microenvironment, which may introduce bias and impact the interpretation of the results. Additionally, while this study identified immune cell subsets associated with OP and their potential mechanisms of action, the specific molecular pathways through which these subsets promote OP development remain to be fully elucidated. Further research is needed to clarify the functions of these subsets and their precise roles in OP formation. Finally, the absence of long-term follow-up data complicates the assessment of dynamic changes in immune cell subsets and their long-term effects on the progression of OP.
Conclusion
In conclusion, we identified several immune cell subpopulations that may play a role in promoting the formation of OP and further elucidated the transcription factors regulating these subpopulations. We hypothesize that the development of OP may be associated with immune inflammatory responses mediated by these subpopulations. These findings offer potential candidate molecules and cells for future OP research and may aid in the development of new therapeutic strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
All authors contributed to the study conception and design. Bioinformatics analysis were performed by Weiwei Yang, Yulin Wang and Ke Mo. The first draft of the manuscript was written by Wenyang Chen and Xiangtao Xie, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Project of Guangxi Science and Technology Department (No.AA24010007), Guangxi Natural Science Foundation (2023GXNSFAA026407), Liuzhou Science and Technology Project (No.2022SB014).
Data availability
All data generated or analysed during this study are included in this published article.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Weiwei Yang and Yulin Wang contributed equally.
Contributor Information
Wenyang Chen, Email: chenwenyang295@163.com.
Xiangtao Xie, Email: xiexiangtao813@163.com.
References
- 1.Nih Consensus Development Panel on Osteoporosis Prevention & Therapy, D. Osteoporosis prevention, diagnosis, and therapy. JAMA285 (6), 785–795 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Sambrook, P., Cooper, C. & Osteoporosis Lancet ;367(9527):2010–2018. (2006). [DOI] [PubMed] [Google Scholar]
- 3.Adachi, J. D. et al. The influence of osteoporotic fractures on health-related quality of life in community-dwelling men and women across Canada. Osteoporos. Int.12 (11), 903–908 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. P. Long-term treatment of postmenopausal osteoporosis. Endocrinol. Metab. (Seoul). 36 (3), 544–552 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClung, M. R. Role of bone-forming agents in the management of osteoporosis. Aging Clin. Exp. Res.33 (4), 775–791 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. P. Antiresorptives: safety concerns-clinical perspective. Toxicol. Pathol.45 (7), 859–863 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Khan, A. A. et al. Diagnosis and management of osteonecrosis of the jaw: a systematic review and international consensus. J. Bone Min. Res.30 (1), 3–23 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Shane, E. et al. Atypical subtrochanteric and diaphyseal femoral fractures: report of a task force of the American Society for Bone and Mineral Research. J. Bone Min. Res.25 (11), 2267–2294 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Arron, J. R. & Choi, Y. Bone versus immune system. Nature408 (6812), 535–536 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Boyce, B. F. & Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys.473 (2), 139–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava, R. K., Dar, H. Y. & Mishra, P. K. Immunoporosis: immunology of osteoporosis-role of T cells. Front. Immunol.9, 657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava, R. K. & Sapra, L. The rising era of immunoporosis: role of Immune System in the pathophysiology of osteoporosis. J. Inflamm. Res.15, 1667–1698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mundy, G. R. Osteoporosis and inflammation. Nutr. Rev.65 (12 Pt 2), S147–S151 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Zupan, J., Jeras, M. & Marc, J. Osteoimmunology and the influence of pro-inflammatory cytokines on osteoclasts. Biochem. Med. (Zagreb). 23 (1), 43–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing, L. et al. NF-kappaB p50 and p52 expression is not required for RANK-expressing osteoclast progenitor formation but is essential for RANK- and cytokine-mediated osteoclastogenesis. J. Bone Min. Res.17 (7), 1200–1210 (2002). [DOI] [PubMed] [Google Scholar]
- 16.von Gunten, S. & Simon, H. U. Linking glucocorticoid-induced osteoporosis to osteoimmunology. Cell. Death Dis.11 (12), 1026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, V. & Haffner-Luntzer, M. Interaction between bone and immune cells: implications for postmenopausal osteoporosis. Semin Cell. Dev. Biol.123, 14–21 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Vaananen, H. K. & Harkonen, P. L. Estrogen and bone metabolism. Maturitas23 (Suppl), S65–S69 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol.14 (10), 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Zhang, W. et al. Immunoporosis: role of immune system in the pathophysiology of different types of osteoporosis. Front. Endocrinol. (Lausanne). 13, 965258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sims, N. A. & Walsh, N. C. Intercellular cross-talk among bone cells: new factors and pathways. Curr. Osteoporos. Rep.10 (2), 109–117 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Yang, W., He, Q., Hu, Z. & Xie, X. FOXO4 may be a biomarker of postmenopausal osteoporosis. Int. J. Gen. Med.15, 749–762 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu, X. et al. Single-cell RNA sequencing of human femoral head in vivo. Aging (Albany NY). 13 (11), 15595–15619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirraco, R. P., Reis, R. L. & Marques, A. P. Effect of monocytes/macrophages on the early osteogenic differentiation of hBMSCs. J. Tissue Eng. Regen Med.7 (5), 392–400 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Li, Y. et al. B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo. Blood109 (9), 3839–3848 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colucci, S. et al. T cells support osteoclastogenesis in an in vitro model derived from human multiple myeloma bone disease: the role of the OPG/TRAIL interaction. Blood104 (12), 3722–3730 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Manabe, N. et al. Connection between B lymphocyte and osteoclast differentiation pathways. J. Immunol.167 (5), 2625–2631 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol.18 (1), 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Chen, B., Zhu, L., Yang, S. & Su, W. Unraveling the heterogeneity and Ontogeny of dendritic cells using single-cell RNA sequencing. Front. Immunol.12, 711329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, H., Ye, F. & Guo, G. Revolutionizing immunology with single-cell RNA sequencing. Cell. Mol. Immunol.16 (3), 242–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36 (5), 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. (2018). [DOI] [PubMed]
- 33.Chai, R. C., Single-Cell, R. N. A. & Sequencing Unravelling the bone one cell at a time. Curr. Osteoporos. Rep.20 (5), 356–362 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald, M. M. et al. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell184 (7), 1940 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y. et al. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthr. Cartil.29 (9), 1324–1334 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Zhou, B. O., Yue, R., Murphy, M. M., Peyer, J. G. & Morrison, S. J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell. Stem Cell.15 (2), 154–168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y. et al. MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell. Death Dis.9 (11), 1107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS16 (5), 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell184 (3), 792–809e23 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32 (4), 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc.15 (7), 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y. et al. iTALK: an R Package to Characterize and Illustrate Intercellular Communication. bioRxiv. :507871. (2019).
