Abstract
Red light optogenetic systems are in high demand for the precise control of gene expression for gene- and cell-based therapies. Here, we report a red/far-red light-inducible photoswitch (REDLIP) system based on the chimeric photosensory protein FnBphP (Fn-REDLIP) or PnBphP (Pn-REDLIP) and their interaction partner LDB3, which enables efficient dynamic regulation of gene expression with a timescale of seconds without exogenous administration of a chromophore in mammals. We use the REDLIP system to establish the REDLIP-mediated CRISPR-dCas9 (REDLIPcas) system, enabling optogenetic activation of endogenous target genes in mammalian cells and mice. The REDLIP system is small enough to support packaging into adeno-associated viruses (AAVs), facilitating its therapeutic application. Demonstrating its capacity to treat metabolic diseases, we show that an AAV-delivered Fn-REDLIP system achieved optogenetic control of insulin expression to effectively lower blood glucose levels in type 1 diabetes model mice and control an anti-obesity therapeutic protein (thymic stromal lymphopoietin, TSLP) to reduce body weight in obesity model mice. REDLIP is a compact and sensitive optogenetic tool for reversible and non-invasive control that can facilitate basic biological and biomedical research.
Subject terms: Optogenetics, Synthetic biology, CRISPR-Cas9 genome editing
Red light optogenetic systems are in high demand for therapeutic applications. Here, the authors introduce REDLIP, a red-light-inducible system that enables rapid gene regulation without external chromophores, effectively modulating insulin and anti-obesity protein expression in disease models.
Introduction
Given the increasing demand for precision medicine, developing technologies that can precisely coordinate the release kinetics of therapeutic agents has been an area of active research1–6. Many precise control systems have been designed to sense and respond to chemical cues7–11 and physical cues, including light12–17, magnetic and mechanical forces18, temperature19, and electricity20,21. Light has features including high spatiotemporal resolution, excellent adjustability and reversibility, and optogenetics has been widely used for the light-mediated manipulation of biological processes in living organisms, which has paved the way for precision interventions in gene- and cell-based therapies22,23.
For many optogenetic applications, red/far-red light is preferable to other wavelengths24,25, due to its advantages, including deep tissue penetration, low light scattering, and low photo-cytotoxicity23. Progress has recently been made toward developing red/far-red light-responsive optogenetic tools for the precise, traceless, and remote control of therapeutic gene expression in many experimental disease models13,14,24–29. Various red-shifted photoresponsive proteins have been characterized and engineered to provide flexible options for optogenetic applications based on red light (RL) illumination. The RL-responsive systems based on plant phytochrome B and phytochrome interacting factor 3 (PIF3) or 6 (PIF6) from Arabidopsis thaliana have been applied for transcriptional control30,31, cell signaling32,33, and protein localization34. However, these systems are limited by the requirement of supplying an exogenous chromophore and by their low packaging capacity for use with adeno-associated virus (AAV) vectors23. Our recently reported red/far-red light-mediated and minimized ΔPhyA-based photoswitch (REDMAP) system showed fast ON/OFF kinetics, and a relatively small construct size suitable for AAV packaging24. However, its applicability, especially for long-term controllable gene therapy, is still limited by the need for phycocyanobilin (PCB), which must be provided through exogenous administration or engineering cells to introduce PCB biosynthesis enzymes24.
Another class of bacteriophytochrome-based optogenetic systems that do not require exogenous supplementation of biliverdin (BV) has been developed for optogenetic manipulation in mammalian cells and mice14,35–37. Our previously reported bacterial phytochrome BphS optical controllable system exhibited strong transcriptional activation but required continuous 4–6 h of far-red light (FRL) illumination14,28. Moreover, its multiple circuit components exceed the AAV packaging size. Recently, BphP1/PpsR2 (engineered QPAS1) derived from Rhodopseudomonas palustris35 and IsPadC-photosensory core module (PCM) from Idiomarina sp. A28L36 have been developed as near-infrared (NIR) optogenetic systems to precisely control gene expression in a timescale of minutes. However, they are limited by low transcriptional (about 6-fold) activation in deep tissues of mice35,36. Recently, MagRed system based on DrBphP derived from Deinococcus radiodurans and its photo-state-specific binder (named Aff6_V18FΔN) that uses split-protein reassembly to control gene expression with RL illumination38. However, this system has low transcriptional activation in mammalian cells and requires repetitive pulses of illumination over 24 h38, indicating the low possibility for controllable gene- and cell-based therapy in vivo. A recently reported BICYCL-Green system employs a cyanobacteriochrome GAF domain from Acaryochloris marina (AM1_C0023g2)37. This system relies on green light to terminate transgene expression, potentially posing a drawback for in vivo applications37.
In this study, we seek to develop a red/far-red light-inducible photoswitch (REDLIP) system based on the chimeric photosensory proteins PnBphP (Pn-REDLIP) or FnBphP (Fn-REDLIP) and their common interaction partner LDB3. We first engineer PnBphP or FnBphP by fusing the N-terminal extension (NTE) of phytochrome A (PhyA)39 or the fungal phytochrome FphA derived from Aspergillus nidulans40 to the N-terminus of DrBphP-PCM, where NTE was reported to exert a stabilizing effect on the active FRL-absorbing state (Pfr)40,41. Under RL (660 nm) illumination, PnBphP or FnBphP interacts with their common interaction partner LDB3 to allow for the precise control of gene expression; when illuminated with FRL (780 nm), FnBphP or PnBphP dissociate from LDB3 to terminate transgene expression42. These genetically encoded REDLIP systems require no external BV, exhibit fast ON/OFF kinetics (10 s RL illumination and 1 min FRL illumination), strong activation of target gene expression (FnBphP: 65-fold; PnBphP: 106-fold), and a dose-dependent response to red/far-red light. We also demonstrate that the REDLIP-mediated CRISPR-dCas9 (REDLIPcas) system enables high induction (FnBphP: 328-fold; PnBphP: 1158-fold) of multiple endogenous genes under 10 s RL illumination. Moreover, the Fn-REDLIP components can be delivered to mouse muscles by AAV vectors and are used to induce RL-dependent luciferase reporter expression in mice. We also demonstrate that an AAV-delivered Fn-REDLIP system could dynamically regulate insulin expression to lower blood glucose in streptozotocin (STZ)-induced type 1 diabetic (T1D) model mice and to control thymic stromal lymphopoietin (TSLP) expression43 to treat obesity in high-fat diet (HFD)-fed model mice. Our study thus illustrates a powerful tool for the optogenetic control of transgene expression and user-defined endogenous gene activation, enabling reversible and non-invasive control that can facilitate basic biological and biomedical research.
Results
Design, construction, and optimization of the REDLIP system
To generate a red/far-red light-inducible photoswitch (REDLIP) system, we focused on the bacterial phytochrome photoreceptor DrBphP from Deinococcus radiodurans. DrBphP uses BV to respond to light signals, and since this molecule is ubiquitous and endogenous in mammalian cells and animals, there is no need to administer an exogenous chromophore35,38,42. Based on a previous study reporting that PhyA’s NTE influences photoresponse properties by stabilizing the active Pfr state39,40,44, we initially engineered chimeric photosensory proteins BphP (PnBphP or FnBphP) by fusing the NTE of phytochrome A (PhyA) from Arabidopsis thaliana or the fungal phytochrome FphA from Aspergillus nidulans to the N-terminus of DrBphP-PCM.
Next, we constructed three types of REDLIP systems (Pn-REDLIP, Fn-REDLIP, Dr-REDLIP), in which the PnBphP, FnBphP, or the original DrBphP was fused to Gal4, a yeast DNA binding domain (Gal4 DBD), to form a hybrid DNA binding protein (Gal4 DBD-PnBphP, Gal4 DBD-FnBphP, or Gal4 DBD-DrBphP). Their common interacting partner LDB3 was fused to a hybrid trans-activator p65-HSF1 [the 65-kDa trans-activator subunit of NF-κB (p65) and heat shock factor 1 (HSF1) transactivation domain] to form the light-inducible trans-activator LDB3-p65-HSF1. Upon RL illumination, PnBphP, FnBphP, or DrBphP interacts with LDB3, enabling it to bind to an RL-specific chimeric promoter (PRL, 5×UAS-PTATA) via the Gal4 DBD domain to initiate transgene expression. Upon exposure to FRL, the BphPs dissociate from LDB3, thereby terminating target transgene expression (Fig. 1a, b). In addition, we evaluated the impact of the different BphPs fused to Gal4 DBD and found the Fn-REDLIP system efficiently activated SEAP reporter expression upon RL illumination, and the Pn-REDLIP system significantly decreased leakiness in the dark compared to the Dr-REDLIP system (Supplementary Fig. 1).
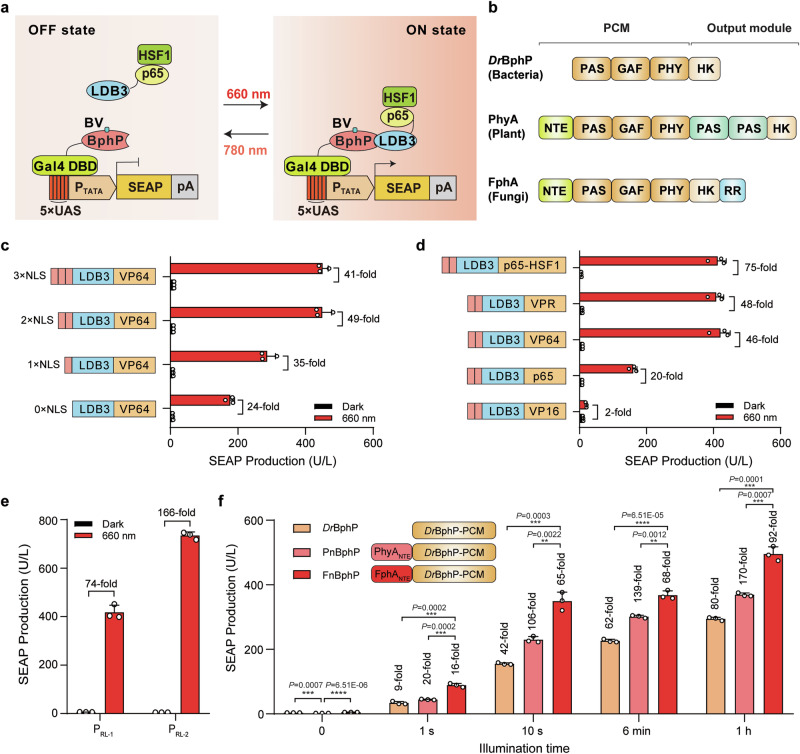
Fig. 1. Construction and optimization of the REDLIP system.
a Schematic showing the design of the REDLIP system for controlling gene expression. b The domain organization of three phytochrome-related proteins: DrBphP, PhyA, and FphA. The photosensory core module (PCM) includes the N-terminal extension (NTE), Period/ARNT/Single-minded-cGMP phosphodiesterase/adenylyl cyclase/FhlA (PAS–GAF) core, and the phytochrome-specific (PHY) domain. The C-terminal output module comprises a variable domain. HK: histidine kinase; RR: response regulator. c Comparison of the performance of varying nuclear localization signal (NLS) copy numbers in the red light (RL)-dependent trans-activator (LDB3-p65-HSF1). HEK-293T cells were co-transfected with the Gal4 DBD-FnBphP vector (pQL326), the SEAP reporter expression vector (pDL6), and the light-inducible trans-activator containing different NLS copy numbers (pQL243, pQL242, pQL232, or pQL207), then illuminated with RL (2.0 mW/cm²) for 24 h. SEAP production in the culture supernatant was quantified 24 h after illumination. d Screening of different transcriptional activators fused to LDB3. HEK-293T cells were co-transfected with pQL326, pDL6, and either pQL242, pQL251, pQL250, pQL252, or pNX12, followed by illumination as described in (c). SEAP production was quantified 24 h after illumination. e Optimization of RL-responsive PRL-X-driven SEAP expression. HEK-293T cells were transfected with pQL326, pNX12, and either pDL6 or pYZ430, followed by illumination as described in (c). SEAP production was quantified 24 h after illumination. f Comparison of the performance of different chimeric BphP constructs fused to the Gal4 DBD, illuminated with RL (2.0 mW/cm2) for 1 s, 10 s, 6 min, and 1 h. HEK-293T cells were co-transfected with pYZ430, pNX12, and either pQL217, pQL325, or pQL326, and then illuminated with RL for the indicated times (0–1 h). SEAP production was quantified 24 h after illumination, with fold change compared to the 0-hour time point. Data in (c–f) are presented as means ± SD; n = 3 independent experiments. P values in (f) were calculated by one-way ANOVA with multiple comparisons. Detailed descriptions of the genetic constructs and transfection mixtures are provided in Supplementary Data 1 and Supplementary Data 3. **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
To optimize the induction profiles of the Fn-REDLIP system, we fused the VP64 activator [(a tetrameric repeat of the minimal Herpes simplex-derived trans-activator VP16 (herpes simplex viral protein 16)] with 1 or 2 or 3 copies of nuclear localization signal (NLS); upon RL illumination, VP64 with 2 copies of NLS (2 NLS) enhanced the fold induction (49-fold induction) compared to the dark control sample (Fig. 1c). We also examined the effects of LDB3 fused with five different trans-activators, including VP16, p65, VP64, VP64-p65-Rta (VPR), and p65-HSF1: LDB3 fused with p65-HSF1 produced the highest induction efficiency (75-fold) of SEAP expression upon RL illumination (Fig. 1d). For further improvement, we tested two RL specific chimeric promoters (PRL-1, 5×UAS-PhCMVmin; PRL-2, 5×UAS-PTATA) driving SEAP expression and found that SEAP expression under the control of the PRL-2 (5×UAS-PTATA) promoter resulted in a high induction (166-fold induction) (Fig. 1e). We also observed that 10 s of RL at 2.0 mW/cm2 induced gene expression, achieving ~60% of the efficiency obtained from 1 h of illumination (Fig. 1f). Compared to the Dr-REDLIP system, the Fn-REDLIP system showed the highest gene expression under RL illumination, while the Pn-REDLIP system exhibited the lowest extent of leaky activity in the dark and the highest induction efficiency (Fig. 1f). As such, the Pn-REDLIP system could be preferable for applications with stringent background requirements.