- 43.Bai, R. J., Li, Y. S. & Zhang, F. J. Osteopontin, a bridge links osteoarthritis and osteoporosis. Front. Endocrinol. (Lausanne). 13, 1012508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pieters, B. C. H. et al. Macrophage-derived extracellular vesicles as carriers of alarmins and their potential involvement in bone homeostasis. Front. Immunol.10, 1901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara, N. & Kobayashi, K. Macrophages in inflammation. Curr. Drug Targets Inflamm. Allergy. 4 (3), 281–286 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Valimaki, E. et al. Calpain activity is essential for ATP-Driven unconventional vesicle-mediated protein secretion and inflammasome activation in human macrophages. J. Immunol.197 (8), 3315–3325 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Batoon, L. et al. Osteal macrophages support osteoclast-mediated resorption and contribute to bone pathology in a postmenopausal osteoporosis mouse model. J. Bone Min. Res.36 (11), 2214–2228 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Hansen, M. S. et al. Transcriptional reprogramming during human osteoclast differentiation identifies regulators of osteoclast activity. Bone Res.12 (1), 5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitano, V. J. F. et al. LDL uptake-dependent phosphatidylethanolamine translocation to the cell surface promotes fusion of osteoclast-like cells. J. Cell. Sci. ;133(10). (2020). [DOI] [PubMed]
- 50.Chang, G. R. et al. Kefir peptides attenuate atherosclerotic vascular calcification and osteoporosis in atherogenic diet-fed ApoE (-/-) knockout mice. Front. Cell. Dev. Biol.11, 1158812 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, X., Zhang, Q., Chen, G. & Luo, D. Multi-omics Analysis showed the clinical value of Gene signatures of C1QC(+) and SPP1(+) TAMs in Cervical Cancer. Front. Immunol.12, 694801 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, Q. et al. Single-cell RNA sequencing reveals the heterogeneity of Tumor-Associated Macrophage in Non-small Cell Lung Cancer and differences between sexes. Front. Immunol.12, 756722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, Z. et al. Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunology11 (1), 2085432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao, Z. et al. RSAD2 is an effective target for high-yield vaccine production in MDCK cells. Viruses ;14(11). (2022). [DOI] [PMC free article] [PubMed]
- 55.Dong, Y. et al. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell. Signal.34, 55–65 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Zheng, X. et al. Downregulation of HINFP induces senescence-associated secretory phenotype to promote metastasis in a non-cell-autonomous manner in bladder cancer. Oncogene41 (28), 3587–3598 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Mukaida, N., Sasaki, S. I. & Baba, T. CCL4 signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol.1231, 23–32 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Romero, J. M. et al. A four-chemokine signature is Associated with a T-cell-inflamed phenotype in primary and metastatic pancreatic Cancer. Clin. Cancer Res.26 (8), 1997–2010 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Gopal, R., Selvarasu, K., Pandian, P. P. & Ganesan, K. Integrative transcriptome analysis of liver cancer profiles identifies upstream regulators and clinical significance of ACSM3 gene expression. Cell. Oncol. (Dordr). 40 (3), 219–233 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Anand, S. et al. From modulation of cellular plasticity to potentiation of therapeutic resistance: new and emerging roles of MYB transcription factors in human malignancies. Cancer Metastasis Rev.43 (1), 409–421 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas, M. D., Kremer, C. S., Ravichandran, K. S., Rajewsky, K. & Bender, T. P. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity23 (3), 275–286 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Fahl, S. P., Crittenden, R. B., Allman, D. & Bender, T. P. c-Myb is required for pro-B cell differentiation. J. Immunol.183 (9), 5582–5592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greig, K. T. et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood115 (14), 2796–2805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timblin, G. A., Xie, L., Tjian, R. & Schlissel, M. S. Dual mechanism of rag gene repression by c-Myb during Pre-B Cell Proliferation. Mol. Cell. Biol. ;37(12). (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.