To confirm the nuclear translocation of the photoreceptor BphP under RL illumination, we fused a hybrid DNA-binding protein (Gal4 DBD-FnBphP) to EGFP (EGFP-Gal4 DBD-FnBphP), while its interacting partner, LDB3, was fused to mCherry (LDB3-mCherry) (Supplementary Fig. 2a, b). We observed that no EGFP signals were evident in the nucleus of cells expressing only EGFP-Gal4 DBD-FnBphP, both in darkness and after RL illumination. However, a clear EGFP signal in the nucleus was detected in cells co-expressing EGFP-Gal4 DBD-FnBphP and LDB3-mCherry under RL illumination, as evidenced by the overlap of green and red fluorescence, resulting in a yellow signal (Supplementary Fig. 2c). These results suggest that the nuclear translocation of FnBphP is mediated by its interaction with LDB3 upon RL exposure.
Characterization of the Fn-REDLIP and Pn-REDLIP systems
We assessed the performance of the Fn-REDLIP and Pn-REDLIP systems in HEK-293T cells. First, we evaluated the kinetics of the two REDLIP systems and found that the SEAP reporter gene expression depended on the time and intensity of RL illumination (Fig. 2a, b, and Supplementary Fig. 3a, b) without a noticeable background increase over time under dark conditions; there was no increase in the SEAP signal within 24 h of stopping RL illumination (Fig. 2c and Supplementary Fig. 3c). Remarkably, two REDLIP systems were more sensitive to RL: RL illumination for only 10 s at 2 mW/cm2 resulted in a substantial increase in SEAP expression, achieving ~60% of the maximum by 24 h of RL illumination. Moreover, the Fn-REDLIP system can induce reporter SEAP expression (350 U/L) at levels similar to that of our previous REDMAP system24, but more than that of the Pn-REDLIP system (SEAP production, 200 U/L) and other red/far-red light-inducible systems (based on BphP1/PpsR235, BphS14, PhyB/PIF334, and MagRed38) (Supplementary Fig. 4a–c). SEAP signals from the two REDLIP systems were observed under RL illumination, while only minimal SEAP expression was observed under ambient light; note that no induced gene expression was observed upon exposure to ultraviolet light, blue light, green light, or far-red light (Fig. 2d and Supplementary Fig. 3d). The results support the chromatic specificity (660 nm wavelength) of the Fn-REDLIP and Pn-REDLIP systems.
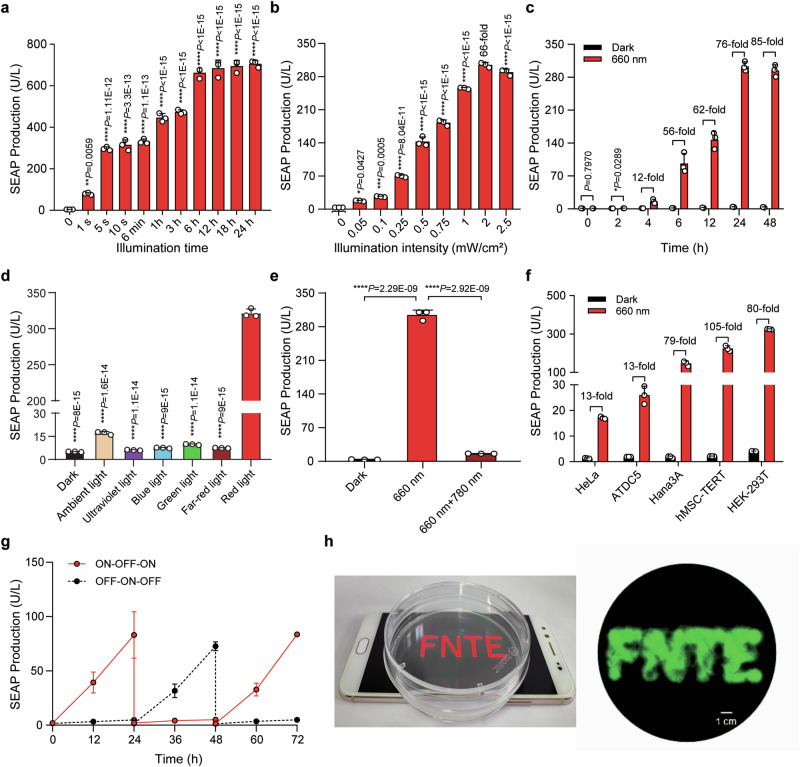
Fig. 2. Characterization of the Fn-REDLIP system performance.
a Assessment of illumination time-dependent Fn-REDLIP-mediated transgene expression kinetics. HEK-293T cells were co-transfected with pQL326, pNX12, and pYZ430, then illuminated with RL (2.0 mW/cm2) for the indicated durations. SEAP production was quantified 24 h after initial illumination. b Illumination intensity-dependent Fn-REDLIP-mediated transgene expression kinetics. HEK-293T cells, transfected as described in (a), were illuminated with RL at the indicated intensities for 10 s. SEAP production was quantified 24 h after illumination. c Quantification of Fn-REDLIP-mediated transgene expression kinetics. HEK-293T cells, transfected as described in (a), were illuminated with RL (2.0 mW/cm2) for 10 s. After illumination, SEAP production in the culture supernatant was profiled at the indicated time periods. d Chromatic specificity of the Fn-REDLIP system. HEK-293T cells, transfected as described in (a), were illuminated with ultraviolet light, blue light, green light, far-red light, or red light at 2.0 mW/cm2 or ambient light (750 Lux) for 10 s. SEAP production was quantified 24 h after illumination. e The switch ON/OFF performance of the Fn-REDLIP system. Three groups of HEK-293T cells, co-transfected as described in (a). Twenty-four hours after transfection, cells were illuminated with RL for 10 s and switched to the dark condition or 780 nm illumination at 2.0 mW/cm2 for 2 min for the “660 nm + 780 nm” group. As a control, cells were kept in the dark throughout the experiment. SEAP production was quantified 24 h after 660 nm or 780 nm illumination. f Fn-REDLIP-mediated SEAP expression in the indicated mammalian cell lines. g Reversibility of Fn-REDLIP-mediated transgene expression. h Spatial control of Fn-REDLIP-mediated transgene expression. Scale bar, 1 cm. Data in (a–g) are presented as means ± SD; n = 3 independent experiments. P values in (a, b, d, e) were calculated by one-way ANOVA with multiple comparisons. P values in (c) were calculated using a two-tailed unpaired t-test. Detailed descriptions of the genetic constructs and transfection mixtures are provided in Supplementary Data 1 and Supplementary Data 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
We next examined whether the Fn-REDLIP and Pn-REDLIP systems can be switched ON with RL and OFF with FRL. HEK-293T cells transfected with Fn-REDLIP or Pn-REDLIP systems were exposed to RL (660 nm, 2.0 mW/cm2) for 10 s to induce SEAP expression, followed immediately by exposure to FRL (780 nm, 1.0 mW/cm2) for 2 min. An obvious SEAP signal was only observed in cells exposed to RL illumination; no signal was evident for cells first exposed to RL and then to FRL (Fig. 2e and Supplementary Fig. 3e). Further, exposure to various durations and intensities of FRL (780 nm) showed that a 0.5 min exposure time at 1.0 mW/cm2 was sufficient to switch OFF approximately 90% of the maximum gene expression observed in the sample exposed to RL (660 nm, 2.0 mW/cm2) for 10 s (Supplementary Fig. 5a–e). We also found that the two REDLIP systems were functional in five different mammalian cell types (Fig. 2f and Supplementary Fig. 3f). Further, the Fn-REDLIP and Pn-REDLIP systems enabled reversible control of transgene expression (Fig. 2g and Supplementary Fig. 3g) and exhibited spatial transgene activation upon local illumination (Fig. 2h and Supplementary Fig. 3h). Thus, the Fn-REDLIP and Pn-REDLIP systems represent the optogenetic tools for robust, fast, wavelength-specific, adjustable, and reversible transgene activation.
REDLIP-controlled CRISPR-dCas9 systems for activation of endogenous gene transcription
After demonstrating that the REDLIP system induced highly efficient exogenous gene transcription under RL illumination, we investigated whether the system can control endogenous gene activation. Taking advantage of the synergistic activation mediator (SAM) system45, which has been confirmed to enhance dCas9-targeted endogenous gene activation by introducing sgRNA-bearing MS2 coat protein (MS2) RNA aptamers, we generated an RL-induced trans-activator by fusing MS2 to p65-HSF1, which can be recruited by sgRNA bearing MS2 aptamers and the dCas9 complex to induce gene transcription. When FRL is turned on, the chimeric BphPs dissociate from LDB3, inactivating CRISPR-dCas9 targeted endogenous gene transcription (Fig. 3a and Supplementary Fig. 6a).
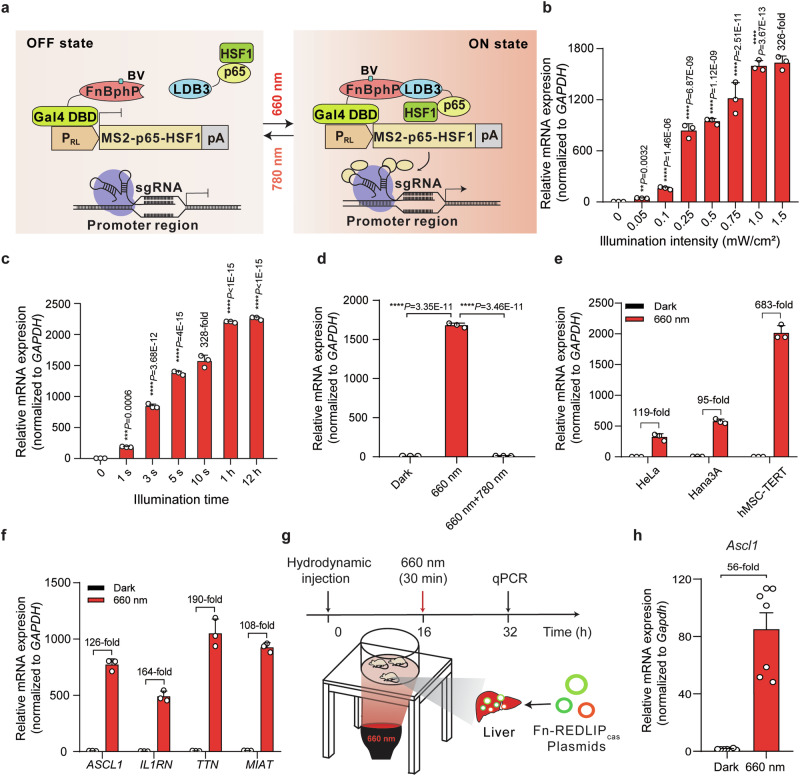
Fig. 3. Fn-REDLIP-controlled CRISPR-dCas9 system (Fn-REDLIPcas) for endogenous gene activation in mammalian cells and mice.
a Schematic design of the Fn-REDLIPcas system for transcriptional activation. b Illumination intensity-dependent activation of endogenous gene transcription by the Fn-REDLIPcas system. HEK-293T cells were co-transfected with pQL326, pNX12, pSZ69, a PRL-driven MS2-p65-HSF1 expression vector (pDQ100), and two sgRNAs targeting the RHOXF2 locus (pSZ105 and pSZ106) at a 15:15:1:10:5:5 (w/w/w/w/w/w) ratio, followed by RL illumination at the indicated intensities (0 to 1.5 mW/cm2) for 10 s. c Illumination time-dependent activation of endogenous gene transcription by Fn-REDLIPcas system. HEK-293T cells were co-transfected as described in (b) and illuminated with RL (1.0 mW/cm2) for varying durations (0 to 12 h). d Switch ON/OFF performance of the Fn-REDLIPcas system. Three groups of HEK-293T cells were transfected as described in (b). Twenty-four hours post-transfection, cells were illuminated with RL for 10 s followed by either a dark condition or 780 nm illumination at 2.0 mW/cm2 for 2 min. As a control, cells were kept in the dark throughout the experiment. Endogenous RHOXF2 levels were quantified by qPCR at 24 h after 660 nm or 780 nm illumination. P values in (b–d) were calculated by one-way ANOVA with multiple comparisons. e The Fn-REDLIPcas-mediated endogenous gene activation in the indicated mammalian cell lines. Three mammalian cell lines were co-transfected as described in (b) and illuminated with RL (1.0 mW/cm2) for 10 s. f The Fn-REDLIPcas-mediated activation of different endogenous genes. Data in (b–f) are presented as relative mRNA expression levels quantified by qPCR 24 h after illumination. All data are represented as means ± SD; n = 3 independent experiments. g Schematic representation of the experimental procedure and time schedule for Fn-REDLIPcas-mediated activation of endogenous genes in mouse livers. h qPCR analysis of the Ascl1 activation with the Fn-REDLIPcas system in mice. Data in (h) are presented as means ± SEM (n = 7 mice). Detailed descriptions of the genetic constructs and transfection mixtures are provided in Supplementary Data 1 and Supplementary Data 3. **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
To assess the REDLIP-mediated CRISPR-dCas9 systems (Fn-REDLIPcas and Pn-REDLIPcas) for activating transcription of the Rhox homeobox family member 2 (RHOXF2) gene, we co-transfected HEK-293T cells with the Fn-REDLIPcas or the Pn-REDLIPcas system and two sgRNAs targeting the RHOXF2 locus. We found that the transcriptional activation of RHOXF2 depended on the intensity and time of RL illumination (Fig. 3b, c, and Supplementary Fig. 6b, c). Note that a very short (10 s) illumination efficiently activated RHOXF2 transcription using the Pn-REDLIPcas system (1158-fold induction) with no apparent leaky activity in the dark or using the Fn-REDLIPcas system (328-fold induction) with a higher level of gene transcription (Supplementary Fig. 6c and Fig. 3c). Moreover, our two REDLIPcas systems could activate RHOXF2 transcription under RL illumination and deactivate transcription under RL and then FRL illumination (Fig. 3d and Supplementary Fig. 6d).
Supporting the broad application potential of the two REDLIPcas systems, we transfected the two REDLIPcas systems into three different cell lines, respectively, and found that they can activate endogenous RHOXF2 in the tested cells (Fig. 3e and Supplementary Fig. 6e). In addition, we successfully deployed them to activate the transcription of multiple endogenous genes using four pairs of sgRNAs, with each pair targeting the promoter of a single gene, including achaete-scute homolog 1 (ASCL1), interleukin 1 receptor antagonist (IL1RN), titin (TTN), or myocardial infarction associated transcript (MIAT) (Fig. 3f and Supplementary Fig. 6f).
We next examined whether the REDLIPcas system can activate endogenous gene transcription in mice under RL illumination. We transiently transfected the Fn-REDLIPcas system and two sgRNAs targeting the ASCL1 locus into C57BL/6 mice using hydrodynamic injection via the tail vein. Sixteen hours after injection, the abdomens of the transfected mice were illuminated with RL for 30 min (660 nm, 20 mW/cm2) (Fig. 3g). qPCR showed that the Ascl1 level was significantly up-regulated (56-fold) in illuminated mouse livers compared to dark control mouse livers (Fig. 3h). These results demonstrate that the Fn-REDLIPcas system can activate the transcription of user-defined endogenous genes in mammalian cells and mice upon illumination with a noninvasive light emitting diode (LED).
In addition, we constructed a Fn-REDLIP-controlled CRISPR-dCas9 system by fusing FnBphP or its interacting partner LDB3 to either the N- or C-terminus of the hybrid trans-activator p65-HSF1 and dCas9 protein (Supplementary Fig. 7a). We tested all fusion configurations in eight different combinations using a SEAP reporter in HEK-293T cells (Supplementary Fig. 7b) and found that only one combination (dCas9-FnBphP and LDB3-p65-HSF1) showed a 5-fold induction upon RL illumination (Supplementary Fig. 7c). We hypothesize that steric hindrance could hinder the binding interactions between dCas9-FnBphP and LDB3-p65-HSF, resulting in lower transcriptional activation efficiency due to spatial conformation challenges.
To further improve activation efficiency, we designed a second REDLIPcas system, fusing FnBphP or LDB3 to either the N- or C-terminus of p65-HSF1 and the MS2 coat protein (Supplementary Fig. 7d, e). Among the configurations, one combination (MS2-FnBphP and LDB3-p65-HSF1) achieved the highest induction (14-fold) upon RL illumination (Supplementary Fig. 7f).
We then optimized and compared different linker lengths between MS2 and FnBphP, but observed no increase in SEAP reporter gene activation induction efficiency with RL illumination (Supplementary Fig. 8a). Previous studies have explored the use of FUSn (N-terminal IDR of human oncogene, which can form transcription factor droplet46) fusions with transcriptional activators to enhance gene activation through liquid-liquid phase separation, resulting in negligible off-target effects47,48. Accordingly, we optimized the system by fusing LDB3 with various transactivators, including p65-HSF1, FUSn-p65-HSF1, and FUSn-VP64. LDB3 fused with FUSn-VP64 exhibited the highest induction efficiency, achieving a 22-fold increase in SEAP reporter activity and a 238-fold increase in endogenous RHOXF2 gene expression upon RL illumination (Supplementary Fig. 8b, c).
Optogenetic control of gene expression using AAV-delivered Fn-REDLIP system in mice
For gene transcription activation in vivo, we also transiently transfected wild-type C57BL/6 mice with Gal4 DBD-FnBphP or Gal4 DBD-PnBphP or Gal4 DBD-DrBphP and LDB3-p65-HSF1 and luciferase reporter plasmids (5×UAS-PTATA-Luciferase-pA) using hydrodynamic injection via the tail vein. Sixteen hours after injection, the transfected mice were illuminated with RL (660 nm, 20 mW/cm2) for different time periods (0 to 60 min) (Fig. 4a and Supplementary Fig. 9a). Luciferase reporter expression was measured using an in vivo imaging system: mice illuminated with RL exhibited significantly higher luciferase expression compared with the dark control mice. Moreover, the in vivo transgene expression induced by these systems was exposure-time dependent (Fig. 4b, c, Supplementary Figs. 9b, c and 10a–c). Strikingly, RL illumination for 1 min was sufficient to induce luciferase expression in the Fn-REDLIP or Pn-REDLIP system-equipped mice. In contrast, a bioluminescence signal was observed upon RL illumination for a much longer illumination time (30 min) in mice bearing the Dr-REDLIP system. Moreover, the mice injected with the Fn-REDLIP system showed stronger bioluminescence signal intensities in livers compared to mice injected with the Pn-REDLIP system, and the mice injected with the Dr-REDLIP system (Fig. 4c and Supplementary Fig. 9b, c). These results demonstrate that the Fn-REDLIP system can efficiently regulate transgene expression in deep organ tissues of mice, giving it potential for applications in vivo.
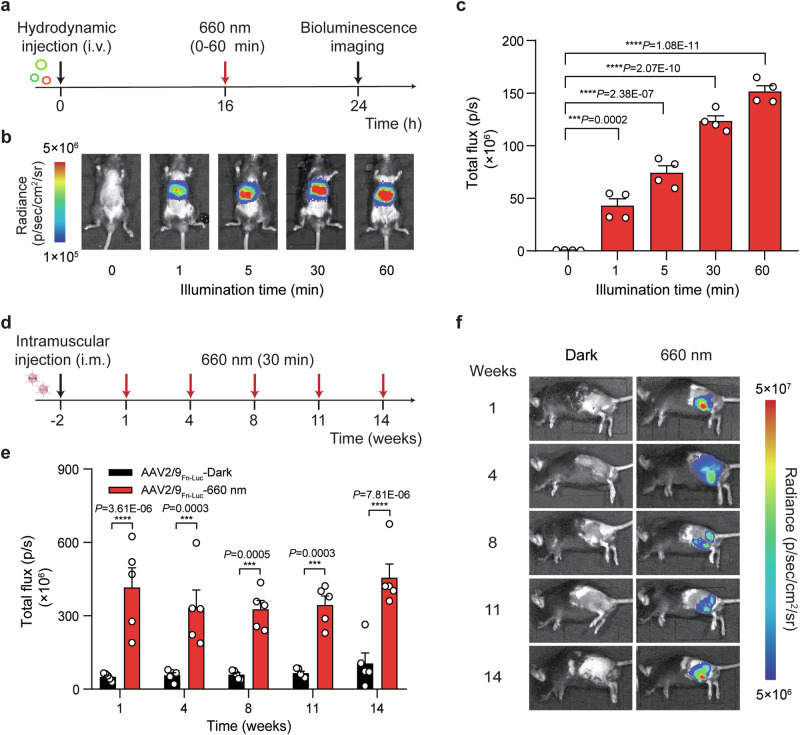
Fig. 4. Fn-REDLIP-mediated transgene expression in mice.
a Schematic representation of the experimental procedure and the time schedule for Fn-REDLIP-mediated transgene expression in mouse livers. b, c Exposure time-dependent Fn-REDLIP-mediated transgene expression kinetics in mice. Mice were hydrodynamically injected (tail vein) with Fn-REDLIP-encoding plasmids. At 16 h post-injection, the mice were illuminated with RL (660 nm, 20 mW/cm2) for the indicated time periods (0–60 min). The bioluminescence signal was quantified 8 h after illumination using an in vivo imaging system. d–f AAV-serotype of 2/9 delivery of the Fn-REDLIP-mediated transgene expression in mice. d Schematic representation of the experimental procedure and time schedule for the AAV2/9 delivery of Fn-REDLIP-mediated transgene expression in the mouse left gastrocnemius muscle. Mice were intramuscularly injected with a mixture of AAV vectors encoding the Fn-REDLIP system [(pQL382 (ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR) and pNX137 (ITR-PEMS-LDB3-p65-HSF1-pA-ITR)] and the luciferase reporter pQL271 (ITR-PRL-Luciferase-pA-ITR) at an AAV titer of 2 × 1011 Vector Genomes (vg). After 2 weeks, mice were illuminated at an intensity of 20 mW/cm2 for 30 min once every three or four weeks. e Bioluminescence was quantified 8 h after illumination. f Bioluminescence measurements of long-term activated luciferase expression of the AAV-delivered Fn-REDLIP system in mice under RL illumination. Data in (c, e) are presented as means ± SEM (c, n = 4 mice; e, n = 5 mice). P values in (c) were calculated by one-way ANOVA with multiple comparisons. P values in (e) were calculated using a two-tailed unpaired t-test. ***P < 0.001, ****P < 0.0001. Detailed bioluminescence images of the mice are provided in Supplementary Figs. 10a, 12. Source data are provided as a Source Data file.
AAV vectors are frequently used in biological research and for gene therapies in clinical settings, due to their low immunogenic potential and their low oncogenic risk from host-genome integration and long-term gene expression in tissues and organs49. We selected muscle-tropic AAV serotype 2/9 vectors as a mouse delivery method for our Fn-REDLIP system. We deployed our approach in two separate AAV vectors: the constitutively expressed RL-responsive sensor Gal4 DBD-FnBphP (2571 bp) or Gal4 DBD-PnBphP (2289 bp) and the trans-activator LDB3-p65-HSF1 (1542 bp). The mice were intramuscularly injected in the left gastrocnemius muscle with a mixture of these two AAV vectors and the RL-responsive luciferase reporter AAV vector. Two weeks after AAV transduction (Fig. 4d and Supplementary Fig. 11a), we illuminated the mice at an intensity of 20 mW/cm2 for 30 min once every three or four weeks. Bioluminescence imaging at different time points showed that the mice given this AAV-packaged Fn-REDLIP or Pn-REDLIP system with RL illumination can induce luciferase reporter expression for up to 3.5 months (Fig. 4e, f, and Supplementary Figs. 11b, c, and 12). The AAV2/9Luc group (reporter only) had significantly decreased luciferase expression levels compared to the AAV2/9Pn-Luc-dark group; this apparent leakage was marginal when considered against the strong induction upon RL illumination (Supplementary Fig. 11b, c). Collectively, these results indicate that the AAV-packaged Fn-REDLIP and Pn-REDLIP systems enable non-invasive and long-term regulation of gene expression in vivo.
Optogenetic control of glucose homeostasis using an AAV-delivered Fn-REDLIP system in T1D mice
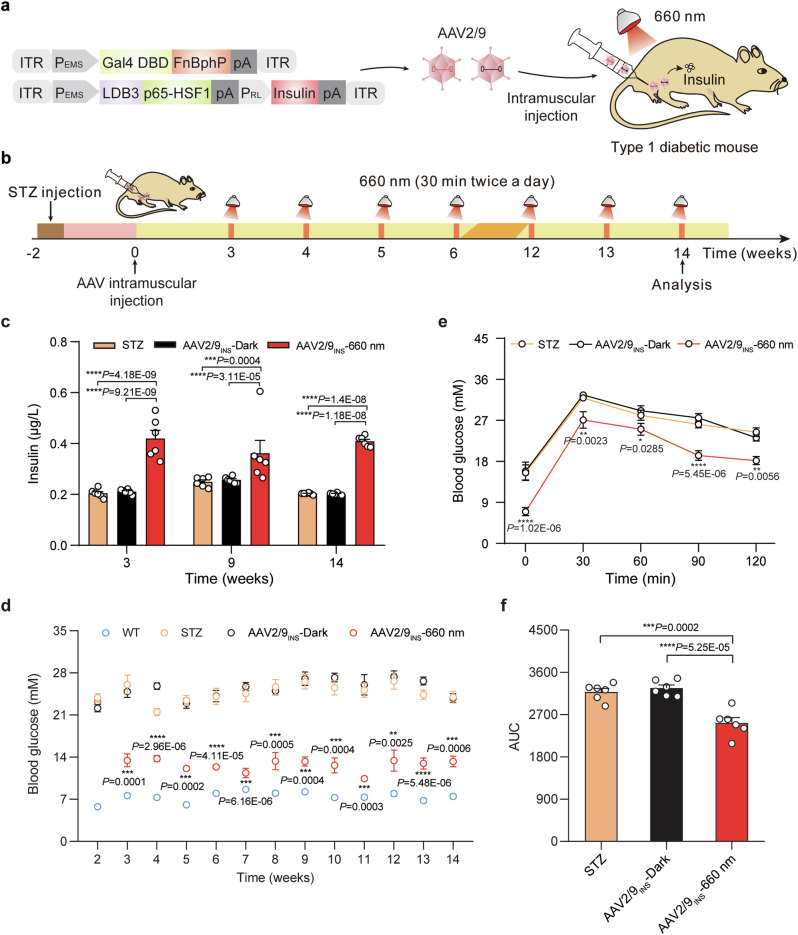
We tested the Fn-REDLIP system for the on-demand production of therapeutic agents in vivo by deploying our approach in two separate AAV-2/9 vectors: the constitutively expressed RL-responsive sensor Gal4 DBD-FnBphP and a single vector which concatenated the constructs for the hybrid trans-activator module LDB3-p65-HSF1 and the PRL-driven insulin expression module. We then transduced T1D mice with a mixture of these two AAV vectors by intramuscular injection in the left gastrocnemius muscle (AAV2/9INS), and two weeks after AAV transduction, mice were illuminated with RL (660 nm, 20 mW/cm2) for 30 min twice a day or kept in the dark (Fig. 5a, b). RL illumination of the Fn-REDLIP AAV-transduced mice (AAV2/9INS-660 nm) resulted in significantly elevated insulin levels compared to dark control AAV-transduced mice (AAV2/9INS-Dark) and to the STZ-induced T1D control mice (STZ) (Fig. 5c). Moreover, the RL-induced insulin reduced blood glucose levels for up to 12 weeks, while the STZ group and AAV2/9INS-Dark group had continuously high blood glucose levels (Fig. 5d). An intraperitoneal glucose tolerance test (IPGTT) showed a significant improvement in glucose tolerance in the AAV2/9INS-660 nm group (Fig. 5e, f). These results demonstrate that the AAV-delivered Fn-REDLIP system regulates insulin expression to achieve long-term blood glucose control in T1D mice.
Fig. 5. An AAV-delivered Fn-REDLIP system for long-term insulin expression to control blood glucose homeostasis in T1D mice.
a Schematic representation of the genetic configuration of the AAV2/9 vectors used for the Fn-REDLIP system in T1D model mice experiments. b Schematic representation of the experimental procedure and timeline for AAV-delivered Fn-REDLIP-mediated insulin expression in T1D mice. T1D mice were intramuscularly injected into the left gastrocnemius muscle with a mixture of AAV vectors encoding the Fn-REDLIP system, including pQL382 (ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR) and the concatenated vector pQL388 (ITR-PEMS-LDB3-p65-HSF1-pA::PRL-EGFP-2A-insulin-pA-ITR), at an AAV titer of 2 × 1011 vg. After 2 weeks, the injected T1D mice were illuminated with or without RL at an intensity of 20 mW/cm2 for 30 min twice a day at the indicated time point (AAV2/9INS-660 nm or AAV2/9INS-Dark). Control groups included non-model wild-type mice (WT group) and STZ-induced T1D control mice (STZ group). c, d The AAV-delivered Fn-REDLIP system for long-term blood glucose control in T1D mice. T1D mice were injected as described in (b) and subsequently illuminated with RL (660 nm, 20 mW/cm2, 30 min) twice daily. Blood insulin levels (c) were profiled weekly using a mouse insulin ELISA kit, and blood glucose levels (d) were profiled weekly using a blood glucose meter. e An intraperitoneal glucose tolerance test (IPGTT) was conducted at week 14 after AAV injection. f Area under the curve (AUC) analysis of the IPGTT data in (e). Data in (c–f ) are presented as means ± SEM (n = 6 mice). P values were calculated by one-way ANOVA with multiple comparisons. P values in (d, e) were calculated by comparing the AAV2/9INS-Dark group with the AAV2/9INS-660 nm group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
We next assessed the capacity for turning insulin expression on and off in vivo. T1D mice were intravenously injected with a mixture of AAV-2/8 vectors (targeting the livers) encoding the Fn-REDLIP system (Supplementary Fig. 13a). After 2 weeks, the injected T1D mice were randomly divided into three groups: the dark control group (Dark), RL illumination group (660 nm), RL/FRL illumination group (660 nm + 780 nm) (Supplementary Fig. 13b, c). Compared to the dark control mice, the mice in RL illumination group received RL (660 nm, 20 mW/cm2) for 30 min twice a day (Supplementary Fig. 13b), while mice in RL/FRL group were first exposed to RL and transitioned to FRL (780 nm, 20 mW/cm2) at the indicated time (Supplementary Fig. 13c). After 24 h of exposure, both RL and the combination of RL/FRL induced a significant increase in circulating insulin levels, resulting in a dramatic reduction in blood glucose levels. In contrast, the dark control mice exhibited no change in insulin and blood glucose levels (Supplementary Fig. 13d, e), indicating that the change in insulin and blood glucose depends entirely on RL and RL/FRL exposure. Moreover, after a subsequent 48 h interval, the insulin and blood glucose returned to the same levels as in the dark control mice (Supplementary Fig. 13d, e), indicating the tunability of this system. Further, a second round of RL and RL/FRL exposure efficiently induced the increase of insulin and reduction of blood glucose levels in the RL and RL/FRL group (Supplementary Fig. 13d, e). These results indicate that this response could be turned on/off by adjusting the illumination exposure to achieve the desired insulin and blood glucose levels in T1D mice.
Optogenetic control of TSLP expression using an AAV-delivered Fn-REDLIP system in HFD-induced obesity model mice
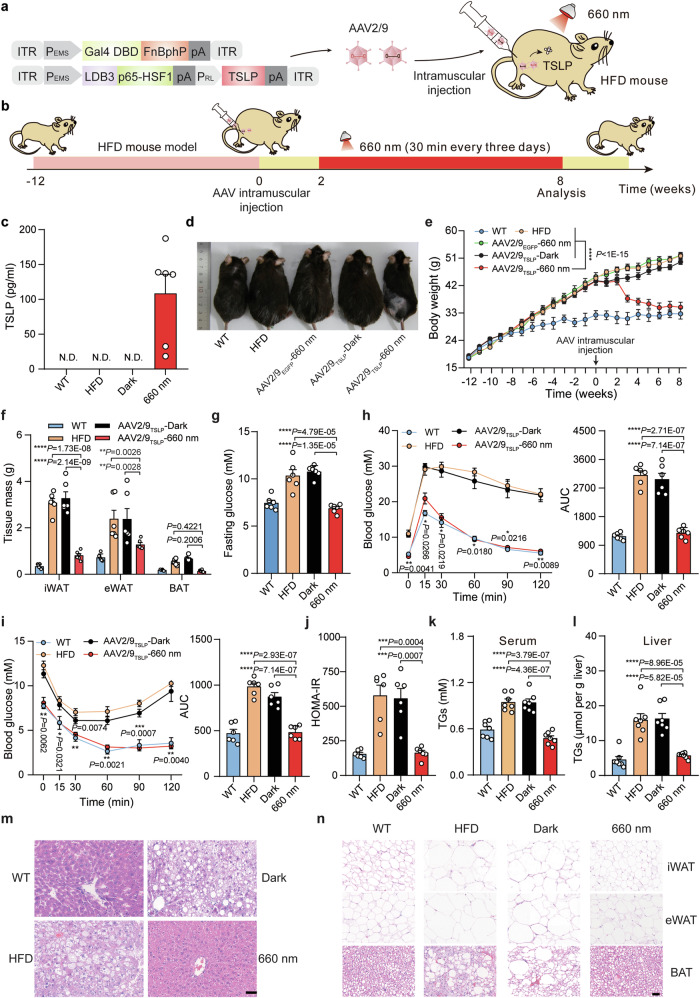
We next developed an Fn-REDLIP system for an in vivo gene therapy application to control the expression of thymic stromal lymphopoietin (TSLP), which has been recently reported to protect against obesity and obesity-related complications43. However, the overexpression of TSLP can cause airway inflammation and hyperreactivity50,51. We constructed a relatively simple system comprising two AAV-2/9 vectors encoding the constitutively expressed RL-responsive sensor Gal4 DBD-FnBphP and the concatenated constructs for the hybrid trans-activator LDB3-p65-HSF1 and the PRL-driven TSLP expression module (Fig. 6a). We intramuscularly injected HFD-induced obesity model mice in the left gastrocnemius muscle with these two AAV vectors or two control AAVs containing RL-responsive sensor Gal4 DBD-FnBphP and the concatenated hybrid trans-activator vector LDB3-p65-HSF1 and the PRL-driven EGFP expression module (AAV2/9TSLP or AAV2/9EGFP). After two weeks, the injected HFD mice were illuminated at an intensity of 20 mW/cm2 for 30 min once every three days until week 6 (AAV2/9TSLP-660 nm) or were kept in the dark (AAV2/9TSLP-Dark) (Fig. 6b). AAV2/9TSLP-660 nm group had significantly increased TSLP levels compared to AAV2/9TSLP-Dark group, to AAV2/9EGFP-660 nm group, and untreated HFD control mice (Fig. 6c).
Fig. 6. An AAV-delivered Fn-REDLIP system for long-term TSLP expression to control body weight in HFD mice.
a Schematic representation of the genetic configuration of the AAV2/9 vectors for the Fn-REDLIP system used for the HFD obesity model mice experiment. b Schematic representation of the experimental procedure and time schedule for AAV-delivered Fn-REDLIP-mediated TSLP transgene expression in HFD mice. HFD mice were intramuscularly injected in the left gastrocnemius muscle with a mixture of AAV vectors encoding the Fn-REDLIP system iteration containing pQL382 and the concatenated vector pQL383 at an AAV titer of 2 × 1011 vg. After 2 weeks, the injected HFD mice were illuminated with or without RL illumination (20 mW/cm2, 30 min) every three days for 6 weeks (AAV2/9TSLP-660 nm or AAV2/9TSLP-Dark). The examined controls included non-model wild-type control mice (WT group), HFD control mice (HFD group), and HFD mice transduced with control EGFP AAV vectors with RL illumination (AAV2/9EGFP-660 nm). c The levels of TSLP expression in serum were quantified using a mouse TSLP ELISA kit. N.D., not detected. d Representative images of the AAV2/9TSLP-660 nm group at 8 weeks. The examined controls included the AAV2/9TSLP-Dark group, WT group, HFD group, and AAV2/9EGFP-660 nm group. e–l Analysis of metabolic parameters of HFD mice treated with the AAV-delivered Fn-REDLIP system, including (e) Body weight; (f) adipose tissue (eWAT, iWAT, and BAT) weights; (g) Fasting blood glucose; (h) IPGTT (left) and AUC analysis of the IPGTT data (right); (i) ITT (left) and AUC analysis of the ITT data (right); (j) HOMA-IR; (k) Serum TG, and (l) Liver TG. m, n Representative H&E staining of the liver (m) and adipose (n) tissues. Scale bars = 50 µm. Three times the experiment was repeated with similar results. Data in (c) and (e–l) are presented as means ± SEM (c, f–j, n = 6 mice; e, k, l, n = 7 mice). P values were calculated by one-way ANOVA with multiple comparisons. P values in (h, i) were calculated by comparing the AAV2/9TSLP-Dark group with the AAV2/9TSLP-660 nm group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Strikingly, after receiving the RL illumination for two weeks, the AAV2/9TSLP-660 nm group had significantly reduced body weight compared to the AAV2/9TSLP-Dark group, the AAV2/9EGFP-660 nm group, and the untreated HFD control mice; indeed, these mice had body weights that did not differ from wild type control mice at 8 weeks after the initial AAV transduction followed by illumination (Fig. 6d, e). Moreover, this treatment group displayed significantly decreased weights of adipose tissues (including epididymal white adipose tissue (eWAT), subcutaneous inguinal white adipose tissue (iWAT), and brown adipose tissue (BAT)) compared to the AAV2/9TSLP-Dark group, WT control mice, and untreated HFD control mice (Fig. 6f). A battery of metabolic tests, including a fasting blood glucose test (Fig. 6g), an IPGTT (Fig. 6h), an insulin tolerance test (ITT) (Fig. 6i), a homeostatic model assessment of insulin resistance (HOMA-IR) test (Fig. 6j), as well as measuring serum and liver triglycerides (TGs) (Fig. 6k, l), consistently showed significant decreases in the AAV2/9TSLP-660 nm group compared to the relevant controls; these parameters did not differ from wild type control mice 8 weeks after the initial AAV transduction followed by illumination.
Hematoxylin and eosin (H&E) staining revealed that the AAV2/9TSLP-660 nm group exhibited significantly smaller adipose droplets, decreased severity of hepatic steatosis in the liver (Fig. 6m), and smaller adipocyte sizes in adipose tissues (eWAT, iWAT, and BAT) (Fig. 6n). Notably, long-term observations of mice given this AAV-delivered Fn-REDLIP system showed no obvious adverse effects, including blood biochemistry [alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), creatinine (CRE)], hematology [albumin/globulin ratio (A/G), counts for red blood cells (RBC), hemoglobin (HGB), platelet (PLT), total white blood cells (WBC), percentage of blood lymphocytes (LYMPH), blood monocytes (MONO), blood eosinophil (EO)], serum inflammatory cytokine (serum IL-6 and serum TNF-α), and immunoglobulin (serum IgG) (Supplementary Figs. 14, 15). These results demonstrate that the AAV-delivered Fn-REDLIP system enables precise optogenetic control of the therapeutic protein to implement effective gene therapy in animals.
We also achieved dosage control of TSLP with Fn-REDLIP in mouse livers. Briefly, intravenously injected HFD mice with a mixture of AAV2/8TSLP vectors and exposed mice to different RL (20 mW/cm2) illumination time (0, 5, 30 min) every three days for 4 weeks (Supplementary Fig. 16a, b). The mice of the 30 min exposure group had a significantly higher TSLP levels. They showed the expected attendant reduction in body weight (Supplementary Fig. 16c–e). These results demonstrate that the AAV-delivered Fn-REDLIP system enables dose-dependent regulation of transgene expression through varying durations of light exposure in deep tissues of mice.
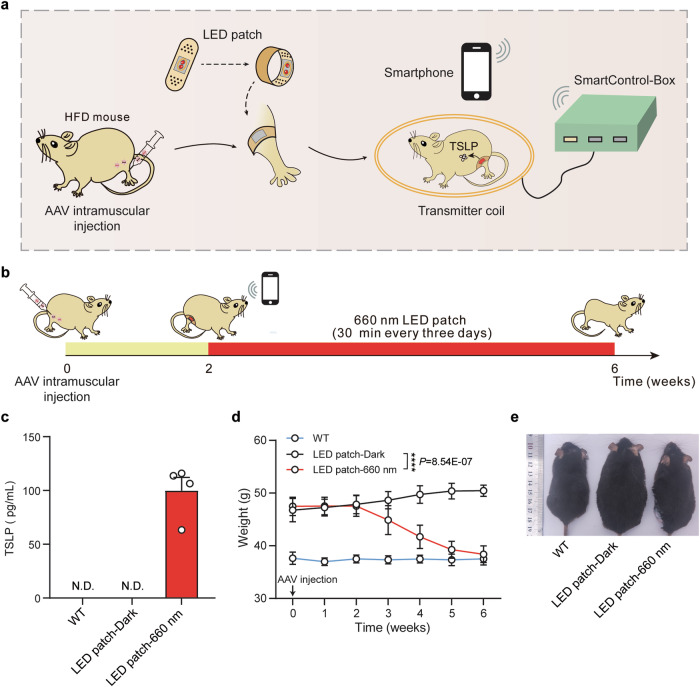
Regarding the design of practical application of the REDLIP system, we conducted in vivo experiments where AAV-delivered Fn-REDLIP mediated TSLP transgene expression in the muscles of HFD mice under an LED patch controlled by a smartphone (Supplementary Fig. 17, Fig. 7a). HFD mice were injected intramuscularly in the gastrocnemius muscle with a mixture of AAV vectors encoding the Fn-REDLIP system. After two weeks, the mice were illuminated with or without RL using a smartphone-controlled LED patch (Fig. 7b), preventing unexpected activation by ambient light. Following two weeks of RL illumination, mice in the LED patch-660 nm group showed significantly increased TSLP levels (Fig. 7c) and reduced body weight compared to the LED patch-Dark group and untreated HFD control mice (Fig. 7d, e). These results demonstrate that applying an LED patch on the skin can control drug expression on demand for treating diseases.
Fig. 7. AAV-delivered, Fn-REDLIP-mediated expression of TSLP in the muscles of HFD mice controlled by an LED patch operated via smartphone.
a Schematic of smartphone controlled AAV-delivered Fn-REDLIP-mediated TSLP transgene expression in HFD mice muscles. HFD mice were injected intramuscularly in the gastrocnemius muscle with AAV encoding the Fn-REDLIP system and illuminated with or without RL (20 mW/cm2) using the smartphone-controlled LED patch. The ECNU-TeleMed app controlled the transmitter coils through the SmartControl-Box, enabling RL-responsive Fn-REDLIP to induce TSLP expression. b Schematic and timeline of the experimental procedure for AAVTSLP-delivered Fn-REDLIP-mediated TSLP expression in HFD mice. HFD mice were injected with a mixture of AAV vectors pQL382 (ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR) and pQL383 (ITR-PEMS-LDB3-p65-HSF1-pA-PRL-TSLP-pA-ITR) at a titer of 2 × 1011 vg. After 2 weeks, mice were illuminated with or without RL for 30 min every three days for 6 weeks using the smartphone-controlled LED patch. c TSLP levels in serum were quantified using a mouse TSLP ELISA kit. N.D., not detected. d Analysis of body weight in HFD mice treated with the AAV-delivered Fn-REDLIP system. e Representative images of the LED patch-660 nm group at 6 weeks. Controls included the LED patch-Dark group, and non-model wild-type control mice (WT group). Data in (c, d) are presented as means ± SEM (n = 4 mice). P values in (d) were calculated by one-way ANOVA with multiple comparisons. ****P < 0.0001. Source data are provided as a Source Data file.
Discussion
Optogenetics allows for the traceless and remote control of living cells, tissues, and organisms with tailored functions. Moreover, optogenetics is moving from basic research toward therapeutic applications14,52–54. In this study, we have successfully developed the REDLIP system (Fn-REDLIP and Pn-REDLIP) based on the chimeric photosensitive protein BphPs and their binding partner LDB3, which enables control of gene expression in a light-intensity and light-duration dependent manner, as well as provides reversible features in response to light. The Fn-REDLIP system components can be delivered by AAV to control in vivo transgene expression.
Compared with existing red/far-red light-responsive optogenetic systems, the REDLIP activation requires a shorter illumination time (10 s of exposure to RL), fast ON/OFF kinetics (10 s ON and 1 min OFF), and strong activation of gene expression (> 100-fold). Moreover, our system does not require users to introduce exogenous chromophore, whereas other red/far-red optogenetic systems based on PhyB/PIF6, PhyA/FHY1, or PhyA/FHL each require users to supply an exogenous chromophore24,30 or produce the chromophore through co-expression of biosynthesis genes32,55. In addition, the Fn-REDLIPcas and Pn-REDLIPcas systems activation also requires a shorter illumination time (10 s) and minutes (30 min) in mammalian cells (> 326-fold) and mice (> 56-fold), respectively, while the recently reported Red-CPTS38 requires repetitive pulses of illumination over 24 h. We also anticipate that the REDLIP system can be adaptable to the Cre-loxP system to achieve RL-activatable Cre-mediated DNA recombination, which can be used for gene insertion, deletion, and inversion in various animal models and the creation of transgenic mouse models.
Translational optogenetics would benefit from a safe and effective gene delivery method. AAV vectors have recently gained popularity as gene delivery vehicles with acceptable safety, low immunogenicity, and long-term gene expression potential56. The small size of the Fn-REDLIP system enables it to be packaged into AAV vectors to achieve RL-inducible protein expression. We have demonstrated that the Fn-REDLIP system can be delivered to the skeletal muscles/livers of mice by AAV and enables the light-regulated expression of insulin in diabetes model mice to reduce blood glucose or expression of TSLP in HFD mice to control body weight. The REDLIP system is well-suited to treat diseases that require long-term yet controllable expression of therapeutic proteins.
There is still room for improvement with regard to the delivery method of our system. Developments in capsid design for AAV variants57–59 and non-viral vectors22,60,61 can advance gene delivery strategies, which could help facilitate the delivery of our system into targeted tissues. Moreover, more compact systems and existing or new miniature engineered photoreceptors will be developed to simplify their applications in vivo. Additionally, to avoid potential gene expression leakiness induced by ambient light, REDLIP-mediated gene therapy could be administered via subcutaneous muscle delivery using an LED patch attached to the skin, effectively shielding it from ambient light exposure62. The versatility of our system should enable it to control various protein-based drugs, including enzymes, peptide hormones, and antibodies. For instance, the REDLIP system could be used to induce the expression of urate oxidase to treat gout5 or the expression of parathyroid hormone (PTH) to treat hyperparathyroidism63.
Collectively, the REDLIP system represents an optogenetic tool allowing for efficient gene expression without exogenous administration of a chromophore in mammals in a timescale of seconds with the capacity for deep penetration and invasiveness. We expect that our AAV-delivered REDLIP system will accelerate the progression of optogenetic therapies toward clinical applications.
Methods
Ethical statement
The animal experiments were performed according to the protocol approved by the East China Normal University (ECNU) Animal Care and Use Committee. They were in accordance with the Ministry of Science and Technology of the People’s Republic of China on Animal Care Guidelines. The protocol was approved by the ECNU Animal Care and Use Committee (protocol ID: m20220505, R + RB20210101). All mice were euthanized after the termination of the experiments.
Plasmid construction
Construction details of the plasmids are provided in Supplementary Data 1. The amino acid and DNA sequences for REDLIP components in this study are listed in Supplementary Data 2. The plasmids were constructed by Gibson assembly according to the manufacturer’s instruction (MultiS One Step Cloning Kit, Catalog no. C113-01, Vazyme). DNA sequencing (Shanghai Saiheng Biotechnology) confirmed the sequences.
Cell culture and transfection
Human embryonic kidney cells (HEK-293T, CRL-11268, ATCC), telomerase-immortalized human mesenchymal stem cells (hMSC-TERT, which was obtained from Professor Dr. Martin Fussenegger Department of Biosystems Science and Engineering, ETH Zurich.), human cervical adenocarcinoma cells (HeLa, CCL-2, ATCC), HEK-293-derived Hana3A cells engineered for constitutive expression of RTP1, RTP2, REEP1, and Gαoλϕ64; mouse chondroblast cells (ATDC5, CRL-3419, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Catalog no. 12100061, Gibco) supplemented with 10% (v/v) fetal bovine serum (Catalog no. FBSSA500-S, AusGeneX), and 1% (v/v) penicillin/streptomycin solution (Catalog. no. ST488-1/ST488-2, Beyotime). All the cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2, and were regularly tested for the absence of Mycoplasma and bacterial contamination. The concentration and viability of the cell lines were evaluated using a Countess II Automated cell counter (AMEP4746, Life Technologies).
All cells except for ATDC5 were transfected using an optimized polyethyleneimine (PEI)-based protocol. Briefly, 6 × 104 cells per well were plated into a 24-well plate and cultured for 18 h. The cells were subsequently co-transfected with corresponding plasmid mixtures for 6 h with 50 μL PEI (Catalog no. 24765, Polysciences; molecular weight 40,000, stock solution 1 mg/mL in ddH2O; PEI and DNA at a mass ratio of 3:1 for HEK-293T, hMSC-TERT, and Hana3A and at a mass ratio of 5:1 for HeLa.). For the ATDC5 cells, 6 × 104 cells per well were plated into a 24-well plate, cultured for 18 h, and co-transfected with corresponding plasmid mixtures using Lipofectamine™ 3000 (Catalog. no. L3000015, Thermo Fisher Scientific) according to the manufacturer’s instructions.
REDLIP-mediated transgene expression in mammalian cells
For light-controlled exogenous transgene activation experiments, 6 × 104 cells were plated in a 24-well plate and cultivated for 18 h. The cells were subsequently co-transfected with corresponding plasmid mixtures for 6 h (Supplementary Data 3, 4). The cell culture plate was then placed below a custom-designed 12 × 8 red light LED array (660 nm; Shenzhen Bested Opto-electronic; each LED centered above a single well), and the cells were exposed to red light (660 nm, 2 mW/cm2, 10 s, unless indicated specifically). The light intensity was measured at a wavelength of 660 nm or 780 nm using an optical power meter (Q8230, Advantest). SEAP production levels in the culture medium were quantified 24 h after illumination.
SEAP assay
A p-nitrophenylphosphate-based light absorbance time course assay was used to quantify the production of human placental SEAP. Briefly, 100 μL cell culture supernatants were heat-inactivated at 65 °C for 30 min. 120 μL of substrate solution [100 μL of 2 × SEAP buffer (pH 9.8) containing 20 mM L-homoarginine hydrochloride (Catalog no. A602842, Sangon Biotech), 1 mM MgCl2 (Catalog no. A610328, Sangon Biotech), 21% (w/w) diethanolamine (Catalog no. A600162, Sangon Biotech), and 20 μL of p-NPP substrate solution (Catalog no. 333338-18-4, Sangon Biotech) containing 120 mM p-nitrophenylphosphate] was added to 80 μL heat-inactivated supernatants. The time course of the absorbance at 405 nm was measured by the Synergy H1 hybrid multi-mode microplate reader (BioTek Instruments) with Gen5 software (version 2.04).
Confocal fluorescence imaging
HEK-293T cells were plated at 2 × 105 cells per well in a 6-cm confocal dish and cultured for 18 h before transfection with plasmids (pQL500 or pcDNA3.1, pQL501, pYZ430). Six hours post-transfection, the cell culture media was replaced with normal growth media. After 24 h, one group of cells was illuminated with 660 nm RL for 24 h, while the other group was kept in the dark. Cells were imaged by a Leica TCS SP8 CARS confocal microscope (TCS SP8 CARS, Leica).
The ON/OFF performance of the Pn-REDLIP and Fn-REDLIP systems in HEK-293T cells
HEK-293T cells were plated into a 24-well plate (6 × 104 per well), cultivated to 70–90% confluence, and co-transfected with a total of 375 ng of the plasmids encoding the Pn-REDLIP system [pNX12, pQL325, and pYZ430 at a 2:2:1 (w/w/w) ratio] or a total of 375 ng of plasmids encoding the Fn-REDLIP system [pNX12, pQL326, and pYZ430 at a 2:2:1 (w/w/w) ratio]. Subsequently, the cells were illuminated with RL (660 nm, 1.0 mW/cm2) for 3 s (ON) or kept in the dark (OFF). The culture medium was refreshed every 24 h under green light (530 nm) with concomitant reversal of illumination conditions, and SEAP production was quantified every 12 h for 72 h.
Spatial regulation of transgene expression and fluorescence imaging
HEK-293T cells were plated into a 10 cm dish (3 × 106) and co-transfected a total of 12.5 μg of the plasmids encoding the Pn-REDLIP system [pNX12 (PhCMV-2NLS-LDB3-p65-HSF1-pA), pQL325 (PhCMV-Gal4 DBD-PnBphP-pA), and pDQ63(PRL-EGFP-pA) at a 2:2:1 (w/w/w) ratio] or a total of 12.5 μg of the plasmids encoding the Fn-REDLIP system [pNX12, pQL326 (PhCMV-Gal4 DBD-FnBphP-pA), and pDQ63 at a 2:2:1 (w/w/w) ratio]. Twenty-four hours after transfection, the cells were placed on the display of a Vivo X9 smartphone with the pattern “PNTE” or “FNTE” and were illuminated with RL for 15 min (display brightness set to 100%, 40 μW/cm2). Fluorescence images of EGFP expression were acquired 24 h after illumination using ChemiScope 4300 Pro imaging equipment (Clinx).
Construction of the Fn-REDLIP-controlled CRISPR-dCas9 system for direct activation of gene expression
For REDLIPcas-mediated gene transcription by fusing FnBphP or its interacting partner LDB3 to dCas9 protein, the chimeric FnBphP or LDB3 was fused to either the N- or C-terminus of dCas9, creating a fusion light sensor domain (dCas9-FnBphP/-LDB3 or FnBphP-/LDB3-dCas9) driven by the human cytomegalovirus promoter (PhCMV). LDB3 or FnBphP was fused to either the N- or C-terminus of the transcriptional activation domain (p65-HSF1), creating an RL-dependent trans-activator (LDB3-/FnBphP-p65-HSF1 or p65-HSF1-LDB3/-FnBphP) driven by PhCMV. When exposed to RL (660 nm), the trans-activator (p65-HSF1) binds specifically to the sgRNA-dCas9 complex to activate target gene transcription. FRL (780 nm) illumination switches the REDLIPcas system back to an inactive state.
For REDLIPcas-mediated gene transcription by fusing FnBphP or LDB3 to the MS2 coat protein, the chimeric FnBphP or LDB3 was fused to either the N- or C-terminus of MS2, creating a fusion light sensor domain (MS2-FnBphP/-LDB3 or FnBphP-/LDB3-MS2) driven by PhCMV. FnBphP or LDB3 was fused to either the N- or C-terminus of the transcriptional activation domain (p65-HSF1), creating an RL-dependent trans-activator (FnBphP-/LDB3-p65-HSF1 or p65-HSF-FnBphP/-LDB3) driven by PhCMV. When exposed to RL (660 nm), the hybrid transcriptional activator is recruited by the MS2 box of the constitutively expressed sgRNA-dCas9, forming a transcriptional complex to initiate transgene expression. FRL (780 nm) illumination switches the REDLIPcas system back to an inactive state.
For endogenous gene activation experiments, 6×104 HEK-293T cells were co-transfected with pSZ69, pQL508, two sgRNAs targeting the RHOXF2 locus (pSZ105, PU6-sgRNA1RHOXF2-pA; pSZ106, PU6-sgRNA2RHOXF2-pA), and either pQL236 or pQL527. The cells were then illuminated with RL (660 nm) at 1.5 mW/cm2 for 24 h. Endogenous RHOXF2 levels were quantified by qPCR at 24 h after RL illumination.
qPCR analysis
According to the manufacturer’s instructions, cells and tissue samples were harvested for total RNA isolation using an RNAiso Plus Kit (Catalog no. 9108, Takara). A total of 1 μg RNA was reverse transcribed into cDNA using a HiScript® II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Catalog no. R212-01, Vazyme) according to the manufacturer’s instructions. qPCR was performed on a Real-Time PCR Instrument (LightCycler®96, Roche) using ChamQ Universal SYBR qPCR Master Mix (Catalog no. Q711-02, Vazyme) to detect the target genes. The amplification conditions of qPCR were as follows: 95 °C for 10 min followed by 40 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 10 min. The qPCR primers used in this study are listed in Supplementary Table 1. All samples were normalized to the human/mouse housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh), and the results were expressed as a relative mRNA expression level using the standard 2−ΔΔCT method.
Animals
All animals were approved by the Institutional Animal Care and Use Committee of Shanghai and conducted in accordance with the National Research Council Guide for Care and Use of Laboratory Animals. The experimental animals included 4- or 6-week-old or body weight ~25 g C57BL/6 male mice. Mice were reared in East China Normal University Laboratory Animal Center and kept with a standard alternating 12 h light/12 h dark cycle at a room temperature of 23 °C ± 1 °C, humidity of 60% ± 5% during the construction of T1D and HFD mouse models and before AAV transduction. After AAV transduction, the mice in the dark group were consistently maintained in darkness except for specific procedures such as blood sampling, changing water bottles, and cleaning cages, which were conducted under green light (530 nm).
The REDLIP-mediated transgene expression in mammalian cells and mice
For REDLIP-mediated transgene expression in mammalian cells, 6 × 104 cells were plated in a 24-well plate and cultivated to 70–90% confluency during transfection. For the Pn-REDLIP system, the cells were co-transfected with a total of 375 ng of the Pn-REDLIP system-encoding plasmids [pNX12 (PhCMV-2NLS-LDB3-p65-HSF1-pA), pYZ430 (PRL-SEAP-pA) and pQL325 (PhCMV-Gal4 DBD-PnBphP-pA) at a 2:1:2 (w/w/w) ratio]. For the Fn-REDLIP system, the cells were co-transfected with a total of 375 ng of the Fn-REDLIP system-encoding plasmids [pNX12, pYZ430, and pQL326 (PhCMV-Gal4 DBD-FnBphP-pA) at a 2:1:2 (w/w/w) ratio]. Six hours after transfection, the cell culture medium was replaced with a fresh medium. Twenty-four hours after transfection, cell culture plates were placed below a custom-designed 12 × 8 red light LED array (660 nm; Shenzhen Bested Opto-electronic; each LED centered above a single well), and the cells were illuminated with RL (660 nm, 2.0 mW/cm2, 10 s, unless explicitly indicated). The light intensity was determined by an 8230E optical power meter (ADC Corporation). SEAP production was quantified 24 h after illumination. The ambient light intensity (~750 Lux) was determined by a digital illuminometer (DL333204, Ningbo Deli Tools Co., Ltd.).
For REDLIP-mediated transgene expression in mice, the mice (C57BL/6, male, 6-week-old) were hydrodynamically injected with 2 mL (10% of the body weight in grams) Ringer’s solution (0.147 M NaCl, 1.13 M CaCl2, 4 × 10-3 M KCl) containing plasmids encoding the Dr-REDLIP, Pn-REDLIP, or Fn-REDLIP. Each mouse was hydrodynamically injected with 375 μg of the plasmid DNA encoding the Dr-REDLIP [pQL236 (PhCMV-3NLS-LDB3-p65-HSF1-pA), pQL217(PhCMV-Gal4 DBD-DrBphP-pA), and pYZ450 (PRL-Luciferase-pA) at a 2:2:1 (w/w/w) ratio] or a total of 375 μg of the Pn-REDLIP [pQL236, pQL325, and pYZ450 at a 2:2:1 (w/w/w) ratio] or a total of 375 μg of the Fn-REDLIP [pQL236, pQL326, and pYZ450 at a 2:2:1 (w/w/w) ratio]. Sixteen hours after plasmid injection, mice were illuminated with RL (660 nm LED, 20 mW/cm2) for different time periods (0 to 2 h). Eight hours after initial illumination, bioluminescence images of the mice were obtained using an IVIS Lumina II in vivo imaging system (Perkin Elmer), and analyzed with the Living Image software (version 4.3.1).
In vivo bioluminescence and imaging
Each mouse was intraperitoneally injected with 15 mg/mL D-Luciferin solution (150 mg/kg; Catalog no. luc001, Shanghai Sciencelight Biology Science & Technology) and anesthetized with 2% isoflurane (Catalog no. R510-22-10, RWD Life Science) dissolved in oxygen using an economical animal anesthesia machine (HSIV-S, Raymain). Ten minutes after Luciferin injection, bioluminescence images of the mice were taken using the IVIS Lumina II in vivo imaging system (Perkin Elmer). Regions of interest were quantified using the Living Image software (version 4.3.1).
The Pn-REDLIPcas and Fn-REDLIPcas systems for activation of endogenous gene transcription in mammalian cells and mice
For REDLIPcas-mediated endogenous gene transcription in mammalian cells, the different mammalian cells (HEK-293T, hMSC-TERT, HeLa, and Hana3A) were co-transfected with a total of 510 ng of the plasmids encoding the Pn-REDLIPcas [pNX12, pQL325, pDQ100 (PRL-MS2-p65-HSF1-pA), pSZ69 (PhCMV-dCas9-pA), and the corresponding two sgRNAs (Supplementary Table 2) at a 15:15:1:10:5:5 (w/w/w/w/w/w) ratio], or total of 510 ng of the plasmids encoding the Fn-REDLIPcas [pNX12, pQL326, pDQ100, pSZ69, and the corresponding two sgRNAs (Supplementary Table 2) at a 15:15:1:10:5:5 (w/w/w/w/w/w) ratio]. The negative control cells were transfected with the empty pcDNA3.1(+) vector. Twenty-four hours after transfection, the cells were illuminated for different time periods (660 nm, 0–10 s) and light intensities (660 nm, 0 to 2.0 mW/cm2). Cells were then collected, and total RNA was extracted for qPCR analysis 24 h after illumination.
For REDLIPcas-mediated endogenous gene transcription in mice, the mice (C57BL/6, male, 6-week-old) were hydrodynamically injected with 2 mL (10% of the body weight in grams) Ringer’s solution containing a total of 410 μg of the plasmid DNA encoding the Fn-REDLIPcas [pQL236, pQL326, pYZ561 (PU6-sgRNA1Ascl1-pA::PU6-sgRNA2Ascl1-pA::PhCMV-dCas9-pA), and pDQ100 at a 15:15:10:1 (w/w/w/w) ratio] via tail vein. Sixteen hours after plasmid injection, mice were illuminated with RL (660 nm LED, 20 mW/cm2, 30 min). Negative control mice were hydrodynamically injected with Ringer’s solution. At 16 h following illumination, the mice were sacrificed, and liver tissues were collected for qPCR analysis of Ascl1 expression. qPCR primers used in this study are listed in Supplementary Table 1. All samples were normalized to housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh) values, and the results are expressed as the relative mRNA level normalized to that in the negative control using the standard 2−ΔΔCT method.
Construction of mouse model of type 1 diabetes (T1D)
The type 1 diabetes mouse model (T1D) was induced by streptozotocin (STZ) injection. Briefly, adult male mice (C57BL/6, with body weight ~25 g) were fasted for 16 h and intraperitoneally injected with STZ (Catalog no. S0130, Merck; 50 mg/kg in 0.1 M citrate buffer, pH 4) every day for five days. Two weeks after the initial injection, fasted mice with hyperglycemia over 16.7 mmol/L glucose were considered diabetic and used for further animal experiments.
AAV production
For the AAV-delivered Fn-REDLIP/Pn-REDLIP system by intramuscular injection, the adeno-associated virus vector (serotype 2/9) was selected to package the following: RL-responsive sensor Gal4 DBD-FnBphP or Gal4 DBD-PnBphP expression vector (pQL382, ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR; pNX257, ITR-PEMS-Gal4 DBD-PnBphP-pA-ITR), the hybrid trans-activator LDB3-p65-HSF1 expression vector (pNX137, ITR-PEMS-LDB3-p65-HSF1-pA-ITR), the light-inducible luciferase reporter pQL271 (ITR-PRL-Luciferase-pA-ITR), the single vector concatenated the constructs for the expression of the trans-activator LDB3-p65-HSF1 module, and the PRL-driven insulin expression module (pQL388, ITR-PEMS-LDB3-p65-HSF1-pA::PRL-EGFP-2A-insulin-pA-ITR), the PRL-driven TSLP expression module (pQL383, ITR-PEMS-LDB3-p65-HSF1-pA::PRL-TSLP-pA-ITR), or the PRL-driven EGFP expression module (pNX221, ITR-PEMS-LDB3-p65-HSF1-pA::PRL-EGFP-pA-ITR). These AAVs were produced by Shanghai Taitool Bioscience, purified AAVs were titrated using quantitative PCR and concentrated in PBS to ~2 × 1013 vector genomes (vg) per mL.
For AAV-delivered Fn-REDLIP system by intravenous injection, the adeno-associated viral vector (serotype 2/8, targeting the livers) was selected to package the following: RL-responsive sensor Gal4 DBD-FnBphP expression vector (pNX177, ITR-PhCMV-Gal4 DBD-FnBphP-pA-ITR), the single vector concatenated the constructs for the expression of the trans-activator LDB3-p65-HSF1 module, the PRL-driven insulin expression module (pQL318, ITR-PhCMV-LDB3-p65-HSF1-pA::PRL-EGFP-2A-insulin-pA-ITR), and the PRL-driven TSLP expression module (pNX166, ITR-PhCMV-LDB3-p65-HSF1-pA::PRL-TSLP-pA-ITR). These AAVs were produced by Shanghai Taitool Bioscience, purified AAVs were titrated using quantitative PCR and concentrated in PBS to ~2 × 1013 vector genomes (vg) per mL.
Optogenetic control of luciferase expression using AAV-delivered Fn-REDLIP/Pn-REDLIP system in mice
Wild-type male C57BL/6 mice (6-week-old) were randomly divided into two groups and were intramuscularly injected into the left gastrocnemius muscle using an Omnican® 40 (0.3 mm × 8 mm) syringe with a mixture of 50 μL AAVs (serotype 2/9) containing three AAV vectors: the Gal4 DBD-FnBphP (pQL382, ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR, 2 × 1011 vg) or Gal4 DBD-PnBphP (pNX257, ITR-PEMS-Gal4 DBD-PnBphP-pA-ITR, 2 × 1011 vg) expression vector, the hybrid trans-activator LDB3-p65-HSF1 expression vector (pNX137, ITR-PEMS-LDB3-p65-HSF1-pA-ITR, 2 × 1011 vg) and the luciferase reporter pYZ450 (ITR-PRL-Luciferase-pA-ITR, 2 × 1011 vg). The control group received an intramuscular injection (left gastrocnemius muscle) of the RL-responsive luciferase reporter AAV vector (AAV2/9Luc, 2 × 1011 vg). Two weeks after injection, the mice were illuminated with RL at an intensity of 20 mW/cm2 for 30 min once every three or four weeks. Eight hours after illumination, bioluminescence images of the mice were obtained using an IVIS Lumina II in vivo imaging system (Perkin Elmer), and analyzed with the Living Image software (version 4.3.1).
Optogenetic control of blood glucose homeostasis using an AAV-delivered Fn-REDLIP system in T1D mice
For AAV-delivered Fn-REDLIP system controlling intramuscular insulin expression experiment, T1D mice were intramuscularly injected in the left gastrocnemius muscle with a mixture of 50 μL AAVs (serotype 2/9) packaging two AAV vectors: the Gal4 DBD-FnBphP expression vector (pQL382, ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR, 2 × 1011 vg) and a single vector which concatenated the constructs for the hybrid trans-activator LDB3-p65-HSF1 module and the PRL-driven insulin (pQL388, ITR-PEMS-LDB3-p65-HSF1-pA::PRL-EGFP-2A-insulin-pA-ITR, 2 × 1011vg) expression module. Two weeks after transduction, the injected T1D mice were illuminated with or without red light at an intensity of 20 mW/cm2 for 30 min twice a day for 12 weeks. Twenty-four hours after first illumination, blood insulin was profiled in the 3rd, 9th, and 14th week using a mouse insulin ELISA kit, and blood glucose was profiled once a week using a blood glucose meter. The intraperitoneal glucose tolerance test (IGTT) was performed in the 14th week after the AAV injection.
For the AAV-delivered Fn-REDLIP system controlling hepatic insulin expression experiment, T1D mice were intravenously injected with a mixture of 50 μL AAVs (serotype 2/8) packaging two AAV vectors: the Gal4 DBD-FnBphP expression vector (pNX177, ITR-PhCMV-Gal4 DBD-FnBphP-pA-ITR, 2×1011 vg) and the single vector concatenated the constructs for the expression of the trans-activator LDB3-p65-HSF1 module and the PRL-driven insulin expression module (pQL318, ITR-PhCMV-LDB3-p65-HSF1-pA::PRL-EGFP-2A-insulin-pA-ITR, 2 × 1011 vg). After 2 weeks, the injected T1D mice were randomly divided into three groups: i) Dark group: T1D mice kept in the dark; ii) 660 nm group: T1D mice exposed to RL (660 nm, 20 mW/cm2) for 30 min twice a day; iii) 660 nm+780 nm group: T1D mice exposed to RL (660 nm, 20 mW/cm2) for 30 min twice a day (ON), followed by exposure to FRL (780 nm, 20 mW/cm2) for 30 min twice a day (OFF) after a 3-hour interval. Each group of AAV-injected T1D mice underwent the procedure every 3 days for a total of 6 days. Starting 24 h after the first RL illumination, blood insulin was profiled on days 1, 2, 4, 5, and 7 using a mouse insulin ELISA kit; blood glucose was profiled at the same time points using a blood glucose meter.
Optogenetic control of TSLP expression using an AAV-delivered Fn-REDLIP system in HED mice
For the AAV-delivered Fn-REDLIP system controlling intramuscular TSLP expression experiment, the HFD mouse model was created by feeding a high-fat diet. The male mice (C57BL/6, 4-week-old) were fed high-fat diets consisting of 60 kcal% fat (Catalog no. D12492, Research Diets) for 13 weeks until the body weight exceeded 40 g. Subsequently, the HFD mice were intramuscularly injected in the left gastrocnemius muscle with a mixture of 50 μL AAVs (serotype 2/9) packaging two AAV vectors: the Gal4 DBD-FnBphP expression vector (pQL382, ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR, 2 × 1011 vg) and the single vector concatenated the constructs for the expression of the trans-activator LDB3-p65-HSF1 module and the PRL-driven TSLP module (pQL383, ITR-PEMS-LDB3-p65-HSF1-pA::PRL-TSLP-pA-ITR, 2 × 1011 vg) or the PRL-driven EGFP module (pNX221, ITR-PEMS-LDB3-p65-HSF1::pA-PRL-EGFP-pA-ITR, 2 × 1011 vg). Two weeks after the AAV transduction, the injected HFD mice were illuminated with or without RL illumination at an intensity of 20 mW/cm2 for 30 min every three days for 6 weeks. The examined controls included non-model wild-type control mice, HFD control mice, and HFD mice transduced with control EGFP AAVs with RL illumination. Twenty-four hours after illumination, blood samples were collected from mouse retro-orbital sinus, transferred to ethylenediaminetetraacetic acid (EDTA) coated mini vacutainer tubes (Catalog no. BD-68784, BD Biosciences), and allowed to clot at 37 °C for 30 min and then 4 °C for 2 h. The clotted blood was centrifuged at 1000 × g for 10 min to get the serum, and serum TSLP levels were measured using a mouse TSLP ELISA Kit. Mice were weighed weekly, and the metabolic parameters were assessed, including fasting blood glucose, IPGTT, ITT, HOMA-IR, serum TG, and liver TG. The H&E staining of liver and adipose tissues was analyzed on the 8th week after the AAV transduction.
For the AAV-delivered Fn-REDLIP system controlling hepatic TSLP expression experiment, HFD mice were intravenously injected with a mixture of 50 μL AAVs (serotype 2/8) packaging two AAV vectors: the Gal4 DBD-FnBphP expression vector (pNX177, ITR-PhCMV-Gal4 DBD-FnBphP-pA-ITR, 2 × 1011 vg) and the single vector concatenated the constructs for the expression of the trans-activator LDB3-p65-HSF1 module and the PRL-driven TSLP expression module (pNX166, ITR-PhCMV-LDB3-p65-HSF1-pA::PRL-TSLP-pA-ITR, 2 × 1011 vg). Two weeks after the AAV transduction, the injected HFD mice were illuminated with RL at an intensity of 20 mW/cm2 for 5 or 30 min every three days for 4 weeks. The examined controls include non-model wild-type control mice (WT group), untreated HFD control mice (HFD group), and the AAV-injected HFD mice without illumination (AAV2/8TSLP-Dark). Twenty-four hours after illumination, blood samples were collected, and serum TSLP levels were measured using a mouse TSLP ELISA Kit. All the mice were weighed weekly.
Smartphone-controlled SmartController design and fabrication
SmartController was fabricated according to our previous study14. Briefly, a SmartControl-Box containing a 16-bit MSP430 (MSP430F1611, Texas Instruments Inc.) and a 443-MHz radio signal receiver and transmitter (Huawei Technologies Co. Ltd.) was used to drive an autonomous electromagnetic emission circuit. This circuit included a low dropout (LDO) linear regulator chip (TPS79733, Texas Instruments Inc., USA), a power amplifier circuit (0 to 24 V), and a home-made transmitting circular coil capable of generating a 180 KHz electromagnetic sine wave signal. This setup wirelessly powered custom-designed receiver LEDs, integrating receiver coils, an SMD 0805 tantalum resonance capacitor (15 nF), and two red LEDs (Shenzhen Bested Opto-electronic). The SmartControl-Box was controlled by the ECNU-TeleMed app.
AAV-delivered Fn-REDLIP-mediated TSLP transgene expression in HFD mice muscles under an LED patch controlled by a smartphone
HFD mice were intramuscularly injected in the gastrocnemius muscle with a mixture of AAVs encoding the Fn-REDLIP system, containing pQL382 (ITR-PEMS-Gal4 DBD-FnBphP-pA-ITR) and the concatenated vector pQL383 (ITR-PEMS-LDB3-p65-HSF1-pA-PRL-TSLP-pA-ITR) at a titer of 2 × 1011 vg. After 2 weeks, the injected HFD mice were illuminated with or without RL illumination (20 mW/cm2) for 30 min every three days for 4 weeks using a LED patch controlled by the ECNU-TeleMed smartphone app14. The app controlled the transmitter coils through the SmartControl-Box, enabling RL-responsive Fn-REDLIP to induce TSLP transgene expression.
The enzyme-linked immunosorbent assay (ELISA)
Insulin and TSLP in mouse serum were quantified using the mouse insulin ELISA Kit (Catalog no. 10-1247-01, Mercodia) and the mouse TSLP ELISA Kit (Catalog no. MTLP00, R&D), respectively. The levels of interleukin 6 (IL-6), and tumor necrosis factor α (TNF-α) were quantified using the mouse IL-6 ELISA Kit (Catalog no. EK206, Multi Sciences), and the mouse TNF-α ELISA Kit (Catalog no. EK282, Multi Sciences). IgG in mouse serum was quantified using the mouse IgG ELISA Kit (Catalog no. EK271, Multi Sciences). All samples for the ELISA assay were tested according to the manufacturer’s instructions.
Triglycerides measurement
To measure triglycerides (TGs) in liver and serum, mouse liver and serum samples were collected 8 weeks after AAV injection. Liver tissues (20–100 mg) were homogenized with 9 μL absolute ethanol (Catalog no. A500737, Sangon Biotech) per mg liver tissue, smashed at 4 °C using a low-temperature freezing grinding instrument (JXFSTPRP-CL, Thundersci), and centrifugated at 1000 × g for 10 min at 4 °C. Blood was collected from the mouse eye socket and centrifugated at 1000 × g for 15 min to obtain serum. Then, the supernatants of liver tissues and serum TG levels were measured by a triglyceride assay kit (Catalog no. A110-2-1, Nanjing Jian Cheng Bioengineering Institute) according to the manufacturer’s instructions.
Intraperitoneal glucose tolerance test (IPGTT) in mice
Mice were fasted overnight for 16 h, and intraperitoneally injected with 10 μL 20% w/v D-glucose dissolved in 0.85% NaCl solution per g of body weight. Blood glucose levels were measured from tail vein blood samples at 0, 15, 30, 60, 90, and 120 min after D-glucose injection using a handheld glucometer (Exactive Easy III, MicroTech Medical). The 0 min sample was used to determine the fasting plasma glucose level. The trapezoidal rule determined the area under the curve (AUC) for IPGTT.
Insulin tolerance test (ITT) in mice
Mice were fasted for 4 h and intraperitoneally injected with 0.75 U/kg insulin solution (Catalog no.11061-68-0, Sigma). Blood samples were obtained via tail vein at 0, 15, 30, 60, 90, and 120 min after insulin injection, and the plasma insulin levels were determined by ELISA (Catalog no.10-1247-01, Mercodia) assay. The HOMA-IR (Homeostatic Model Assessment for Insulin Resistance) index was calculated using the standard formula: HOMA-IR index = fasting concentration of insulin (µIU/mL) × fasting concentration of glucose (mM)/22.5.
Complete blood count and kidney function analysis
Mice were euthanized, and their whole blood was collected and immediately analyzed for complete blood count using the Sysmex XT-2000iV hematology analyzer (Sysmex). The parameters of hepatic function include alanine aminotransferase (ALT), aspartate aminotransferase (AST), and albumin/globulin ratio (A/G). The parameters of kidney function, including creatinine (CRE) and blood urea nitrogen (BUN), were measured using an automatic biochemical analyzer BX-3010 (Sysmex). Plasma and serum samples were simultaneously analyzed for standard biochemical analytes.
Hematoxylin and eosin (H&E) staining of liver and adipose tissues
The liver and adipose tissue samples were collected and fixed in 4% paraformaldehyde (Catalog no. G1101, Servicebio) overnight at room temperature. The fixed samples were gently dehydrated by immersing them in a graded series of alcohol solutions, cleaned in xylene, embedded in paraffin, and sliced into 4 µm-thick sections with a rotary microtome (Leica RM2235, Manual Rotary Microtome). These sections were stained with an H&E Staining Kit (Catalog no. G1005, Servicebio) according to the manufacturer’s instructions and were observed using an upright microscope (BX53, Olympus).
Statistics & reproducibility
Unless otherwise mentioned, all in vitro data represent means ± SD of three independent biological replicates. We were blinded to group allocation during data collection and analysis. For the animal experiments, each treatment group consisted of randomly selected mice (n = 4–7). Comparisons between groups were performed using a two-tailed unpaired t-test or one-way ANOVA with multiple comparisons, and the results are expressed as means ± SEM. Differences were considered statistically significant at *P < 0.05, very significant at **P < 0.01, and extremely significant at ***P < 0.001. Neither animals nor samples were excluded from the study. GraphPad Prism 8 software was used for statistical analysis. n and P values are described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was financially supported by grants from the National Natural Science Foundation of China (no.32250010, no.32261160373, and no.32430064), the National Key R&D Program of China, Synthetic Biology Research (no. 2019YFA0904500), the Science and Technology Commission of Shanghai Municipality (no. 23HC1410100 and 22N31900300), and the Fundamental Research Funds for the Central Universities to H.Y. This work was also partially supported by National Key R&D Program of China (no. 2019YFA0110802), the National Natural Science Foundation of China (no. 32171414), the Natural Science Foundation of Shanghai (no. 23ZR1419500), and the Nature Science Foundation of Chongqing, China (no. CSTB2022NSCQ-MSX0461) to M.W. We also thank the support from the CAS Youth Interdisciplinary Team and the ECNU Multifunctional Platform for Innovation (011) for supporting the mice experiments and the Instruments Sharing Platform of the School of Life Sciences, ECNU.
Author contributions
H.Y. conceived the project. H.Y., L.Q., M.W., and L.N. designed the experiments, analyzed the results, and wrote the manuscript. L.Q., L.N., Z.W., D.K., and G.Y. performed the experimental work. H.Y., L.Q., M.W., L.N., and Z.W. designed, analyzed, and interpreted the experiments. All authors edited and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data associated with this study are available in the paper or the Supplementary Information. The data generated in this study are provided in the Source Data file. Sequences for plasmids used in the study are available in the Supplementary Data file. All genetic components associated with this paper are available with a material transfer agreement and can be obtained from H.Y. (hfye@bio.ecnu.edu.cn). Source data are provided with this paper.
Competing interests
H.Y., L.Q., L.N., and Z.W. are inventors of patent applications (Chinese patent application number 202210607216.9) submitted by ECNU that cover the optogenetic REDLIP system. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Longliang Qiao, Lingxue Niu, Meiyan Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54781-2.
References
- 1.Guan, N. et al. Engineering of optogenetic devices for biomedical applications in mammalian synthetic biology. Eng. Biol.6, 35–49 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye, H. et al. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc. Natl Acad. Sci. USA110, 141–146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie, M. et al. β-cell-mimetic designer cells provide closed-loop glycemic control. Science354, 1296–1301 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ye, H. et al. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng.1, 0005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kemmer, C. et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol.28, 355–360 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Rössger, K. et al. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat. Commun.4, 2825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bojar, D. et al. Caffeine-inducible gene switches controlling experimental diabetes. Nat. Commun.9, 2318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, L. et al. Engineering genetic devices for in vivo control of therapeutic T cell activity triggered by the dietary molecule resveratrol. Proc. Natl Acad. Sci. USA118, e2106612118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin, J. et al. A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Sci. Transl. Med.11, eaav8826 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. et al. Genetic-code-expanded cell-based therapy for treating diabetes in mice. Nat. Chem. Biol.18, 47–55 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Wang, Y. et al. A versatile genetic control system in mammalian cells and mice responsive to clinically licensed sodium ferulate. Sci. Adv.6, eabb9484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, Y. et al. Optogenetic control of chemokine receptor signal and T-cell migration. Proc. Natl Acad. Sci. USA111, 6371–6376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, Y. et al. Engineering a far-red light-activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci. Adv.6, eabb1777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao, J. et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci. Transl. Med.9, eaal2298 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Nguyen, N. T. et al. Nano-optogenetic engineering of CAR T cells for precision immunotherapy with enhanced safety. Nat. Nanotechnol.16, 1424–1434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, X. et al. A far-red light-inducible CRISPR-Cas12a platform for remote-controlled genome editing and gene activation. Sci. Adv.7, eabh2358 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu, J. et al. A non-invasive far-red light-induced split-Cre recombinase system for controllable genome engineering in mice. Nat. Commun.11, 3708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler, M. A. et al. Genetically targeted magnetic control of the nervous system. Nat. Neurosci.19, 756–761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefanov, B. A. et al. Genetically encoded protein thermometer enables precise electrothermal control of transgene expression. Adv. Sci.8, e2101813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczyk, K. et al. Electrogenetic cellular insulin release for real-time glycemic control in type 1 diabetic mice. Science368, 993–1001 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Zhao, H. et al. Autonomous push button-controlled rapid insulin release from a piezoelectrically activated subcutaneous cell implant. Sci. Adv.8, eabm4389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal, A. et al. Towards translational optogenetics. Nat. Biomed. Eng.7, 349–369 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Tan, P. et al. Optophysiology: Illuminating cell physiology with optogenetics. Physiol. Rev.102, 1263–1325 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, Y. et al. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat. Biotechnol.40, 262–272 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Leopold, A. V. et al. A general approach for engineering RTKs optically controlled with far-red light. Nat. Methods19, 871–880 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Yu, Y. et al. Optogenetic-controlled immunotherapeutic designer cells for post-surgical cancer immunotherapy. Nat. Commun.13, 6357 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Z. et al. A self-powered optogenetic system for implantable blood glucose control. Research2022, 9864734 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, G. et al. Far-red light-activated human islet-like designer cells enable sustained fine-tuned secretion of insulin for glucose control. Mol. Ther.30, 341–354 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansouri, M. et al. Light-controlled mammalian cells and their therapeutic applications in synthetic biology. Adv. Sci.6, 1800952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller, K. et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res.41, e77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golonka, D. et al. Deconstructing and repurposing the light-regulated interplay between Arabidopsis phytochromes and interacting factors. Commun. Biol.2, 448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uda, Y. et al. Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc. Natl Acad. Sci. USA114, 11962–11967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goglia, A. G. et al. Optogenetic control of Ras/Erk signaling using the Phy-PIF system. Methods Mol. Biol.1636, 3–20 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Beyer, H. M. et al. Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth. Biol.4, 951–958 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Kaberniuk, A. A. et al. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods13, 591–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaberniuk, A. A. et al. Single-component near-infrared optogenetic systems for gene transcription regulation. Nat. Commun.12, 3859 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang, J. et al. Engineering of bidirectional, cyanobacteriochrome-based light-inducible dimers (BICYCL)s. Nat. Methods20, 432–441 (2023). [DOI] [PubMed] [Google Scholar]
- 38.Kuwasaki, Y. et al. A red light-responsive photoswitch for deep tissue optogenetics. Nat. Biotechnol.40, 1672–1679 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Nagano, S. et al. Structural insights into photoactivation and signalling in plant phytochromes. Nat. Plants6, 581–588 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Brandt, S. et al. The fungal phytochrome FphA from Aspergillus nidulans. J. Biol. Chem.283, 34605–34614 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takala, H. et al. Tips and turns of bacteriophytochrome photoactivation. Photochem. Photobiol. Sci.19, 1488–1510 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Huang, Z. et al. Creating red light-switchable protein dimerization systems as genetically encoded actuators with high specificity. ACS Synth. Biol.9, 3322–3333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choa, R. et al. Thymic stromal lymphopoietin induces adipose loss through sebum hypersecretion. Science373, eabd2893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gourinchas, G. et al. Influence of the N-terminal segment and the PHY-tongue element on light-regulation in bacteriophytochromes. J. Biol. Chem.294, 4498–4510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell160, 339–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider, N. et al. Liquid-liquid phase separation of light-inducible transcription factors increases transcription activation in mammalian cells and mice. Sci. Adv.7, eabd3568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, R. et al. Specific multivalent molecules boost CRISPR-mediated transcriptional activation. Nat. Commun.15, 7222 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma, S. et al. Phase-separated DropCRISPRa platform for efficient gene activation in mammalian cells and mice. Nucleic Acids Res.51, 5271–5284 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, B. et al. Thymic stromal lymphopoietin as a key initiator of allergic airway inflammation in mice. Nat. Immunol.6, 1047–1053 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Al-Shami, A. et al. A role for TSLP in the development of inflammation in an asthma model. J. Exp. Med.202, 829–839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, Z. et al. Emerging molecular technologies for light-mediated modulation of pancreatic beta-cell function. Mol. Metab.64, 101552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He, L. et al. Circularly permuted LOV2 as a modular photoswitch for optogenetic engineering. Nat. Chem. Biol.17, 915–923 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Gauvain, G. et al. Optogenetic therapy: high spatiotemporal resolution and pattern discrimination compatible with vision restoration in non-human primates. Commun. Biol.4, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uda, Y. et al. Improvement of phycocyanobilin synthesis for genetically encoded phytochrome-based optogenetics. ACS Chem. Biol.15, 2896–2906 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Wang, D. et al. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov.18, 358–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goertsen, D. et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci.25, 106–115 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Tabebordbar, M. et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell184, 4919–4938.e4922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adachi, K. et al. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun.5, 3075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin, H. et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet.15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Gee, P. et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun.11, 1334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mansouri, M. et al. Smart-watch-programmed green-light-operated percutaneous control of therapeutic transgenes. Nat. Commun.12, 3388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu, Y. et al. An optogenetic approach for regulating human parathyroid hormone secretion. Nat. Commun.13, 771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito, H. et al. RTP family members induce functional expression of mammalian odorant receptors. Cell119, 679–691 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data associated with this study are available in the paper or the Supplementary Information. The data generated in this study are provided in the Source Data file. Sequences for plasmids used in the study are available in the Supplementary Data file. All genetic components associated with this paper are available with a material transfer agreement and can be obtained from H.Y. (hfye@bio.ecnu.edu.cn). Source data are provided with this paper.